Abstract
Background
Many Firmicutes bacteria, including solvent-producing clostridia such as Clostridium acetobutylicum, are able to utilize xylose, an abundant carbon source in nature. Nevertheless, homology searches failed to recognize all the genes for the complete xylose and xyloside utilization pathway in most of them. Moreover, the regulatory mechanisms of xylose catabolism in many Firmicutes except Bacillus spp. still remained unclear.
Results
A comparative genomic approach was used to reconstruct the xylose and xyloside utilization pathway and analyze its regulatory mechanisms in 24 genomes of the Firmicutes. A novel xylose isomerase that is not homologous to previously characterized xylose isomerase, was identified in C. acetobutylicum and several other Clostridia species. The candidate genes for the xylulokinase, xylose transporters, and the transcriptional regulator of xylose metabolism (XylR), were unambiguously assigned in all of the analyzed species based on the analysis of conserved chromosomal gene clustering and regulons. The predicted functions of these genes in C. acetobutylicum were experimentally confirmed through a combination of genetic and biochemical techniques. XylR regulons were reconstructed by identification and comparative analysis of XylR-binding sites upstream of xylose and xyloside utilization genes. A novel XylR-binding DNA motif, which is exceptionally distinct from the DNA motif known for Bacillus XylR, was identified in three Clostridiales species and experimentally validated in C. acetobutylicum by an electrophoretic mobility shift assay.
Conclusions
This study provided comprehensive insights to the xylose catabolism and its regulation in diverse Firmicutes bacteria especially Clostridia species, and paved ways for improving xylose utilization capability in C. acetobutylicum by genetic engineering in the future.
Background
The Firmicutes (Bacilli/Clostridia) are a diverse group of Gram-positive bacteria that includes a large number of species that produce lactic acid, acetone, butanol, and ethanol through fermentation of a variety of carbon sources. Many of these bacteria were originally isolated from the plant environments such as garden soil, fruits, and vegetables [1,2]. Among them, Clostridium acetobutylicum is one of the best-studied clostridia and was used to develop an industrial fermentation process for producing solvents [3,4]. This strain is known to utilize a broad range of monosaccharides, disaccharides, starches, and other substrates such as whey and xylan [5,6].
Xylan and xyloglucan, the major hemicellulose components of plant cell walls, are two of the most abundant polysaccharides in nature and play an important role in supplying carbon and energy to a variety of organisms [7,8]. Depolymerization of xylan and xyloglucan produces β- and α-xylosides, respectively, that are transported into the cell and further degraded into D-xylose [9]. D-Xylose is finally transformed to the common metabolic intermediate xylulose 5-phosphate. Genetics and regulation of the xylanolytic machinery have been studied in some model species of Bacilli class, such as Bacillus subtilis [10] and Lactococcus lactis [11], but so far not in Clostridia.
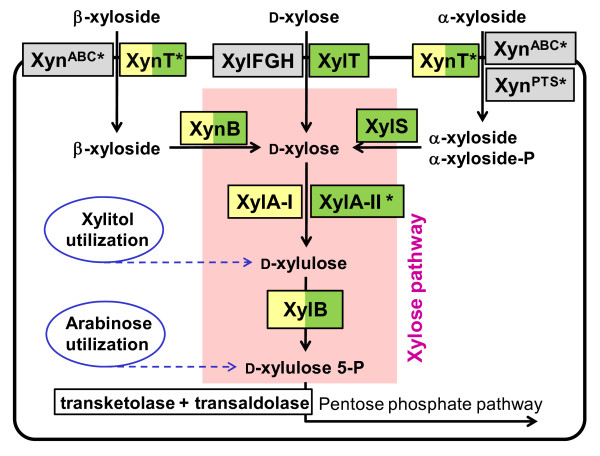
In bacteria, the transformation of D-xylose to xylulose 5-P is catalyzed via consecutive isomerization to D-xylulose and phosphorylation reactions (Figure 1). This two-step biochemical pathway appears to be conserved in both non-xylanolytic bacteria (such as Escherichia coli) and xylanolytic bacteria such as B. subtilis [12,13]. In B. subtilis, the genes involved in the xylose and xyloside utilization pathway are clustered into two operons, xylAB and xynTB (Figure 2). Their expression is negatively regulated at the transcriptional level by the regulator XylR [14]. Due to the lack of a xylose uptake system, B. subtilis is unable to grow with xylose as a sole carbon source [15].
Figure 1.
Reconstruction of the xylose and xyloside utilization pathway in Firmicutes. Functional roles present in C. acetobutylicum and B. subtilis are shown on green and yellow backgrounds, respectively. Those present in other bacteria of the same lineage (but not in C. acetobutylicum and B. subtilis) are shown on a gray background. Tentatively predicted functional roles are marked by asterisks. Solid arrows denote enzymatic reactions and transport, and broken arrows denote links to other catabolic pathways (utilization of arabinose and xylitol) that are not analyzed in this study.
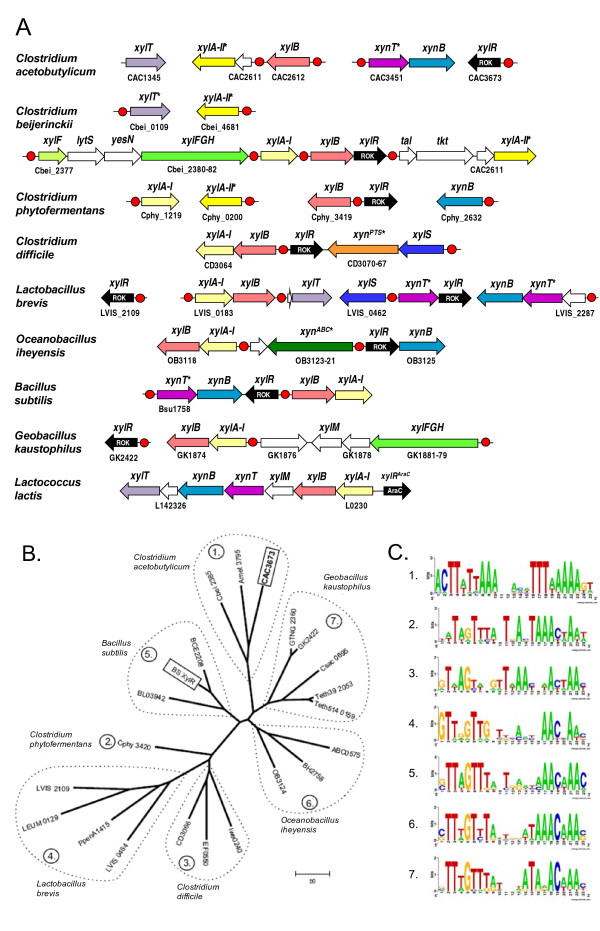
Figure 2.
Genomic context of genes associated with xylose and xyloside utilization in Firmicutes. (A) Examples of chromosomal clusters and putative regulons containing genes involved in xylose and xyloside utilization. Candidate regulatory sites of XylR from ROK family are shown by red circles. Genes predicted by genome context analysis are marked by asterisks. Homologous genes are marked by matching colors. (B) Phylogenetic tree and (C) DNA recognition motifs of XylR including all known and predicted xylose regulators from Firmicutes. XylR proteins recognizing the same DNA motif are grouped, and the corresponding motif sequence logos are shown in (C).
Several Clostridium species have been shown to metabolize D-xylose by early studies and our preliminary analysis [16,17]. However, the initial genomic survey of C. acetobutylicum ATCC 824 identified only the gene encoding xylulokinase in the xylose pathway, whereas the ortholog of the xylA gene encoding xylose isomerase was not found [4]. Although several genes of xyloside metabolism are annotated in public databases (e.g. GenBank or KEGG), some of these annotations are imprecise and have not been consistently projected across all the sequenced clostridia. Moreover, our current knowledge of transcriptional regulation of xylose utilization pathway in Gram-positive bacteria was limited to Bacillus spp. This prompted us to perform a detailed analysis of xylose utilization and its regulatory mechanisms in the species of Bacilli and Clostridia classes by combining comparative genomic analyses with genetic and biochemical techniques.
In this study, we used a subsystems-based comparative genomic analysis [18,19] to explore the xylose and xyloside utilization machinery in the Firmicutes species with completely sequenced genomes. A novel xylose isomerase (named XylA-II) that is not homologous to previously characterized XylA, was identified in several Clostridia species (e.g. CAC2610). In C. acetobutylicum the xylose utilization pathway also includes a xylulokinase (XylB, CAC2612), a xylose proton-symporter (XylT, CAC1345), and a transcriptional regulator (XylR, CAC3673). The predicted functions of these genes in C. acetobutylicum were experimentally confirmed through a combination of genetic and biochemical techniques. We conclusively showed that the identified gene xylA-II encodes a fully functional xylose isomerase that catalyze the transformation of D-xylose to D-xylulose. In addition, we have also tentatively identified several other genes likely associated with the utilization of β- or α-xyloside.
Many of these genes occurred in operons that formed a predicted regulon controlled by XylR. Comparative analysis of upstream regions of the xylose and xyloside utilization genes allowed identification of candidate DNA motifs for various groups of XylR regulators and reconstruction of XylR regulons. A novel XylR-binding DNA motif, which is exceptionally distinct from the DNA motif known for Bacillus XylR, was identified in three Clostridiales species and experimentally validated in C. acetobutylicum by an electrophoretic mobility shift assay (EMSA).
Results
(i) Comparative genomics of xylose and xyloside utilization in Firmicutes
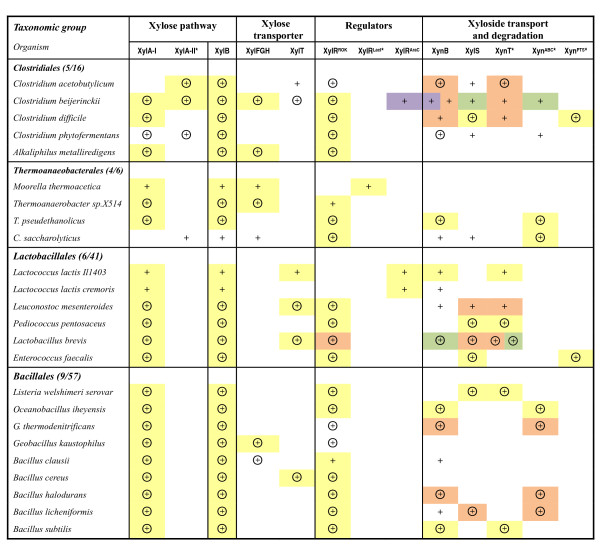
The subsystems-based approach was used to assess metabolic potential of Firmicutes species with completely sequenced genomes in utilization of xylose and its oligomeric precursors. For the twenty-four species that possess the xylose pathway genes, we reconstructed the xylose and xyloside utilization pathway and analyzed its regulatory mechanisms. The detailed results of this analysis are captured in the SEED subsystem available on line and in Additional file 1. The key results are illustrated in Figure 3 and Table 1 and contain both previously known and novel features predicted using the genome context analysis. Some of these features are briefly highlighted below where we focus mostly on novel findings and conjectures.
Figure 3.
Occurrence and features of genes involved in xylose and xyloside utilization pathway in Firmicutes. Species with completely sequenced genomes in Bacilli/Clostridia classes are shown as rows. The presence of genes for the respective functional roles is shown by "+". Genes clustered on the chromosome are marked by the same background color. Candidate XylR regulon members are circled. Tentatively predicted functional roles are marked by asterisks. The number of species with xylose pathway divided by the total number of genomes in each taxonomic group is shown in parenthesis.
Table 1.
Predicted functional assignments in the xylose and xyloside utilization pathway
| Proteina | Example gene ID | Predicted functional role | Annotation in GenBank | Phylogenetic distribution |
|---|---|---|---|---|
| XylA-II | CAC2610 | Xylose isomerase | Fucose isomerase related protein | Clostridium, C. saccharolyticus |
| XylB | CAC2612 | Xylulokinase | Xylulokinase (with CAC1344) | Bacilli, Clostridia |
| XylT | Cbei_0109 | Xylose H+-symporter | Sugar transporter | Clostridium, Lactobacillales |
| XylR | CAC3673 | Xylose regulator | XylR regulator (with CAC0933) | Bacilli, Clostridia |
| XynT | CAC3451 | Xyloside Na+(H+)-symporter | Sugar Na+(H+)-symporter | Clostridium, Lactobacillales, Bacillales |
| XynABC | OB3123-3121 | Xyloside ABC transporter | Sugar ABC transporter | Clostridium, Thermoanaeobacterales, Bacillaceae |
| XynPTS | CD3070-3067 | Xyloside PTS transporter | PTS system | C. difficile, E. faecalis |
a Predicted functional assignments that were experimentally verified in C. acetobutylicum are marked by bold type and underlined.
XylR regulon
The transcriptional factor XylR belongs to ROK (Repressor, Open reading frame, Kinase) protein family and has an N-terminal DNA-binding domain and a C-terminal sugar kinase-like domain [20]. Based on homology supported by chromosomal clustering with xylose pathway genes, we identified XylR orthologs in most genomes of Firmicutes (Figure 3 and Additional file 1). Although XylR has been characterized in Bacillus spp. as a repressor of the xyl operon encoding xylose pathway genes [12], the annotations of XylR homologs in public databases are incomplete and imprecise due to the presence of functionally divergent paralogs and limited experimental data on their characterization. For example, xylR is assigned to four genes in C. acetobutylicum genome (i.e. CAC3673, CAC0933, CAC1086, and CAC0183) in the public databases (e.g. KEGG). Only CAC3673 was deemed to xylose regulator in this study, because it displayed a closest homology with the characterized XylR from B. subtilis (31% vs. <22% identity). Moreover, the close orthologs of CAC3673 in Clostridium beijerinckii and Alkaliphilus metalliredigens (Cbei_2385 and Amet_3795) are clustered on the chromosome with xylose pathway genes (Figure 3).
The phylogenetic tree was constructed for 22 representative XylR proteins from Firmicutes including two paralogs in Lactobacillus brevis, where at least 7 branches were observed on the phylogenetic tree of XylR proteins (Figure 2B). The lowest pairwise sequence similarity between different groups of XylR proteins was 21% (between XylR from C. acetobutylicum and Leuconostoc mesenteroides). XylR from C. acetobutylicum was in the same group with orthologous proteins from C. beijerinckii, and A. metalliredigens. This group of clostridial XylR proteins stands alone from XylR orthologs in other Clostridium such as C. phytofermentans and C. difficile.
Although the XylR regulons in Bacilli/Clostridia have been analyzed previously [21], a rapidly growing number of complete genomes in these classes allowed significant improvement of the accuracy of XylR-binding DNA motifs and expansion of XylR regulons. Analysis of upstream regions of XylR-controlled genes and their orthologs in XylR-encoding genomes resulted in identification of the group-specific XylR-binding DNA motifs that were used to search for additional candidate
XylR-binding sites in the analyzed groups of genomes (Figure 2C). The obtained consensus sequences for XylR-binding sites in the Bacillaceae genomes (groups 5, 6 and 7 on the XylR protein tree) were in accordance with that experimentally determined for B. subtilis XylR [22]. Predicted DNA motifs of XylR regulators from other groups on the tree were partially similar to that from the Bacillaceae. However, the group 1 of XylR regulators from C. acetobutylicum, C. beijerinckii, and A. metalliredigens has an exceptionally different DNA recognition motif, a 25-bp inverted repeat with consensus sequence 5'-ACTTattAAAnnnnnTTTaaAAAgt-3' (Figure 2C).
The identified candidate DNA-binding sites of different groups of XylR regulators were detected in the promoter regions of most xylose utilization genes in Firmicutes (Figure 3 and Additional file 2). The most conserved part of the XylR regulon includes the xylose pathway genes xylA and xylB and transporter genes xylFGH or xylT. The XylR regulon members also include the genes involved in uptake and degradation of α-, and β-xylosides. In addition, the presence of XylR-binding sites upstream of the xylR gene in many species suggests possible autoregulation of its expression. Hence, assignment of the gene xylR (e.g. CAC3673) is further supported by sharing upstream XylR-binding sites with xylose pathway genes. Remarkably, a XylR-binding site in the C. acetobutylicum and several other Clostridium genomes was detected upstream of a hypothetical gene encoding a novel non-orthologous variant of xylose isomerase characterized in this work (see below).
Orthologs of XylR regulator were not found in several species of Firmicutes possessing a complete version of the xylose utilization pathway, i.e., Moorella thermoacetica and L. lactis. In the first species, a hypothetical LacI-type transcriptional factor (Moth_2024) was inferred based on chromosomal clustering with the xylose pathway genes (Figure 3). The xylose operon in L. lactis is known to be controlled by an AraC-type regulator (also called XylR) [23]. Since there are only few genomes encoding these two types of xylose regulator, their DNA recognition motifs could not be determined accurately. Overall, alternative transcriptional regulators of xylose metabolism appear to be present in various species of Firmicutes.
Xyloside uptake and degradation
The analysis of operons and regulons associated with xylose and xyloside utilization subsystem allowed us to accurately annotate and map some previously uncharacterized components of xyloside utilization machinery in Firmicutes. Most genes encoding β- and α-xylosidases (xynB and xylS, respectively) appear to be clustered on the chromosome and/or co-regulated with the xylose pathway genes (Figure 3).
Based on the genome context analysis, we predicted the involvement of three types of transporters, XynT, XynABC, and XynPTS, in the xyloside uptake (Figure 3). XynT belongs to the MFS (Major Facilitator Superfamily) transporter family. The xynT gene was identified as a member of the XylR regulon in C. acetobutylicum (CAC3451) and several Lactobacillales and Bacillales (designated as xynP in B. subtilis by [24]). It is also positionally clustered with xylosidase and xylose pathway genes (Figure 3). An alternative system of xyloside transport via a committed ABC cassette was predicted for Oceanobacillus iheyensis (OB3123-OB3121) and several other species. The XynABC transport system is homologous to an oligosaccharide ABC transporter from the Streptococcus mutans (~24% identity; [25]). The functional prediction is supported by chromosomal clustering and by sharing upstream XylR-binding sites with xylosidase and other xyl genes (Figure 2A). A novel xyloside transporter from the phosphotransferase (PTS) system family was predicted for Enterococcus faecalis and C. difficile. The gene cluster encoding this XynPTS system (e.g. CD3070-CD3067) is a candidate member of the predicted XylR regulons and co-localized with α-xylosidase gene (xylS) in both species (Figure 2A), suggesting involvement of XynPTS in uptake of α-xyloside.
Xylose transport
The ABC-type xylose transporter XylFGH was originally described in E. coli [26]. An orthologous xylose ABC transporter was found in the genomic context of the xylose utilization genes in several Clostridiales, Thermoanaerobacterales, and Bacillales species (e.g. Cbei_2380-Cbei_2382 in C. beijerinckii; see Figure 3). Another xylose transporter belonging to the MFS transporter family, XylT, was described in Bacillus megaterium [27] and L. brevis [28]. Its orthologs in several species of Bacilli/Clostridia have been annotated as xylose proton-symporter in the public databases (e.g. CAC1345 of C. acetobutylicum). In this study XylT was tentatively identified in more species such as C. beijerinckii. This functional assignment is supported by the conserved co-localization on the chromosome and by predicted co-regulation (via upstream XylR-binding sites) with other xyl genes (Figure 3).
Xylose pathway
Xylose isomerase (EC 5.3.1.5) is required for the first reaction of xylose utilization, converting D-xylose into D-xylulose. This enzyme, a product of the xylA gene, was characterized in detail in L. lactis [29], and its orthologs are present in many bacteria including B. subtilis. However, analysis of the xylose utilization subsystem showed that this gene is missing in C. acetobutylicum and Caldicellulosiruptor saccharolyticus that have other components of the xylose utilization pathway. Based on genomic and functional context analysis (Figure 3), we have tentatively identified a candidate gene for an alternative xylose isomerase (termed here XylA-II), which is not homologous to XylA from B. subtilis and other Firmicutes (called here as XylA-I). XylA-II belongs to the fucose isomerase FucI family (PF02952), with a weak ~14% similarity to FucI from E. coli [30]. The xylA-II gene in C. acetobutylicum (CAC2610) and C. beijerinckii (Cbei_2389) is clustered with xylB and preceded by candidate XylR-binding sites (Figure 2A), whereas C. phytofermentans has a separate xylA-II gene preceded by a candidate XylR site.
Xylulokinase (EC 2.7.1.17), XylB, is required for the phosphorylation of D-xylulose yielding D-xylulose 5-P, a key intermediate in the pentose phosphate pathway of the central carbon metabolism. XylB, originally characterized in E. coli [26], is the invariant component of the xylose pathway (Figure 3). However, accurate functional assignment of sugar kinase genes such as xylB is not easy due to the presence of functionally divergent paralogs. For example, both CAC2612 and CAC1344 in C. acetobutylicum genome are annotated as xylulokinase in GenBank based on homology. Here only CAC2612 and its orthologs were identified as XylB based on the genome context evidence as follows: chromosomal clustering with xylose utilization genes and assignment to the XylR regulon (Figure 3).
In summary, the subsystem reconstruction and the genome context analysis in 24 genomes of Firmicutes allowed us to predict the candidate genes for a novel xylose isomerase and a xylulokinase as well as for previously uncharacterized xylose and xyloside transporters and xylose regulator (Table 1). The regulon analysis led to tentative identification of different types of DNA motifs that correspond to the binding sites of transcriptional regulator XylR. In the second part of this study, we have performed experimental validation of the predicted functions of xylA-II, xylB, xylT, and xylR genes as well as the identified XylR-binding sites in C. acetobutylicum.
(ii) Experimental validation
Mutagenesis corroborates predicted xylose pathway genes
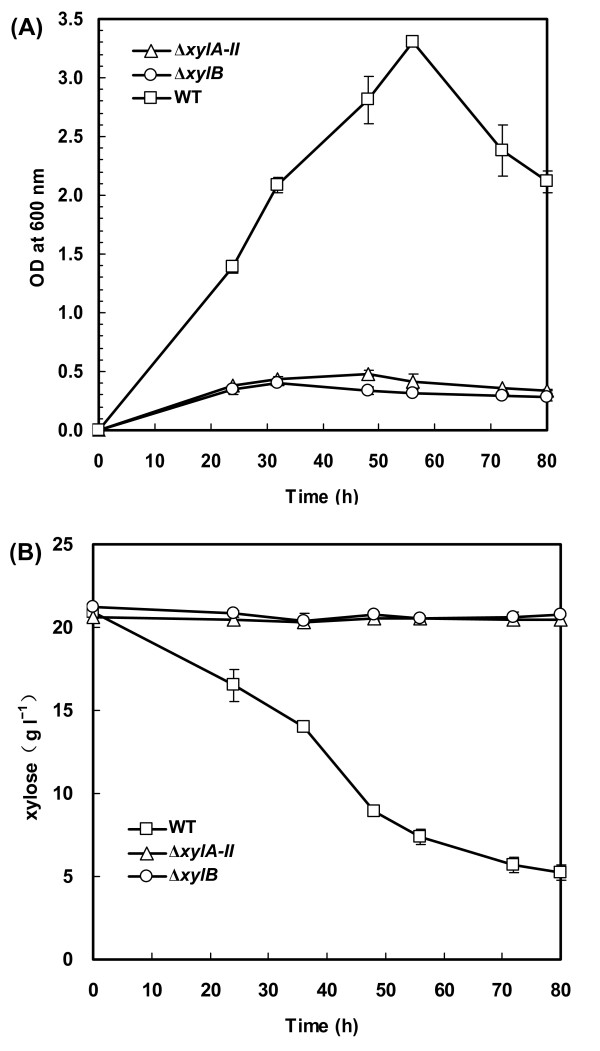
To validate the role of the inferred xylose pathway genes in C. acetobutylicum, we disrupted the respective genes by inserting an intron (confirmed by PCR as shown in Additional file 3) and tested the ability of these mutants to grow on xylose as the sole carbon source. Inactivation of the gene encoding putative xylose isomerase (ΔxylA-II) or xylulokinase (ΔxylB) abolished the growth of the resulting strains on xylose (Figure 4A) whereas their growth on glucose was not impaired (data not shown). The mutants did not consume any xylose after a 80-h incubation in the minimal medium containing 20 g l-1 of xylose (Figure 4B). Therefore, the phenotypes of C. acetobutylicum mutants were consistent with the predictions of the bioinformatics analysis and confirmed the predicted physiological roles of xylA-II and xylB in utilization of xylose.
Figure 4.
Effect of xylA-II or xylB disruption on cell growth and xylose utilization of C. acetobutylicum. Cells were grown in the medium containing 20 g l-1 of D-xylose as the sole carbon source. The optical density at 600 nm (A) and xylose concentration in the medium (B) were monitored. Data shown are means ± standard deviations calculated from triplicate individual experiments.
Heterologous-host complementation supports functional assignments of predicted xylose pathway genes
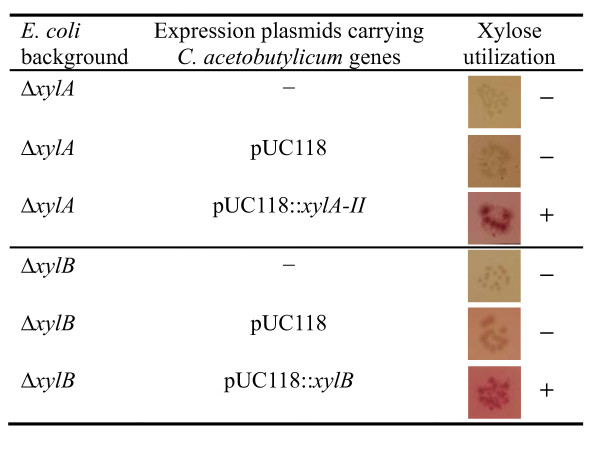
To test whether the identified genes are not only required but also sufficient to perform the predicted functions, we carried out genetic complementation experiments. Plasmid constructs containing C. acetobutylicum genes xylA-II and xylB were introduced into E. coli K-12 mutants deficient in xylose isomerase (ΔxylA) or xylulokinase (ΔxylB). The resulting strains were tested for the ability of xylose utilization using MacConkey agar supplemented with xylose as a carbon source (Figure 5). Expression of the C. acetobutylicum xylA-II gene completely restored the ability of E. coli ΔxylA mutant to utilize xylose. Similarly, expression of xylB from C. acetobutylicum successfully complemented the xylulokinase deficiency and restored the ability of E. coli ΔxylB mutant in xylose utilization (Figure 5). On the other hand, expression of the gene CAC1344, which is also annotated as xylulokinase in GenBank, had no appreciable effect on the impaired xylose utilization of ΔxylB mutant (data not shown). Our results indicate that xylA-II and xylB of C. acetobutylicum encode active enzymes functionally equivalent to xylose isomerase and xylulokinase, respectively.
Figure 5.
Complementation of E. coli mutants deficient in xylose isomerase or xylulokinase by C. acetobutylicum genes. The empty vector was expressed in the same strain as a negative control. The ability of xylose utilization was tested using MacConkey agar supplemented with 10 g l-1 xylose. The colonies of cells with the ability in xylose utilization showed red color, whereas the colonies of cells unable to utilize xylose keep yellow.
In vitro activity of the novel xylose isomerase and xylulokinase
To extend the genetic findings and provide biochemical evidence to the proposed gene assignments, we used the recombinant XylA-II and XylB from C. acetobutylicum, which were overexpressed in E. coli with the N-terminal His6 tag and purified using Ni-NTA affinity chromatography, to test for xylose isomerase and xylulokinase activities, respectively. Expected enzymatic activities of both proteins were verified using the specific assays described in Materials and Methods. The specific activity of the C. acetobutylicum XylA-II was 1.90 ± 0.14 μmol mg-1 min-1, which is comparable with the respective values reported for the enzyme from E. coli (0.87 μmol mg-1 min-1) [31] while lower than that for Bacillus licheniformis XylA (22.2 μmol mg-1 min-1) [32]. The C. acetobutylicum XylB displayed a xylulokinase activity, although the specific activity value (3.04 ± 0.13 μmol mg-1 min-1) is significantly lower than that of E. coli XylB (298 μmol mg-1 min-1) [33]. Therefore, the biochemical activity assays provided an independent verification of the predicted enzymatic activities of XylA-II and XylB from C. acetobutylicum.
Experimental assessment of the predicted xylose transporter gene
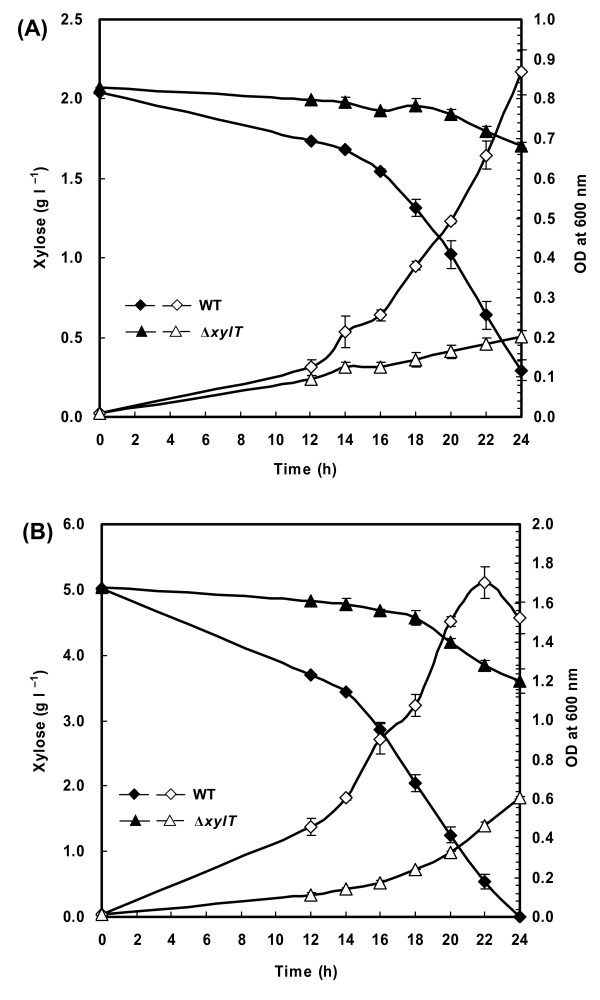
To test the role of the inferred xylose transporter gene in C. acetobutylicum, we disrupted the gene CAC1345 (xylT) by inserting an intron (confirmed by PCR as shown in Additional file 3) and assessed the effect of its inactivation on cell growth and xylose consumption. A significant impairment of cell growth on xylose was observed for ΔxylT mutant compared to the wild-type strain (Figure 6) whereas their growth on glucose was not affected (data not shown). In the medium containing 2 g l-1 of xylose, specific growth rate of ΔxylT mutant (0.06 h-1) was more than three-fold lower than that of wild-type strain (0.20 h-1). After a 20-h incubation on 2 g l-1 of xylose, about 50% of xylose was consumed by the wild-type strain, whereas the ΔxylT mutant consumed less than 8% of xylose (Figure 6A). The difference between the growth curves was smaller at high concentrations of xylose (e.g. 50 g l-1; data not shown). These observations suggest that xylT contributes to the uptake of xylose in C. acetobutylicum. The obtained results also revealed that in addition to XylT, C. acetobutylicum must have another, hitherto-unknown transport system for xylose uptake.
Figure 6.
Effect of xylT disruption on cell growth and xylose utilization of C. acetobutylicum. Cells were grown in the medium containing 2 g l-1 (A) or 5 g l-1 (B) of D-xylose as the sole carbon source. Open symbols, optical density at 600 nm; filled symbols, xylose concentration in the medium. Data shown are means ± standard deviations calculated from triplicate individual experiments.
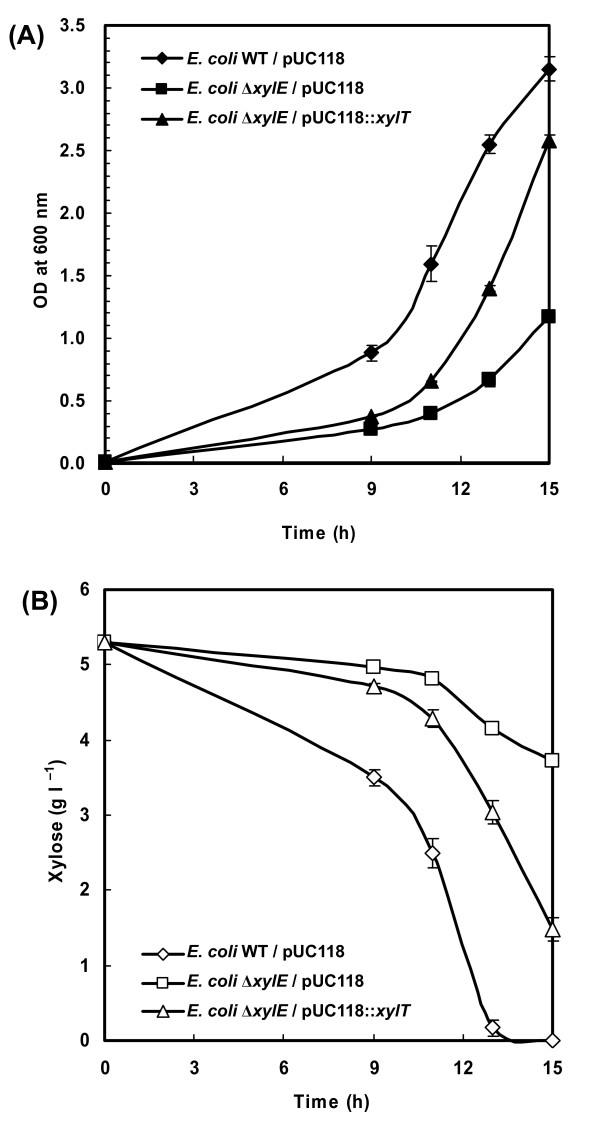
An additional verification of the xylose transporter gene was obtained using the genetic complementation experiment. A plasmid construct containing C. acetobutylicum xylT gene was introduced into E. coli K-12 ΔxylE mutant that lacks xylose proton-symporter while still has the ABC transporter system for xylose (XylFGH). The specific growth rate of this engineered strain (0.36 h-1) was higher than that of the control strain carrying an empty vector plasmid (0.25 h-1) when both strains were grown in the minimal medium containing 5 g l-1 of xylose (Figure 7A). Concomitantly, expression of C. acetobutylicum xylT led to an acceleration in xylose consumption (Figure 7B), indicating that the xylT gene product is able to transport xylose. Overall, the cumulative evidence generated in the two different types of experiments described above provided strong support for the tentative gene assignment of the xylose transporter in C. acetobutylicum.
Figure 7.
An accelerated xylose utilization of E. coli ΔxylE mutant carrying C. acetobutylicum xylT gene. Cells were grown in the minimal medium containing 5 g l-1 of D-xylose as the sole carbon source. The optical density at 600 nm (A) and xylose concentration in the medium (B) were monitored. E. coli ΔxylE mutant strain with an empty vector was used a control. Data shown are means ± standard deviations calculated from triplicate individual experiments.
Experimental testing of xylose regulator binding to predicted DNA targets
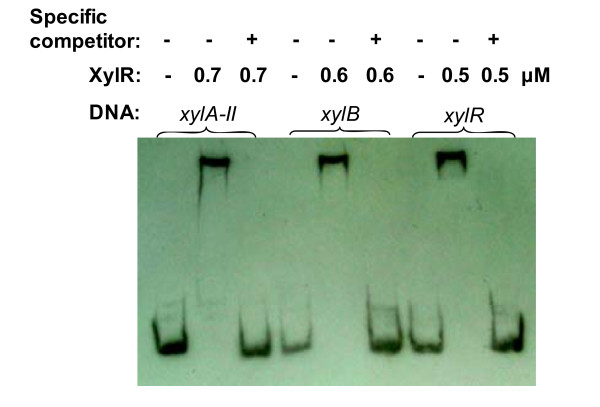
The ability of XylR protein to specifically bind to the predicted DNA sites was tested by EMSA using the purified recombinant XylR protein from C. acetobutylicum. The predicted XylR-binding sites in C. acetobutylicum and several other species represent a distinct DNA motif from that known for Bacillus XylR (as described in the previous section). Three predicted target DNA fragments from the upstream regions of C. acetobutylicum xylA-II, xylB, and xylR genes, respectively, were used in EMSA. A substantial shift of the DNA band was observed in all three cases upon incubation of XylR protein with the target DNA fragments (Figure 8). A typical protein concentration dependence of DNA-binding is illustrated in Additional file 4 showing increasing intensity of the shifted DNA band in the presence of increasing amounts of the XylR protein. The shift of the DNA band for all three target sites was essentially complete at XylR concentration above 0.7 μM. The band shift was suppressed in the presence of 400-fold excess unlabeled DNA fragments but not in the presence of non-specific competitor, salmon sperm DNA (Figure 8). No binding was observed for the negative control DNA segment without XylR-binding DNA motif (see Additional file 4). These observations provided an experimental confirmation of the target XylR-binding sites and XylR-regulated genes tentatively identified by comparative genomic techniques (as described in the previous section).
Figure 8.
EMSA to assess the interactions of C. acetobutylicum XylR with their cognate DNA signals. Each of the three 180-bp target DNA fragments (1 nM) from the upstream region of CAC2611-xylA-II, xylB, and xylR genes, respectively, was incubated for 20 min at 28°C with or without 0.5-0.7 μM of the XylR protein. Specificity of the XylR-DNA interactions was tested by competition with 0.4 μM of non-biotinylated target DNA (specific competitor). Salmon sperm DNA (2 μg) was added to all binding reaction mixtures as a non-specific competitor.
Discussion
The comparative genomics analysis and metabolic reconstruction of the xyloside and xylose metabolic subsystem across a broad range of Firmicutes, including C. acetobutylicum and related species from Bacilli and Clostridia classes, revealed a remarkable pattern of conservation and variation. The role of this subsystem in utilizing exogenous sources of xylose, and its overall topology (Figure 1) including uptake and degradation of xyloside, xylose transport, and the biochemical conversion of xylose to xylulose 5-P, is preserved in all of the analyzed species. However, nearly all these aspects of its implementation are associated with significant variations such as: (i) existence of alternative transcriptional regulators and regulatory DNA signals; (ii) presence or absence of xyloside utilization machinery and variations therein; (iii) alternative mechanisms of xylose uptake; and (iv) nonorthologous gene displacement for a key enzymatic step of the xylose catabolism, xylose isomerase (Figure 3).
Based on the subsystem reconstruction and the genome context analysis, we have identified a novel xylose isomerase, XylA-II that is not homologous to previously characterized XylA. Moreover, the gene encoding xylulokinase, XylB, was unambiguously identified in all of the analyzed Firmicutes species. These functional predictions were experimentally validated in C. acetobutylicum by a combination of genetic and biochemical techniques. The observed low specific activity of XylA-II and XylB from C. acetobutylicum is consistent with the relatively slow growth on xylose, suggesting that these two enzymatic reactions may be rate-limiting steps of xylose utilization in this organism. It is interesting that unlike in C. acetobutylicum and C. saccharolyticus where XylA-II is the only xylose isomerase, both XylA-I and XylA-II are present in C. beijerinckii and C. phytofermentans. The exact interpretation of the observed functional redundancy in these two organisms requires further investigation.
The genes encoding xylose proton-symporter, XylT, were tentatively identified in several genomes of Firmicutes based on analysis of conserved operons and regulons. This prediction was experimentally assessed by mutagenesis of the inferred gene in C. acetobutylicum and by genetic complementation in E. coli. The results allow us to conclude that XylT is involved in xylose transport in C. acetobutylicum, although there may be additional, unknown mechanism for xylose transport. Despite the differences in cell wall structure between Gram-positive and Gram-negative bacteria, XylT from C. acetobutylicum was recognized by the cell sorting machinery of E. coli and functioned properly to complement xylose symporter deficiency. It is important to emphasize that although the performed experiments provided strong support for the predicted functional assignment of XylT, additional studies are necessary to establish its substrate specificity and kinetic parameters, as well as to elucidate the mechanism of XylT-independent xylose uptake.
Based on the genome context analysis, the genes encoding the xylose regulator, XylR, were unambiguously assigned in all of the analyzed species. Comparative analysis of upstream regions of the xylose and xyloside utilization genes allowed identification of candidate XylR-binding sites and reconstruction of XylR regulons. A novel XylR-binding DNA motif, which is exceptionally distinct from the DNA motif known for Bacillus XylR, was identified in C. acetobutylicum, C. beijerinckii, and A. metalliredigens. Experimental validation was performed by EMSA using purified recombinant XylR protein. The results confirmed the proposed gene assignment of the xylose regulator and the predicted XylR-binding sites in C. acetobutylicum. In addition to XylR regulation, expression of xyl genes in Bacillus is also subject to catabolite repression mediated by the transcriptional factor CcpA that binds to the catabolite responsive element (cre) [34]. It has been reported that xylose metabolism in Clostridium was inhibited in the presence of glucose [16]. Studies on the involvement of CcpA and cre in regulation of xyl genes and the contribution of XylR to glucose repression in Clostridium, are now underway.
Since pentose sugars (i.e. xylose and xyloside) are the most abundant carbohydrate in the hemicellulose of lignocellulosic materials such as hardwoods and crop residues, the efficient utilization of pentose sugars offers the opportunity to significantly reduce the cost of solvent fermentation processes. However, before this study very little was known about the machinery for xylose and xyloside utilization in solventogenic clostridia, although some of the strains were shown previously to have this catabolic potential [17]. A subsystem-based approach applied in this study has allowed us to significantly improve the quality of gene annotations and to accurately infer metabolic and regulatory networks associated with xylose and xyloside utilization in Clostridium. Key conjectures about important aspects of this subsystem were validated by focused genetic and biochemical experiments in the model system of C. acetobutylicum ATCC 824, although other functional predictions have yet to be experimentally verified. This study paves the way for genetically engineering the xylose pathway in solventogenic clostridia to enhance its capability of xylose utilization.
Conclusions
We reconstructed the xylose and xyloside utilization pathway and XylR regulons in 24 Firmicutes species by using comparative genomics techniques. It allowed us to discover a novel xylose isomerase that is not homologous to previously characterized xylose isomerase, unambiguously assign the genes encoding the xylulokinase and XylR, and tentatively identify several genes involved in xylose transport and xyloside uptake. The key functional predictions were further experimentally verified in C. acetobutylicum through genetic and biochemical techniques. XylR regulons were reconstructed by identification and comparative analysis of XylR-binding sites upstream of xylose utilization genes. We identified and experimentally validated a novel XylR-binding DNA motif in Clostridiales, which is exceptionally distinct from the DNA motif known for Bacillus XylR. These findings provided an accurate and comprehensive understanding of xylose metabolism and its regulation in the diverse species of Firmicutes.
Methods
Bioinformatics analysis
(i) Genome resources and bioinformatics tools
Complete genomes of bacteria from the Firmicutes analyzed in this study were uploaded from GenBank http://www.ncbi.nlm.nih.gov/Genbank/. These and other related genomes used for comparative analysis are integrated in the SEED genomic database http://theseed.uchicago.edu/FIG/index.cgi. Functional coupling of genes via clustering on the chromosome and distribution of genes in the genomes were analyzed using the integrated SEED tools. For analysis of protein families, we used the ClustalX [35] and PHYLIP [36] programs that construct multiple sequence alignments and maximum likelihood phylogenetic trees, respectively. The SignalX program and the Genome Explorer software were used for identification of conserved DNA motifs of transcriptional regulators and genome scanning for candidate DNA-binding sites, respectively [37]. Sequence logos for regulatory motifs were constructed using WebLogo package version 2.6 http://weblogo.berkeley.edu/[38].
(ii) Subsystem encoding and genome context analysis
A set of subsystems-based genomic annotations and metabolic reconstruction tools implemented in the SEED were used to capture the existing knowledge of xylose utilization pathways and to tentatively project it to a broader collection of bacteria with completely sequenced genomes. This approach was previously applied for the analysis of various metabolic subsystems and for gene and pathway discovery in a broad range of species [39-41]. Briefly, a subsystem is initiated by defining a list of functional roles (enzymes, transporters, regulators) associated with xylose utilization. This information is obtained by review of pathway-reaction-compound information available in public resources such as KEGG http://www.genome.jp/kegg/ and literature related to xylose metabolism, mostly the studies in Bacillus spp. [27,42] and L. lactis [23]. In this study we have focused on the xylose transport and conversion of xylose to xylulose 5-P placed in a broader functional context with feeding (xyloside degradation and uptake). Subsystem expansion from model species to other bacteria is accomplished by addition of increasingly distant genomes and orthology-based projection of gene annotations. The phylogenetic boundaries were limited to the Firmicutes phylum that consist mainly of Bacilli and Clostridia classes. The subsystems-based approach to genome analysis and the extensive use of a genome context (clustering on the chromosome, phylogenetic profiling, and shared regulatory sites) allowed significant improvement of the accuracy of gene functional assignment and pathway reconstruction. The results of this analysis are captured in the SEED subsystem "Xylose utilization" available at http://theseed.uchicago.edu/FIG/subsys.cgi.
(iii) Regulatory signals and regulons
To identify candidate DNA-binding sites of XylR regulators, we started from a set of upstream regions of potentially co-regulated genes in small subgroups of the Bacilli/Clostridia genomes according to the phylogenetic tree of XylR proteins. The training sets included the upstream regions of known XylR targets in B. subtilis and/or their orthologs in other XylR-encoding genomes [22,27,43]. An iterative motif detection procedure implemented in the program SignalX was used to identify common regulatory DNA motifs in the training sets and to construct the motif recognition profiles (for a recent review see [44]). The constructed recognition profiles were used to scan a subset of the Bacilli/Clostridia genomes encoding XylR orthologs from the same subgroup on the phylogenetic tree. Positional nucleotide weights in the recognition profile and Z-scores of candidate sites were calculated as the sum of the respective positional nucleotide weights. Genome scanning for additional candidate XylR-binding sites was performed using the Genome Explorer software. The threshold for the site search was defined as the lowest score observed in the training set. This analysis produced gene sets with candidate regulatory sites in the upstream regions.
Bacterial strains, plasmids, and reagents
E. coli strains DH5α (Invitrogen, Carlsbad, CA) and ER2275 [45] were used for gene cloning, and BL21(DE3) (Gibco-BRL, Rockville, MD) was used for protein overexpression. E. coli K-12 knockout mutants ΔxylA (Δb3565), ΔxylB (Δb3564) and ΔxylE (Δb4031) from the Coli Genetic Stock Center [46] were used for complementation analysis. C. acetobutylicum ATCC 824 wild-type and mutant strains were used for analysis of growth phenotype on xylose. The pSY6 vector [47] was used for gene disruption in C. acetobutylicum, and pUC118 (Novagen) and pET28a (Novagen) vectors were used for protein expression in E. coli. Enzymes for DNA manipulations and PCR were from Fermentas, and plasmid purification kits were from Axygen Biotechnology (Hangzhou, China). Antibiotics, buffer components, and all reagents for enzymatic assays were purchased from Sigma-Aldrich.
Gene disruption in C. acetobutylicum and phenotype analysis
Gene disruption in C. acetobutylicum ATCC 824 was performed by using group II intron-based Targetron technology as described previously [47]. Briefly, respective 350 bp fragments for retargeting introns to insert CAC2610 (xylA-II), CAC2612 (xylB), and CAC1345 (xylT) genes were generated by one-step assembly PCR reaction using the primers shown in Additional file 5 according to TargeTron™ gene knockout system (Sigma). The PCR products were then digested and ligated to a targetron plasmid pSY6, yielding the pSY6-xylA-II, pSY6-xylB, and pSY6-xylT. These plasmids were methylated in vivo in E. coli ER2275 (pAN1) [45] and electroporated into C. acetobutylicum ATCC 824, respectively. The transformants were selected on CGM plates with 50 μg ml-1 erythromycin. The resulting mutants with intron insertion of xyl genes were confirmed by PCR (see Additional file 3).
For phenotype growth assays C. acetobutylicum ATCC 824 wild-type and mutant strains were pre-cultured on CGM medium [48] to late-exponential growth phase, and washed twice using P2 minimal medium [49] without any carbon sources. The cultures were started with the same optical density at 600 nm (OD600 nm~0.04), and performed at 37°C in triplicates in 100 ml of P2 minimal medium supplemented with 2, 5, or 20 g l-1 xylose as the sole carbon source. Cell growth was monitored spectrophotometrically at 600 nm. Xylose was quantified by high-pressure liquid chromatography with a model 1200 instrument (Agilent) equipped with a Waters Sugar-Pak I column and a refractive index detector. Double distilled water was used as the mobile phase at a flow rate of 0.6 ml min-1, and the column was operated at 70°C.
Gene cloning in E. coli and complementation analysis
The full-length coding regions of CAC2610 (xylA-II), CAC2612 (xylB), and CAC1345 (xylT) from C. acetobutylicum ATCC 824 were amplified using the primers shown in Additional file 5. PCR amplification was performed using C. acetobutylicum ATCC 824 genomic DNA. PCR fragments were cloned into the pUC118 expression vector digested by BamHI and PstI. The resulting plasmids were transformed into E. coli ΔxylA, ΔxylB, or ΔxylE knockout mutants for the complementation analysis. The empty vector was expressed in the same strain and used as a negative control.
The xylA-II, xylB, and xylT genes from C. acetobutylicum were expressed constitutively under control of the lac promoter in E. coli ΔxylA, ΔxylB, and ΔxylE, respectively. Briefly, cells were pre-cultured on Luria-Bertani medium to mid-exponential growth phase, washed twice, and diluted to the same optical density (OD600 nm~0.04) using the minimal medium without any carbon sources. Complementation experiments were carried out on modified MacConkey agar plates [50] supplemented with 10 g l-1 xylose or on M9 minimal medium containing 5 g l-1 xylose. After incubation at 37°C for 24 h, the colonies of cells with the ability in xylose utilization showed red color on MacConkey agar plates, whereas the colonies of cells unable to utilize xylose keep yellow.
Protein overexpression and purification
For protein overexpression, the CAC2610 (xylA-II), CAC2612 (xylB), and CAC3673 (xylR) genes were PCR-amplified (primers shown in Additional file 5) and cloned into the expression vector pET28a. The recombinant proteins were overexpressed as N-terminal fusions with a His6 tag in E. coli BL21(DE3). The cells were grown on LB medium to an optical density at 600 nm of 0.8 at 37°C, induced by 0.2 mM isopropyl-β-D-thiogalactopyranoside, and harvested after 12 h shaking at 20°C. Protein purification were performed using a rapid nickel-nitrilotriacetic acid (Ni-NTA) agarose minicolumn protocol as described previously [51]. Briefly, harvested cells were resuspended in 20 mM HEPES buffer (pH 7.0) containing 100 mM NaCl, 0.03% Brij-35, and 2 mM β-mercaptoethanol supplemented with 2 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Sigma-Aldrich). Lysozyme was added to a concentration of 1 mg ml-1, and the cells were lysed by freezing-thawing, followed by sonication. After centrifugation at 16,000 g, the Tris-HCl buffer (pH 8.0) was added to the supernatant (final concentration, 50 mM), and it was loaded onto a Ni-NTA agarose column (0.2 ml). After bound proteins were washed with the starting buffer containing 1 M NaCl and 0.3% Brij-35, they were eluted with 0.3 ml of the starting buffer containing 250 mM imidazole. The buffer was then changed to 20 mM HEPES containing 2 mM DTT, 0.5 mM EDTA and 150 mM NaCl by using Bio-Spin columns (Bio-Rad). In all three cases, soluble proteins were obtained with high yield (>1 mg from a 50-ml culture) and purified to >90% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (see Additional file 6).
In vitro enzymatic assays
(i) Xylose isomerase activity
Xylose isomerase activity was assayed by using the colorimetric method as described previously [52]. Briefly, 4-8 μg of purified enzyme was added to 100 μl of 50 mM sodium phosphate buffer (pH 6.8) containing 1 mM MnCl2, 5 mM D-xylose, and incubated for 20 min at 37°C. Formation of D-xylulose was monitored by using the cysteine-sulfuric acid-carbazole method and reading the absorbance at 540 nm [52]. The concentration of xylulose was determined from a standard curve which was made using different concentrations of xylulose. No activity was detected in the control experiments in which another gene (CAC2612 or CAC3673) was expressed in the same vector and purified in parallel.
(ii) Xylulokinase activity
Xylulokinase activity was assayed by coupling the formation of ADP to the oxidation of NADH to NAD+ via pyruvate kinase and lactate dehydrogenase and monitored at 340 nm. Briefly, 3-6 μg of purified enzyme was added to 500 μl of 50 mM Tris buffer (pH 7.5) containing 10 mM MgSO4, 1.2 mM ATP, 1.2 mM phosphoenolpyruvate, 0.3 mM NADH, 1.2 U of pyruvate kinase, 1.2 U of lactate dehydrogenase, and 1 mM D-xylulose. The change in NADH absorbance was monitored at 340 nm by using a Beckman DU-800 spectrophotometer. A NADH extinction coefficient of 6.22 mM-1 cm-1 was used for rate calculation. No activity was detected in the control experiments in which another gene (CAC2610 or CAC3673) was expressed in the same vector and purified in parallel.
Analysis of XylR-DNA interactions by EMSA
Interaction of purified recombinant XylR protein with its DNA motif was assessed by EMSA. The 180-bp DNA fragments from the upstream region of xyl genes were PCR-amplified from C. acetobutylicum ATCC 824 genomic DNA using the primers shown in Additional file 5. One of the primers was 5'-biotinylated by Sangon Corp. (Shanghai, China). The PCR products were purified with the PCR purification kit, and their concentration was determined spectrophotometrically.
For EMSA the biotin-labeled DNA (1 nM) was incubated with the indicated amount of purified XylR protein in 20 μl of binding buffer containing 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, and 0.05% NP-40. After 20 min incubation at 28°C, the reaction mixture was electrophoresed at 4°C on a 6% native polyacrylamide gel in 0.5× Tris-borate-EDTA for 2 h at 80 V. The gel was electrophoretically transferred onto a nylon membrane (Millipore, Billerica, MA) and fixed by UV cross-linking. Biotin-labeled DNA was detected with the LightShift Chemiluminescent EMSA kit (Pierce, Rockford, IL). Specificity of the XylR-DNA interactions was tested by including a 400-fold molar excess of non-biotinylated target DNA (specific competitor) and 2 μg salmon sperm DNA (non-specific competitor) in binding reaction mixtures.
Authors' contributions
CY and WJ conceived and supervised the research, and wrote the manuscript. YG carried out the mutagenesis and genetic complementation experiments. YD performed the EMSA and biochemical analysis. CR and ZS participated in the growth experiments. DAR contributed to the reconstruction of XylR regulons. WZ contributed to the development of the manuscript. SY contributed to the design of the study. All authors read and approved the final manuscript.
Supplementary Material
Occurrence and features of genes involved in xylose and xyloside utilization in Firmicutes. (A) Distribution of genes involved in xylose and xyloside utilization in Firmicutes. (B) Occurrence of xylose pathway genes in all species of Firmicutes with completely sequenced genomes.
Candidate DNA-binding sites of xylose regulator XylR in the genomes of the Firmicutes. Candidate DNA-binding sites of xylose regulator XylR in the genomes of the Firmicutes.
Confirmation of the respective C. acetobutylicum mutants with inactivated xylA-II, xylB, or xylT genes by PCR. Confirmation of the respective C. acetobutylicum mutants with inactivated xylA-II, xylB, or xylT genes by PCR. The genes were disrupted by inserting an intron.
EMSA to assess the interactions of C. acetobutylicum xylose regulator XylR with its cognate DNA signals. EMSA to assess the interactions of C. acetobutylicum xylose regulator XylR with its cognate DNA signals. Each of the three 180-bp target DNA fragments (1 nM) from the upstream region of CAC2611-xylA-II, xylB, and xylR genes, respectively, was incubated for 20 min at 28°C with increasing concentrations of XylR protein (0-0.7 μM). Salmon sperm DNA (2 μg) was added to all binding reaction mixtures as a non-specific competitor. No binding of XylR was observed for the DNA segment from the upstream region of gene CAC1705 that is unrelated to carbon metabolism and used as a negative control.
Primers used in this study. Primers used in this study.
Purified recombinant xylulokinase (XylB), xylose isomerase (XylA-II), and xylose regulator (XylR) from C. acetobutylicum. Purified recombinant xylulokinase (XylB), xylose isomerase (XylA-II), and xylose regulator (XylR) from C. acetobutylicum. Proteins (1-2 μg each) were purified by Ni-NTA mini-column.
Contributor Information
Yang Gu, Email: ygu02@sibs.ac.cn.
Yi Ding, Email: dingyi@sibs.ac.cn.
Cong Ren, Email: rencong@sibs.ac.cn.
Zhe Sun, Email: peakning2008@163.com.
Dmitry A Rodionov, Email: rodionov@burnham.org.
Weiwen Zhang, Email: Weiwen.Zhang@asu.edu.
Sheng Yang, Email: syang@sibs.ac.cn.
Chen Yang, Email: cyang@cibt.ac.cn.
Weihong Jiang, Email: whjiang@sibs.ac.cn.
Acknowledgements
This work was supported in part by National Basic Research Program of China (973: 2007CB707803 to CY and WJ), One-hundred-Talented-People program (KSCX2-YW-G-029 to CY) and the Knowledge Innovation Program (KSCX2-YW-G-075 to WJ) from the Chinese Academy of Sciences, National High-tech Research and Development Program of China (863: 2007AA05Z407 to YG), and Science and Technology Commission of Shanghai Municipality (08dz1207100 to WJ).
References
- Keis S, Bennett CF, Ward VK, Jones DT. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int J Syst Bacteriol. 1995;45(4):693–705. doi: 10.1099/00207713-45-4-693. [DOI] [PubMed] [Google Scholar]
- Turnbull PCB. In: Medical microbiology. 4. Baron S, editor. The University of Texas Medical Branch at Galveston, Galveston; 1996. Bacillus. [PubMed] [Google Scholar]
- Durre P. Biobutanol: an attractive biofuel. Biotechnol J. 2007;2(12):1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J. et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183(16):4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell WJ. Physiology of carbohydrate to solvent conversion by clostridia. Adv Microb Physiol. 1998;39:31–130. doi: 10.1016/S0065-2911(08)60015-6. [DOI] [PubMed] [Google Scholar]
- Lee SF, Forsberg CW, Gibbins LN. Xylanolytic Activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50(4):1068–1076. doi: 10.1128/aem.50.4.1068-1076.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prade RA. Xylanases: from biology to biotechnology. Biotechnol Genet Eng Rev. 1996;13:101–131. doi: 10.1080/02648725.1996.10647925. [DOI] [PubMed] [Google Scholar]
- Moracci M, Cobucci Ponzano B, Trincone A, Fusco S, De Rosa M, Oost J van Der, Sensen CW, Charlebois RL, Rossi M. Identification and molecular characterization of the first a-xylosidase from an archaeon. J Biol Chem. 2000;275(29):22082–22089. doi: 10.1074/jbc.M910392199. [DOI] [PubMed] [Google Scholar]
- Sunna A, Antranikian G. Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol. 1997;17(1):39–67. doi: 10.3109/07388559709146606. [DOI] [PubMed] [Google Scholar]
- Lindner C, Stulke J, Hecker M. Regulation of xylanolytic enzymes in Bacillus subtilis. Microbiology. 1994;140(Pt 4):753–757. doi: 10.1099/00221287-140-4-753. [DOI] [PubMed] [Google Scholar]
- Erlandson KA, Delamarre SC, Batt CA. Genetic evidence for a defective xylan degradation pathway in Lactococcus lactis. Appl Environ Microbiol. 2001;67(4):1445–1452. doi: 10.1128/AEM.67.4.1445-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner D, Geissendorfer M, Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988;170(7):3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Park C. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol. 1997;179(22):7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer P, Gartner D, Allmansberger R, Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989;171(7):3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel D, Hillen W. A Bacillus subtilis 168 mutant with increased xylose uptake can utilize xylose as sole carbon source. FEMS Microbiol Lett. 1996;135:175–178. doi: 10.1111/j.1574-6968.1996.tb07985.x. [DOI] [Google Scholar]
- Ounine K, Petitdemange H, Raval G, Gay R. Regulation and butanol inhibition of D-xylose and D-glucose uptake in Clostridium acetobutylicum. Appl Environ Microbiol. 1985;49(4):874–878. doi: 10.1128/aem.49.4.874-878.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fond O, Engasser JM, Matta-El-Amouri G, Petitdemange H. The acetone butanol fermentation on glucose and xylose. I. Regulation and kinetics in batch cultures. Biotechnol Bioeng. 1986;28(2):160–166. doi: 10.1002/bit.260280203. [DOI] [PubMed] [Google Scholar]
- Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol. 2003;7(2):238–251. doi: 10.1016/S1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer F, Reizer J, Reizer A, Saier MH Jr. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology. 1994;140(Pt 9):2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Mironov AA, Gelfand MS. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiol Lett. 2001;205(2):305–314. doi: 10.1111/j.1574-6968.2001.tb10965.x. [DOI] [PubMed] [Google Scholar]
- Dahl MK, Degenkolb J, Hillen W. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J Mol Biol. 1994;243(3):413–424. doi: 10.1006/jmbi.1994.1669. [DOI] [PubMed] [Google Scholar]
- Erlandson KA, Park JH, Wissam, El K, Kao HH, Basaran P, Brydges S, Batt CA. Dissolution of xylose metabolism in Lactococcus lactis. Appl Environ Microbiol. 2000;66(9):3974–3980. doi: 10.1128/AEM.66.9.3974-3980.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram R, Wunsche A, Sprehe M, Hillen W. Regulated expression of HPrK/P does not affect carbon catabolite repression of the xyn operon and of rocG in Bacillus subtilis. FEMS Microbiol Lett. 2006;259(1):147–152. doi: 10.1111/j.1574-6968.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Russell RR, Aduse-Opoku J, Sutcliffe IC, Tao L, Ferretti JJ. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem. 1992;267(7):4631–4637. [PubMed] [Google Scholar]
- Lin ECC. Dissimilatory pathways of sugars, polyols, and carboxylates. Washington, D.C.: American Society for Microbiology; 1987. [Google Scholar]
- Schmiedel D, Kintrup M, Kuster E, Hillen W. Regulation of expression, genetic organization and substrate specificity of xylose uptake in Bacillus megaterium. Mol Microbiol. 1997;23(5):1053–1062. doi: 10.1046/j.1365-2958.1997.2881654.x. [DOI] [PubMed] [Google Scholar]
- Chaillou S, Bor YC, Batt CA, Postma PW, Pouwels PH. Molecular cloning and functional expression in Lactobacillus plantarum 80 of xylT, encoding the D-xylose-H+ symporter of Lactobacillus brevis. Appl Environ Microbiol. 1998;64(12):4720–4728. doi: 10.1128/aem.64.12.4720-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Batt CA. Restoration of a defective Lactococcus lactis xylose isomerase. Appl Environ Microbiol. 2004;70(7):4318–4325. doi: 10.1128/AEM.70.7.4318-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann JE, Schulz GE. Structure and mechanism of L-fucose isomerase from Escherichia coli. J Mol Biol. 1997;273(1):256–268. doi: 10.1006/jmbi.1997.1280. [DOI] [PubMed] [Google Scholar]
- Schellenberg GD, Sarthy A, Larson AE, Backer MP, Crabb JW, Lidstrom M, Hall BD, Furlong CE. Xylose isomerase from Escherichia coli. Characterization of the protein and the structural gene. J Biol Chem. 1984;259(11):6826–6832. [PubMed] [Google Scholar]
- Vieille C, Epting KL, Kelly RM, Zeikus JG. Bivalent cations and amino-acid composition contribute to the thermostability of Bacillus licheniformis xylose isomerase. Eur J Biochem. 2001;268(23):6291–6301. doi: 10.1046/j.0014-2956.2001.02587.x. [DOI] [PubMed] [Google Scholar]
- Di Luccio E, Petschacher B, Voegtli J, Chou HT, Stahlberg H, Nidetzky B, Wilson DK. Structural and kinetic studies of induced fit in xylulose kinase from Escherichia coli. J Mol Biol. 2007;365(3):783–798. doi: 10.1016/j.jmb.2006.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus A, Hueck C, Gartner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol. 1994;176(6):1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Gelfand MS, Koonin EV, Mironov AA. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 2000;28(3):695–705. doi: 10.1093/nar/28.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchuk GE, Rodionov DA, Yang C, Li X, Osterman AL, Dervyn E, Geydebrekht OV, Reed SB, Romine MF, Collart FR. et al. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc Natl Acad Sci USA. 2009;106(8):2874–2879. doi: 10.1073/pnas.0806798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Rodionov DA, Li X, Laikova ON, Gelfand MS, Zagnitko OP, Romine MF, Obraztsova AY, Nealson KH, Osterman AL. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J Biol Chem. 2006;281(40):29872–29885. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- Yang C, Rodionov DA, Rodionova IA, Li X, Osterman AL. Glycerate 2-kinase of Thermotoga maritima and genomic reconstruction of related metabolic pathways. J Bacteriol. 2008;190(5):1773–1782. doi: 10.1128/JB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler A, Rygus T, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus licheniformis encoded regulon for xylose utilization. Arch Microbiol. 1991;155(6):526–534. doi: 10.1007/BF00245345. [DOI] [PubMed] [Google Scholar]
- Scheler A, Hillen W. Regulation of xylose utilization in Bacillus licheniformis: Xyl repressor-xyl-operator interaction studied by DNA modification protection and interference. Mol Microbiol. 1994;13(3):505–512. doi: 10.1111/j.1365-2958.1994.tb00445.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev. 2007;107(8):3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelstein LD, Papoutsakis ET. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1993;59(4):1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Hu S, Yang Y, Gu Y, Chen J, Yang Y, Jiang W, Yang S. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 2007;17(11):963–965. doi: 10.1038/cr.2007.91. [DOI] [PubMed] [Google Scholar]
- Wiesenborn DP, Rudolph FB, Papoutsakis ET. Thiolase from Clostridium acetobutylicum ATCC 824 and Its Role in the Synthesis of Acids and Solvents. Appl Environ Microbiol. 1988;54(11):2717–2722. doi: 10.1128/aem.54.11.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer SH, Blaschek HP, Smith TL. Effect of Butanol Challenge and Temperature on Lipid Composition and Membrane Fluidity of Butanol-Tolerant Clostridium acetobutylicum. Appl Environ Microbiol. 1987;53(12):2854–2861. doi: 10.1128/aem.53.12.2854-2861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela MF, Wilson TH, Rodon-Rivera V, Shepherd S, Dehne TA, Rector AC. Mutants of the lactose carrier of Escherichia coli which show altered sugar recognition plus a severe defect in sugar accumulation. J Membr Biol. 2000;174(3):199–205. doi: 10.1007/s002320001044. [DOI] [PubMed] [Google Scholar]
- Osterman AL, Lueder DV, Quick M, Myers D, Canagarajah BJ, Phillips MA. Domain organization and a protease-sensitive loop in eukaryotic ornithine decarboxylase. Biochemistry. 1995;34(41):13431–13436. doi: 10.1021/bi00041a021. [DOI] [PubMed] [Google Scholar]
- Dische Z, Borenfreund E. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J Biol Chem. 1951;192(2):583–587. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Occurrence and features of genes involved in xylose and xyloside utilization in Firmicutes. (A) Distribution of genes involved in xylose and xyloside utilization in Firmicutes. (B) Occurrence of xylose pathway genes in all species of Firmicutes with completely sequenced genomes.
Candidate DNA-binding sites of xylose regulator XylR in the genomes of the Firmicutes. Candidate DNA-binding sites of xylose regulator XylR in the genomes of the Firmicutes.
Confirmation of the respective C. acetobutylicum mutants with inactivated xylA-II, xylB, or xylT genes by PCR. Confirmation of the respective C. acetobutylicum mutants with inactivated xylA-II, xylB, or xylT genes by PCR. The genes were disrupted by inserting an intron.
EMSA to assess the interactions of C. acetobutylicum xylose regulator XylR with its cognate DNA signals. EMSA to assess the interactions of C. acetobutylicum xylose regulator XylR with its cognate DNA signals. Each of the three 180-bp target DNA fragments (1 nM) from the upstream region of CAC2611-xylA-II, xylB, and xylR genes, respectively, was incubated for 20 min at 28°C with increasing concentrations of XylR protein (0-0.7 μM). Salmon sperm DNA (2 μg) was added to all binding reaction mixtures as a non-specific competitor. No binding of XylR was observed for the DNA segment from the upstream region of gene CAC1705 that is unrelated to carbon metabolism and used as a negative control.
Primers used in this study. Primers used in this study.
Purified recombinant xylulokinase (XylB), xylose isomerase (XylA-II), and xylose regulator (XylR) from C. acetobutylicum. Purified recombinant xylulokinase (XylB), xylose isomerase (XylA-II), and xylose regulator (XylR) from C. acetobutylicum. Proteins (1-2 μg each) were purified by Ni-NTA mini-column.