Abstract
C5L2 is a new cellular receptor found to interact with the human anaphylatoxins complement factor C5a and its C-terminal cleavage product C5a des Arg. The classical human C5a receptor (C5aR) preferentially binds C5a, with a 10-100-fold lower affinity for C5a des Arg. In contrast, C5L2 binds both ligands with nearly equal affinity. C5aR presents acidic and tyrosine residues in its N-terminus that interact with the core of C5a while a hydrophobic pocket formed by the transmembrane helices interacts with residues in the C-terminus of C5a. Here, we have investigated the molecular basis for the increased affinity of C5L2 for C5a des Arg. Rat and mouse C5L2 preferentially bound C5a des Arg, whereas rodent C5aR showed much higher affinity for intact C5a. Effective peptidic and non-peptidic ligands for the transmembrane hydrophobic pocket of C5aR were poor inhibitors of ligand binding to C5L2. An antibody raised against the N-terminus of human C5L2 did not affect the binding of C5a to C5L2 but did inhibit C5a des Arg binding. A chimeric C5L2, containing the N-terminus of C5aR, had little effect on the affinity for C5a des Arg. Mutation of acidic and tyrosine residues in the N-terminus of human C5L2 revealed that three residues were critical for C5a des Arg binding but had little involvement in C5a binding. C5L2 thus appears to bind C5a and C5a des Arg by different mechanisms and, unlike C5aR, C5L2 uses critical residues in its N-terminal domain for binding only to C5a des Arg.
Complement fragment 5a (C5a1) is a 74 residue polypeptide that is a multi-functional proinflammatory mediator that causes leukocyte chemoattraction and degranulation, increases vascular permeability and stimulates cytokine secretion (1). The C-terminal Arg is rapidly cleaved in vivo to form C5a des Arg, a plasma-stable metabolite that has a different spectrum of activities to intact C5a (2). The classical receptor for C5a (C5aR) is a member of the G protein-coupled receptor superfamily (3,4) and has high affinity for intact C5a but 10-100-fold lower affinity for C5a des Arg (5). The receptor N–terminus is required for high affinity binding of C5a but not for receptor activation (6,7): a series of acidic and O-sulfated tyrosine residues interact with basic residues in the core of C5a (8,9). A second distinct binding site is formed by charged residues in the second and third extracellular loops and the external faces of the transmembrane helical bundle and hydrophobic residues in the core of the receptor (10). This site is responsible for receptor activation and is the target for agonist and antagonist peptidic mimics of the C-terminus of C5a (11). The second C5a receptor to be discovered, C5L2, does not appear to couple to G proteins and appears to have an anti-inflammatory function, balancing the pro-inflammatory role of C5aR (12-14). The mechanism of the anti-inflammatory effect is not known but may be related to the ligand binding properties of C5L2, which has nearly equal affinities for C5a and C5a des Arg. C5L2 has 41% sequence identity with C5aR (15), with a similar array of acidic and tyrosine residues at the N-terminus and many of the charged and hydrophobic residues in the loops and transmembrane regions of C5aR that are involved in the interaction with the C-terminus of C5a are also conserved in C5L2.
Although recent reports have identified a C5aR agonist peptide as a ligand at C5L2 (16,17), the mechanism of ligand binding to C5L2 has not been reported. In this paper, we describe a systematic analysis of the role of the N-terminus of C5L2 in the interaction with ligands and demonstrate that this domain has a critical role in binding to C5a des Arg but not to C5a.
EXPERIMENTAL PROCEDURES
Cell Culture
RBL-2H3 and CHO cells were routinely cultured in DMEM + 10% (v/v) fetal calf serum supplemented with 400mg/L G-418 for transfected cells, at 37°C, 5% CO2. In experiments where NaClO3 was used to inhibit tyrosine sulfation, cells were grown for 5 days in low sulfate DMEM/F12 (Sigma) supplemented with 10% (v/v) dialyzed fetal calf (Gibco) serum and 10mM NaClO3 (Sigma).
Receptor cloning and expression
Mouse C5L2 cDNA was kindly provided by Dr Hui Tian, Amgen Inc. Human C5L2 was cloned as previously described (15). All constructs were cloned into pEE6 (Celltech) and transfected into CHO cells by standard electroporation protocols. After selection in G418, homogenous populations of cells were produced by two rounds of fluorescence-activated cell sorting using rabbit polyclonal antisera that recognize the N-terminal sequence of human, rat or mouse C5L2 on a Becton-Dickinson Vantage flow cytometer (mouse, human), or cloning by limiting dilution (rat). RBL-2H3 cells expressing mouse C5aR were a generous gift from Dr Jorg Zwirner (Gottingen); CHO cells expressing human C5aR were made as previously described (18). Rat C5aR, cloned from rat liver using sequence Y09613 and rat C5L2 cDNA, a generous gift from Dr Vitaliy Gavrilyuk, (University of Illinois), were subcloned into pEE6 for expression in CHO cells. The cDNA for human C5aR with the mutations D15A, D18A or Y14F in pCDNA3.1 were generous gifts from Dr Carla de Haas (Utrecht). A monoclonal antibody (M2, Sigma) that recognizes an N-terminal FLAG tag sequence was used to sort the highest 50% of transfected cells in two rounds of fluorescence-activated cell sorting.
Production of Peptides, Antibodies and Recombinant Ligands
Antiserum against a synthetic peptide analog of the N-terminus of human C5L2(1-32) was prepared as described previously (19) and affinity purified using the synthetic peptide. Compounds 73 and 74 (non-peptidic antagonists (20)) and 3257 and 3261 (non-peptidic agonists (21)) at the human C5a receptor are described in Supplemental Material.
His6-tagged recombinant ligands and peptides were synthesized as described previously (22).
Construction of Mutants of Human C5L2 N-terminus
Substitution mutants (E9A, D12A, D15A, D18A, D22A, D25A, Y8F, Y10F, Y13F) were made using Stratagene QuikChange kits, according to the manufacturer’s instructions and cloned into pEE6 after authentication by DNA sequencing. A chimeric C5L2, with the N-terminus of hC5aR, substituting residues 1-32 (hC5L2) with 1-37 (hC5aR), was made by overlap extension using the primers shown in Supplemental Material. After authentication by DNA sequencing, chimera C5aRNC5L2 was cloned into pEE6 for transfection of CHO cells.
His6-C5a/C5a des Arg Binding Assays
The binding of ligand to wildtype and mutant C5L2 and C5aR was measured using an indirect immunofluorescence method, as previously described (23,24). Cells transfected with the appropriate receptor (50 000 per well of a 96-well microtitre plate) were incubated with the stated concentrations of His6-tagged C5a or C5a des Arg for 30 min at 4°C, in PBS+0.1% BSA, 0.2% (w/v) NaN3, then washed twice with cold PBS to remove unbound ligand. Anti-His6 antibody (mouse IgG, Qiagen) was then added (50μl at 2μg/ml) and incubated with the cells for 30 min at 4°C. After a further wash in PBS, 50μl of FITC-anti-mouse IgG (Sigma) was used to detect bound ligand using a FACSsort flow cytometer (Becton Dickinson). Binding was calculated as the percentage of positive cells i.e. with a fluorescent intensity higher than control cells measured in the absence of ligand. Receptor affinities are reported as EC50 values, calculated by non-linear regression analyses using GraphPad Prism v4.0 (GraphPad Software, San Diego, CA).
RESULTS
Rodent C5L2 homologs are primarily C5a des Arg binding proteins
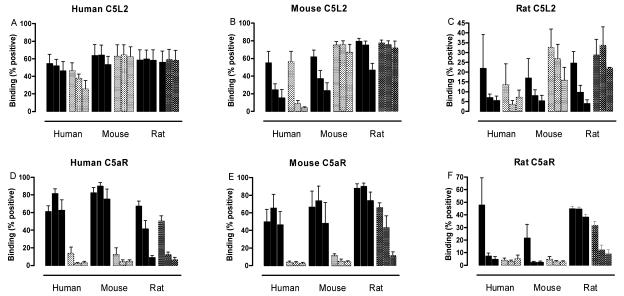
Although human C5L2 has been well characterized, ligand binding by rodent C5L2 has not previously been reported. Rat, mouse and human C5aR and C5L2 were expressed in CHO cells (except for mouse C5aR, which was expressed in RBL cells), and high expressing cell populations were produced either by cell sorting or by cloning. His6-tagged human, rat and mouse C5a and C5a des Arg at a high, intermediate or low dose (5000, 50, 5nM, determined by the previously reported ligand-binding affinities of hC5L2 (15,16) were allowed to bind at 4°C, followed by extensive washing. Surface-bound ligand was then detected by indirect immunofluorescence using an anti-His6 monoclonal antibody. This method measures only the ligand that remains bound after ~1.5 hrs (i.e. that has a slow off-rate) and so C5a des Arg binding to C5aR, for example, appears quite low (Fig 1), and no binding of human C3a or C3a des Arg to human C5L2 can be detected (data not shown). However, the EC50 values for hC5a binding to both hC5L2 (~7nM) and hC5aR (~4nM) (Table 1a) are in close agreement with previously published binding affinities obtained using radiolabelled ligands, ~3nM for both receptors (16). Similarly, hC5a des Arg binding to hC5L2 and hC5aR produced EC50 values of ~36nM and ~527nM (Table 1c), again not dissimilar to affinity values obtained indirectly using radio-competition assays, (~12nM and ~660nM, respectively (16)). Thus, the ligand binding assay employed here is a valid quantitative method for the analysis of receptor affinities, with the advantage that it does not require chemical modification of the ligand and so is used here to facilitate the comparison of ligand binding to receptors from different species.
Figure 1. The binding of C5a and C5a des Arg to human and rodent C5L2 and C5aR.
RBL and CHO cells transfected with human (A, D), mouse (B, E) or rat (C, F) C5L2 (A-C) or C5aR (D - F) were incubated for 30 min with either a high (5000nM), intermediate (50nM) or low (5nM) dose of His6-tagged C5a (solid bars) or His6-tagged C5a de Arg (shaded bars), and binding measured using indirect immunofluorescence, expressed as % of cells with fluorescence higher than cells incubated without ligand. The bars (high to low concentrations shown from left to right) are means +/− SEM of three separate experiments performed in duplicate.
Table 1a. The binding of C5a and C5a des Arg to C5L2 mutated at N-terminal acidic residues.
| Receptor | WT |
E9A |
D12A |
D15A |
D18A |
D22A |
D25A |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | EC50a | pEC50b | nc | EC50 | pEC50 | n | EC50 | pEC50 | n | EC50 | pEC50 | n | EC50 | pEC50 | n | EC50 | pEC50 | n | EC50 | pEC50 | n |
| C5a | 6.92 | 8.16 ±0.05 |
18 | 3.10 | 8.51 ±0.08** |
4 | 9.37 | 8.03 ±0.08ns |
6 | 7.10 | 8.15 ±0.09ns |
4 | 3.88 | 8.41 ±0.10* |
4 | 22.9 | 7.64 ±0.08*** |
4 | 2.59 | 8.59 ±0.14** |
4 |
| C5a desArg | 36.0 | 7.44 ±0.07 |
18 | 94.5 | 7.02 ±0.12* |
4 | 403 | 6.40 ±0.13*** |
6 | 252 | 6.60 ±0.10*** |
4 | 48.6 | 7.31 ±0.07ns |
4 | - | <3 | 4 | 33.4 | 7.48 ±0.15ns |
4 |
EC50 = concentration (nM) resulting in 50% of the maximum binding;
pEC50 = −log10EC50;
n = number of separate experiments performed in duplicate.
pEC50 Significantly different from WT-C5L2:
<5%;
< 0.5%;
< 0.005% (two tailed t test).
Table 1c. The binding of C5a and C5a des Arg to C5aR mutated at N-terminal acidic and tyrosyl residues.
| WT |
D15A |
D18A |
Y14F |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | EC50 a |
pEC50b | nc | EC50 | pEC50 | n | EC50 | pEC50 | n | EC50 | pEC50 | n |
| C5a | 5.52 | 8.26 ±0.05 |
8 | 34.1 | 7.47 ±0.14*** |
4 | 17.0 | 7.71 ±0.06*** |
8 | 41.8 | 7.38 ±0.06*** |
6 |
|
C5a
desArg |
527 | 6.28 ±0.14 |
8 | 2880 | 5.541** ±0.15 |
4 | 4390 | 5.36 ±0.12*** |
8 | 1210 0 |
4.92 ±0.06*** |
6 |
EC50 = concentration (nM) resulting in 50% of the maximum binding;
pEC50 = −log10EC50;
n = number of separate experiments performed in duplicate.
pEC50 significantly different from WT-C5L2:
<5%;
< 0.5%;
< 0.005% (two tailed t test).
It can be seen that C5L2 of all three species has an equal or higher affinity (or slower off-rate) for C5a des Arg compared to C5a. For the rodent species in particular, the lower levels of C5a binding suggests that C5L2 is primarily a C5a des Arg binding protein, with a much lower propensity to bind or retain C5a. Mouse C5L2 is still saturated at 5nM mC5a des Arg, whereas the binding of 5nM mC5a is <50% maximal. When the affinity of mC5L2 was determined more carefully, using a wider range of ligand concentrations, the affinity for mC5a was >4000-fold lower than mC5a des Arg (data not shown). This is in contrast to mC5aR, which clearly shows a preference for mC5a over mC5a des Arg (Fig 1). For rat C5L2, the difference is also marked, with the binding of 5nM rC5a reduced to almost undetectable levels but 5nM rC5a des Arg still at near-maximal binding (Fig 1). The species of C5a/C5a des Arg used also appears to show differences between C5L2 and C5aR. Human C5L2 appears to show promiscuous binding with little preference for same-species ligands, whereas human C5aR shows a higher degree of discrimination. Mouse and rat C5L2, in contrast, bind human C5a and C5a des Arg only weakly but show a clear preference for C5a des Arg over C5a (Fig 1).
A panel of synthetic analogs of the C-terminal of C5a are more potent inhibitors at C5aR than at C5L2
For C5aR, the full agonist potential of C5a is vested in only the C-terminal decapeptide of C5a, which binds in the transmembrane binding pocket. We have used a panel of peptidic and non-peptidic analogs of the C5a C-terminus to compare the characteristics of the transmembrane binding pockets of C5L2 and C5aR. All the analogs were capable of blocking the binding of 50nM His6-hC5a to hC5aR at statistically significant levels (Fig 2A): F-[OP(DCha)WR], YSFKPMPLaR, FKP(DCha)Cha(D-R)-CO2H, FKP(DCha)Cha(D-R)-CONH2, 73, 74, 3261, 3257 (all 100μM), W-54011 and YFKAChaChaL(D-F)R (both 10μM because of limited solubility and toxicity, respectively). In contrast, these inhibitors had very weak effects on His6-hC5a binding to hC5L2 (Fig 2B), and only 3257, 3261 and FKPDChaChaD-R-CONH2 inhibited binding to a significant extent. 3257, 3261, FKPDChaChaD-R-CO2H and FKPDChaChaD-R-CONH2 could partly inhibit His6-hC5a des Arg binding to hC5L2 (Fig 2C). The analogs were all ineffective at mC5L2, although only mC5a des Arg binding was measured as mC5a binding levels were too low for accurate assessment of inhibition (Fig 2D). Thus, despite the conservation of many of the same residues, ligand binding to the transmembrane binding pockets of C5aR and C5L2 is clearly different.
Figure 2. The binding of analogs of the C-terminus of C5a to human C5L2 and C5aR.
CHO cells transfected with either human C5aR (A) human C5L2 (B, C) or mouse C5L2 (D) were pre-incubated with high doses (YFKAChaChaLD-FR, W-54011 10μM, remainder 100μM) of C5a analogs for 15 min, then incubated for 30 min with 50nM His6-tagged human C5a (A, B), human C5a des Arg (C) or mouse C5a des Arg (D) and bound ligand quantified by indirect immunofluorescence, expressed as % of binding to cells in the absence of analogs. The bars are means +/− SEM of 4 -7 separate experiments performed in duplicate. Significance of difference from 100 was assessed by one sample t test, * = p<0.05.
An antibody raised against the N-terminus of human C5L2 inhibits C5a des Arg but not C5a binding
To examine the role of the N-terminus of C5L2 in ligand binding, a rabbit polyclonal anti-serum, prepared using a synthetic peptide with the N-terminal sequence of human C5L2(1-32) (19), was used. The affinity-purified anti-serum was preincubated at 0.28mg/ml with hC5L2, before the binding affinity for C5L2 ligands was measured. For hC5L2, the high affinity binding of His6-hC5a was not affected at all by antibody pre-treatment (p=0.62, Student t test) but the affinity for hC5a des Arg was decreased by more than 370-fold (pEC50 values significantly different, p = 0.0006). The lack of an effect on hC5a binding suggests that the antibody is blocking binding of C5a des Arg by a specific mechanism, possibly by hindering access to key residues in the N-terminal domain of C5L2.
Replacement of the C5L2 N-terminal domain with C5aR sequence does not decrease affinity for C5a des Arg
The inhibition of hC5a des Arg binding by anti-hC5L2 antibody implicates the N-terminus in ligand binding. The role of the N-terminal domain was further investigated by making a chimeric form of hC5L2 with the N-terminus of hC5aR, substituting residues 1-32 (hC5L2) with 1-37 (hC5aR). After transfection and selection of CHO cells expressing similar levels of receptors, the binding affinity of His6-hC5a and hC5a des Arg was measured (Fig 4). The affinity of the chimeric receptor, C5aRNC5L2, for hC5a was changed only slightly (less than 2-fold change relative to both parental receptors). Interestingly, hC5a des Arg binding was also unaffected; the affinity of C5aRNC5L2 was not significantly decreased (<2-fold) relative to hC5L2 although it was clearly higher than the affinity for C5aR (>15-fold). This finding suggests that the N-terminus is not the only determinant of the high affinity of hC5L2 for hC5a des Arg.
Figure 4. The binding of C5a and C5a des Arg to human C5L2, C5aR and an N-terminal chimera.
CHO cells transfected with human C5L2, C5aR and a chimera of human C5L2 with the N-terminal domain of C5aR (C5aRNC5L2) were incubated for 30 min with His6-tagged C5a (A) or C5a des Arg (B), and bound ligand quantified by indirect immunofluorescence, expressed as % of binding to cells in the absence of analogs. The data are means +/− SEM of three separate experiments performed in duplicate.
Mutation of the C5L2 N-terminus primarily affects binding affinity for C5a des Arg
The data from the chimeric receptor suggested that common ligand binding elements were present in the N-termini of both C5aR and C5L2. To try and identify these elements, we measured the binding affinities of C5L2 mutated at residues analogous to those previously shown to be involved in C5a binding by C5aR: 6 acidic (E9A, D12A, D15A, D18A, D22A, D25A) (Fig 5A, B, Table 1a) and 3 tyrosine (Y8F, Y10F, Y13F) (Fig 5C, D, Table 1b) residues in the N-terminus of hC5L2. The affinity for hC5a was either unchanged or actually increased in all cases except D22A, where affinity decreased by ~8-fold. In contrast, the affinity for hC5a des Arg was significantly reduced in several cases: 3-fold (E9A), 11-fold (D12A), 7-fold (D15A), >1000-fold (D22A), 125-fold (Y10F), 39-fold (Y13F). In all cases except D22A the lack of a reduction in affinity for hC5a suggests that this effect on hC5a des Arg binding is specific and not due simply to misfolding of mutant receptors. C5a and C5a des Arg have quite distinct modes of binding to C5L2 that involve different interactions with acidic and tyrosine residues.
Figure 5. The binding of C5a and C5a des Arg to human C5L2 mutated at N-terminal acidic and tyrosyl residues.
CHO cells transfected with wild type (WT, broken line) or mutant human C5L2 (solid lines) were incubated for 30 min with His6-tagged C5a (A, C) or C5a des Arg (B, D), and bound ligand quantified by indirect immunofluorescence, expressed as % of binding of 1μM C5a to cells expressing each mutant C5L2. The data are means +/− SEM of n (see Table 1a) separate experiments performed in duplicate.
Table 1b. The binding of C5a and C5a des Arg to C5L2 mutated at N-terminal tyrosyl residues.
| Receptor | Y8F |
Y10F |
Y13F |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligand | EC50a | pEC50b | nc | EC50 | pEC50 | n | EC50 | pEC50 | n |
| C5a | 4.18 | 8.38 ±0.06* |
8 | 7.70 | 8.11 ±0.08ns |
8 | 5.23 | 8.28 ±0.06ns |
8 |
|
C5a
desArg |
79.7 | 7.10 ±0.10* |
8 | 4490 | 5.35 ±0.44*** |
8 | 1420 | 5.85 ±0.16*** |
8 |
EC50 = concentration (nM) resulting in 50% of the maximum binding;
pEC50 = −log10EC50;
n = number of separate experiments performed in duplicate.
pEC50 Significantly different from WT-C5L2:
<5%;
< 0.5%;
< 0.005% (two tailed t test).
Mutation of the C5aR N-terminus affects binding affinity for both C5a and C5a des Arg
To compare the effects of mutating acidic and tyrosine residues on C5L2 and C5aR, C5a and C5a des Arg binding to CHO cells transfected with human C5aR mutated at D15, D18 or Y14 was measured. Wildtype (WT) C5aR had a 95-fold lower affinity for C5a des Arg than for C5a. All of the mutants had significantly decreased affinities for both C5a and C5a des Arg. Y14F had the largest decreases in affinity, ~76-fold (Table 1c) and 23-fold, for C5a and C5a des Arg, respectively. For D15A, the decreases in affinity were 6- and 5-fold and for D18A, 3- and 8-fold.
Inhibition of sulfotyrosine formation has different effects on ligand binding by C5L2 and C5aR
Incubation of cells with NaClO3 in low-sulfate medium is a specific method for the inhibition of tyrosine sulfation (25). CHO cells expressing either hC5L2 or hC5aR were treated with 10mM NaClO3 and the binding affinities for C5a and C5a des Arg were measured. Receptor expression, determined using specific antibodies, was not significantly affected by this treatment (relative to untreated controls, C5aR expression 115% ± 11; C5L2 expression 73% ± 15, mean ± SEM, n=3). C5a binding to C5aR was substantially inhibited by this treatment (maximal binding was reduced to 24% of untreated control) (Fig 6A) although C5a des Arg binding was almost undetectable for treated and untreated cells. Interestingly, this diminution of the maximal binding was not due to changes in affinity (EC50 ~ 6nM for both untreated and treated cells), suggesting that a minority of the C5aR retained high affinity binding sites after treatment. In contrast, C5a binding to C5L2 was completely unaffected (maximal binding was reduced by <5%) whereas maximal C5a des Arg binding was reduced by ~27% (significantly different from untreated control, p=0.031) but with no change in affinity. This suggests that a majority of C5L2 is unaffected by NaClO3 treatment. Despite this relatively small effect, it is clear that C5a des Arg binding is more sensitive than C5a binding to conditions that are inhibitory for sulfation.
Figure 6. The binding of C5a and C5a des Arg to human C5L2 and C5aR after treatment with sulfation inhibitors.
CHO cells transfected with either C5L2 (A) or C5aR (B) were grown for 5 days in normal medium (+) or low-sulfate DMEM/F12 supplemented with 10mM NaClO3 (−). Cells were incubated with His6-tagged C5a or C5a des Arg and bound ligand quantified by indirect immunofluorescence, expressed as % of 500nM C5a binding to cells. The data are means +/− SEM of three separate experiments performed in duplicate.
DISCUSSION
C5L2 is the most recently discovered anaphylatoxin receptor and is the least well characterized. Although rodent homologs of hC5L2 have been reported (12,16) and a C5L2-nullizygous mouse described (14), the ligand binding properties of these homologs has not yet been reported. We show here that rat and mouse C5L2 have an even stronger preference for C5a des Arg than hC5L2, to such an extent that these receptors might be considered as C5a des Arg rather than C5a receptors. A recent study of the C5L2-nullizygous mouse used only mC5a to stimulate intra-peritoneal accumulation of neutrophils and chemotaxis of mC5L2-deficient bone marrow cells (14) and observed that the loss of C5L2 resulted in an increase in the response to mC5a. It would be interesting to see if any change in responsiveness to the high affinity ligand for mC5L2, mC5a des Arg, occurs in C5L2 nullizygous animals. Our data also suggest that caution should be used when C5a from one species is used to bind to C5L2 or C5aR from another species: The use of human C5a/C5a des Arg in mice could lead to the erroneous conclusion that mC5L2 did not bind these ligands with high affinity.
The ligand binding sites on C5aR have been mapped by a number of methods. Antibodies directed against the N-terminal domain have been shown to inhibit the binding of C5a (26,27) and deletion of the N-terminus also prevents C5a binding (6,7). A chimeric form of C5aR with the N-terminus of the receptor for the closely related anaphylatoxin C3a, C3aR, also loses the ability to bind C5a (28). However, in all of these cases, peptide analogs of the C-terminus of C5a have still been able to activate the receptor, indicating the presence of an additional binding site. These data have lead to the two-site model of receptor activation that proposes a primary high affinity contact between basic residues in the core of C5a and acidic residues in the N-terminus of C5aR, and a secondary interaction between the C-terminus of C5a and a binding pocket formed by hydrophobic residues in the transmembrane domains and charged residues at the bases of the extracellular loops (11). It is interesting to note that C3aR has a similar two-site binding modality, except that the primary site is located on an enlarged second extracellular loop rather than at the N-terminus (28,29). The secondary binding site on C3aR is apparently formed in the same way as the C5aR site, as many of the hydrophobic and charged residues proposed to form this site are conserved and a chimeric C3aR with the N-terminus of C5aR both binds to, and is activated by, C5a. It is therefore very interesting that ligands that might bind at the secondary site of C5L2 have very little ability to block C5a or C5a des Arg binding to C5L2. The lack of effect of the cyclic peptide, F-[OP(DCha)WR], on C5a binding by C5L2 has already been noted (17) and is confirmed here. FKP(DCha)Cha(D-R), an agonist at C5aR that is reported to bind with low affinity to C5L2 (16,17), does not inhibit C5a binding to C5L2 even at 100μM, 4-fold higher than the reported EC50 for C5L2, although we do see a significant inhibition of C5a des Arg binding. The reason for this apparent discrepancy may be the relatively high dose of C5a (50nM) that we employed for this assay compared to the much lower dose used in the previous report (0.1nM). However, we clearly see a significant inhibition of binding to C5aR at this dose of C5a, suggesting that C5L2 has a much lower affinity for FKP(DCha)Cha(D-R) than C5aR. It is interesting to note that of the 6 C5aR agonists tested, 4/6 and 2/6 were capable of inhibiting C5a des Arg and C5a binding to C5L2, respectively, whereas none of the 4 antagonists had any significant effect on either ligand. This suggests that the secondary binding site on C5L2, despite the conservation of residues, might resemble only the agonist-binding conformation of C5aR. It is unclear from our data if the lack of effect of the analogs is due to their low affinity for C5L2 or if the interaction between C5a/C5a des Arg and the secondary site on C5L2 is not critical for high affinity binding. The lack of signaling of C5L2 after ligand binding does suggest that the transmembrane site is not involved in C5a/C5a des Arg binding and that strong interactions with ligand are made in other domains of C5L2.
We also observed a difference between the roles of the N-termini in hC5aR and hC5L2 in ligand binding. In contrast to hC5aR (26), antibodies that bind to the N-terminal domain of hC5L2 could significantly inhibit hC5a des Arg but not C5a binding. Although this could be due to the 9-fold difference in affinity of hC5L2 for hC5a and hC5a des Arg, as measured by immunofluorescence detection of the His6-tag, the apparent affinity shift for hC5a des Arg in the presence of antibody is 373-fold, larger than would be expected on the basis of affinity for hC5a des Arg alone. We also observed a selective inhibition of hC5a des Arg binding to hC5L2 when we treated transfected cells with the chemotaxis inhibitor protein of Staphylococcus aureus (CHIPS) at a concentration that completely inhibited hC5a binding to hC5aR2. CHIPS binds to hC5aR at a site in the N-terminus that involves acidic residues D10, D15 and D18 and is a competitive inhibitor of hC5a binding (30). The differential role of the N-terminus was also observed when a chimeric hC5L2 was produced that expressed the N-terminal domain of hC5aR. The chimeric receptor had an affinity for hC5a des Arg that was not significantly different from hC5L2 but which was significantly higher than hC5aR. As expected, the affinity for hC5a was similar for all three receptors, as was the ratio of affinities for hC5a/hC5a des Arg: for hC5aR, this was 100, whereas hCL2 and C5aRNC5L2 had ratios of 8 and 5, respectively. This suggests that, although the N-terminus is involved in the binding of hC5a des Arg by hC5L2, there is a significant contribution from additional binding sites. For C5aR, residues 1-18 of the N-terminus have been suggested to interact with the extracellular loops to achieve the correct conformation for ligand binding at residues 21-30 of the N-terminus (8). A similar mechanism may occur for C5L2: The antibody epitope is currently unknown but CHIPS binds to residues 1-18 of C5aR (30) and so it is possible that the inhibition of C5a des Arg binding by these molecules is indirect, due to the disruption of a complex binding surface.
As the N-termini of hC5aR and hC5L2 can be exchanged with little effect on ligand binding, it is likely that a similar array of acidic and/or sulfated residues is present in both receptors. The role of the N-terminal acidic and tyrosyl residues in hC5L2 was investigated in more detail by point mutagenesis. Similar studies have previously been performed on hC5aR: hC5aR(D15,16,18,21N) showed a 40-fold decrease in hC5a affinity and hC5aR(D10,15,16,18,21N) showed a 133-fold reduction (7); single mutants of D10N and D27N or a double mutant (D21,27N) had no effect on hC5a binding, whereas the multiple substitutions hC5aR(D10, 15, 16) or hC5aR(D15,16,21,27) showed no detectable hC5a binding (31). Similarly, an NMR study on the hC5aR N-terminus highlighted the importance of residues 21-30 in hC5a binding (8). Thus, we might expect single substitutions to have only small effects on ligand binding by hC5L2. We mutated 6 acidic (5 D and 1 E) residues in the region of the N-terminus analogous to the primary ligand-binding domain of hC5aR and found that only one, D22, caused a significant disruption of hC5a binding (~8-fold decrease in affinity). This residue does not align with a D in hC5aR (N23 is a likely analog) and so the effects of mutation at this position cannot be directly compared in the two receptors. In contrast to the moderate effects on hC5a, four of the mutants caused substantial reductions in the binding of hC5a des Arg: E9A, D12A, D15A and D22A. These mutants had hC5a/C5a des Arg affinity ratios of 30, 43, 81 and >1000, respectively, which were much larger than for WT hC5L2 (~5). Clearly, hC5a des Arg binding has a greater dependence on these acidic residues than hC5a. In contrast, the mutation of two acidic residues on hC5aR, D15 and D18, had similar effects on both hC5a and hC5a des Arg binding. We also mutated tyrosyl residues in hC5L2 that may be modified by the addition of sulfate moieties. In hC5aR, Y11 and Y14 have been shown to be sulfated; the mutant Y11F shows almost complete loss of C5a binding and Y14F shows ~50% loss of binding affinity whereas mutation of Y8 has no effect on either sulfation or ligand binding (9). Here, we confirmed that mutation of Y14 to F could cause a substantial loss of affinity of hC5aR for C5a and also showed that C5a des Arg binding could be similarly affected. In contrast, mutation of Y10F or Y13F on hC5L2 (the analogous residues to Y11 and Y14 of hC5aR) had no significant effect on hC5a binding but could substantially inhibit hC5a des Arg binding (hC5a/hC5a des Arg binding ratios of 583 and 272, respectively). The mutant Y8F had a much smaller effect on binding (ratio of ~19). The possibility that ligand/receptor interactions occur through the aromatic moiety of the Y10 and Y13 is not explored by substitution with phenylalanine. However, as the Y14F substitution in C5aR largely inhibits C5a binding, the importance of these potential aromatic interactions is clearly different for the two receptors. The inhibition of tyrosine sulfation by 16 hours growth in low sulfate medium supplemented with NaClO3 has been shown to significantly inhibit ligand binding to CCR5 (32). C5aR was very sensitive to these conditions whereas C5L2 was much more resistant, even after 5 days growth in NaClO3-containing medium. This resistance suggests a much slower turnover of C5L2 relative to C5aR or that sulfation is largely irrelevant in the formation of the ligand binding site on C5L2. These possibilities are currently under investigation. However, the sensitivity of C5a des Arg binding relative to C5a is further evidence of a greater role of N-terminal residues in C5a des Arg binding.
In conclusion, we have shown that, while ligand binding by C5L2 shares some common features with C5aR, there are also unique properties with the N-termini of these receptors apparently having quite different binding roles. Strikingly, C5L2 appears to use a distinctly different mechanism from C5aR to bind to its two high affinity ligands, C5a and C5a des Arg.
Supplementary Material
Figure 3. The effects of an antibody against the N-terminus of human C5L2 on ligand binding.
CHO cells transfected with human C5L2 were pre-incubated with PBS (filled symbols) or 0.28mg/ml of affinity purified rabbit polyclonal antibody (open symbols) raised against the N-terminus of human C5L2, for 15 min and then incubated for 30 min with His6-tagged human C5a (squares) or C5a des Arg (triangles). Bound ligand was quantified by indirect immunofluorescence and expressed as % of maximal binding to cells, i.e. in the absence of antibody. The data are means +/− SEM of three separate experiments performed in duplicate.
Acknowledgements
This work is supported by Wellcome Trust project grant 072231 (to PNM), ARC DP00210598 (to DPF) and NHMRC 252812 Australian project grants (to DPF and SMT).
Footnotes
- C5a
- complement fragment 5a
- C5aR
- human complement fragment 5a receptor
- CHIPS
- chemotaxis inhibitory protein of Staphylococcus aureus
- Cha
- cyclohexylalainine
- WT
- wildtype
Andrew J. Wright, Adrian Higginbottom, Didier Philippe, Abhishek Upadhyay, Stefan Bagby, Robert C. Read, Lynda J. Partridge and Peter N. Monk. Manuscript in preparation.
References
- 1.Guo RF, Ward PA. Annu Rev Immunol. 2005;23:821–852. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- 2.Eglite S, Pluss K, Dahinden CA. J Immunol. 2000;165(4):2183–2189. doi: 10.4049/jimmunol.165.4.2183. [DOI] [PubMed] [Google Scholar]
- 3.Gerard NP, Gerard C. Nature. 1991;349(6310):614–617. doi: 10.1038/349614a0. [DOI] [PubMed] [Google Scholar]
- 4.Boulay F, Mery L, Tardif M, Brouchon L, Vignais P. Biochemistry. 1991;30(12):2993–2999. doi: 10.1021/bi00226a002. [DOI] [PubMed] [Google Scholar]
- 5.Cain SA, Coughlan T, Monk PN. Biochemistry. 2001;40(46):14047–14052. doi: 10.1021/bi011055w. [DOI] [PubMed] [Google Scholar]
- 6.Mery L, Boulay F. Eur J Haematol. 1993;51(5):282–287. doi: 10.1111/j.1600-0609.1993.tb01609.x. [DOI] [PubMed] [Google Scholar]
- 7.DeMartino JA, Van Riper G, Siciliano SJ, Molineaux CJ, Konteatis ZD, Rosen H, Springer MS. J Biol Chem. 1994;269(20):14446–14450. [PubMed] [Google Scholar]
- 8.Chen Z, Zhang X, Gonnella NC, Pellas TC, Boyar WC, Ni F. J Biol Chem. 1998;273(17):10411–10419. doi: 10.1074/jbc.273.17.10411. [DOI] [PubMed] [Google Scholar]
- 9.Farzan M, Schnitzler CE, Vasilieva N, Leung D, Kuhn J, Gerard C, Gerard NP, Choe H. J Exp Med. 2001;193(9):1059–1066. doi: 10.1084/jem.193.9.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber BO, Meng EC, Dotsch V, Baranski TJ, Bourne HR. J Biol Chem. 2001;276(5):3394–3400. doi: 10.1074/jbc.M007748200. [DOI] [PubMed] [Google Scholar]
- 11.Higginbottom A, Cain SA, Woodruff TM, Proctor LM, Madala PK, Tyndall JD, Taylor SM, Fairlie DP, Monk PN. J Biol Chem. 2005 doi: 10.1074/jbc.M410797200. [DOI] [PubMed] [Google Scholar]
- 12.Gavrilyuk V, Kalinin S, Hilbush BS, Middlecamp A, McGuire S, Pelligrino D, Weinberg G, Feinstein DL. J Neurochem. 2005;92(5):1140–1149. doi: 10.1111/j.1471-4159.2004.02942.x. [DOI] [PubMed] [Google Scholar]
- 13.Gao H, Neff TA, Guo RF, Speyer CL, Sarma JV, Tomlins S, Man Y, Riedemann NC, Hoesel LM, Younkin E, Zetoune FS, Ward PA. Faseb J. 2005;19(8):1003–1005. doi: 10.1096/fj.04-3424fje. [DOI] [PubMed] [Google Scholar]
- 14.Gerard NP, Lu B, Liu P, Craig S, Fujiwara Y, Okinaga S, Gerard C. J Biol Chem. 2005;280(48):39677–39680. doi: 10.1074/jbc.C500287200. [DOI] [PubMed] [Google Scholar]
- 15.Cain SA, Monk PN. J Biol Chem. 2002;277(9):7165–7169. doi: 10.1074/jbc.C100714200. [DOI] [PubMed] [Google Scholar]
- 16.Okinaga S, Slattery D, Humbles A, Zsengeller Z, Morteau O, Kinrade MB, Brodbeck RM, Krause JE, Choe HR, Gerard NP, Gerard C. Biochemistry. 2003;42(31):9406–9415. doi: 10.1021/bi034489v. [DOI] [PubMed] [Google Scholar]
- 17.Otto M, Hawlisch H, Monk PN, Muller M, Klos A, Karp CL, Kohl J. J Biol Chem. 2004;279(1):142–151. doi: 10.1074/jbc.M310078200. [DOI] [PubMed] [Google Scholar]
- 18.Pease JE, Barker MD. Biochem Mol Biol Int. 1993;31(4):719–726. [PubMed] [Google Scholar]
- 19.Kalant D, Cain SA, Maslowska M, Sniderman AD, Cianflone K, Monk PN. J Biol Chem. 2003;278(13):11123–11129. doi: 10.1074/jbc.M206169200. [DOI] [PubMed] [Google Scholar]
- 20.Lanza TJ, Durette PL, Rollins T, Siciliano S, Cianciarulo DN, Kobayashi SV, Caldwell CG, Springer MS, Hagmann WK. J Med Chem. 1992;35(2):252–258. doi: 10.1021/jm00080a008. [DOI] [PubMed] [Google Scholar]
- 21.de Laszlo SE, A. EE, Li B, Ondeyka D, Rivero R, Malkowitz L, Molineaux C, Siciliano SJ, Springer MS, Greenlee WJ, Mantlo N. Bioorganic & Medicinal Chemistry Letters. 1997;7(2):213–218. [Google Scholar]
- 22.Paczkowski NJ, Finch AM, Whitmore JB, Short AJ, Wong AK, Monk PN, Cain SA, Fairlie DP, Taylor SM. Br J Pharmacol. 1999;128(7):1461–1466. doi: 10.1038/sj.bjp.0702938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilken HC, Rogge S, Gotze O, Werfel T, Zwirner J. J Immunol Methods. 1999;226(1-2):139–145. doi: 10.1016/s0022-1759(99)00064-2. [DOI] [PubMed] [Google Scholar]
- 24.Wilken HC, Gotze O, Werfel T, Zwirner J. Immunol Lett. 1999;67(2):141–145. doi: 10.1016/s0165-2478(99)00002-4. [DOI] [PubMed] [Google Scholar]
- 25.Moore KL. J Biol Chem. 2003;278(27):24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 26.Morgan EL, Ember JA, Sanderson SD, Scholz W, Buchner R, Ye RD, Hugli TE. J Immunol. 1993;151(1):377–388. [PubMed] [Google Scholar]
- 27.Oppermann M, Raedt U, Hebell T, Schmidt B, Zimmermann B, Gotze O. J Immunol. 1993;151(7):3785–3794. [PubMed] [Google Scholar]
- 28.Crass T, Ames RS, Sarau HM, Tornetta MA, Foley JJ, Kohl J, Klos A, Bautsch W. J Biol Chem. 1999;274(13):8367–8370. doi: 10.1074/jbc.274.13.8367. [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Ember JA, Chao TH, Fukuoka Y, Ye RD, Hugli TE. Protein Sci. 1999;8(11):2304–2311. doi: 10.1110/ps.8.11.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postma B, Kleibeuker W, Poppelier MJ, Boonstra M, Van Kessel KP, Van Strijp JA, de Haas CJ. J Biol Chem. 2005;280(3):2020–2027. doi: 10.1074/jbc.M412230200. [DOI] [PubMed] [Google Scholar]
- 31.Mery L, Boulay F. J Biol Chem. 1994;269(5):3457–3463. [PubMed] [Google Scholar]
- 32.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Cell. 1999;96(5):667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.