Copy number variants (CNVs) are duplicated or deleted segments of the genome that vary in size from a few bases to several kb and comprise a significant proportion of normal genomic variation1. The role of population-wide CNVs in disease has only recently come under investigation2,3. The chemokine (C-C) motif receptor 5, CCR5, on chromosome 3p21 has been associated with resistance to HIV-1 infection2. One of its ligands, CCL3L1, is encoded by a gene that lies in a CNV on chromosome 17q12,4 which includes another CCR5 ligand, CCL4L1, (Supplementary Figure 1) both of which have been reported to be associated with HIV-1/AIDS susceptibility2,5,6. CCR5 is associated with type 1 diabetes (T1D)7, and hence we hypothesised that CCL3L1 was also associated with T1D.
A reliable method for determining copy number at the 17q12 locus is required, as not only has the region been implicated in HIV-1/AIDS susceptibility, it has also been reported to influence disease progression with and without antiretroviral therapy8,9 and has been suggested as an informative approach to optimizing the design and evaluation of HIV-1 vaccine trials and prevention programs10. Quantitative real-time PCR (QPCR) is considered the gold-standard method for assessing copy number at individual CNV loci and was employed by the original CCL3L1-HIV-1/AIDs study2 as well as by subsequent studies of this CNV11. However, the CCL3L1 HIV-1 association has not been independently replicated, with all reported positive associations originating with almost the same case sample set and control sample set12. Consequently, there may be experimental biases in the current QPCR CCL3L1 assay and its scoring.
In T1D the median effect of susceptibility loci is below an odds ratio 1.5,13 so thousands of samples are required to test for association, which is also the case for most other complex diseases. Hence, the CCL3L1 assay has to work efficiently and accurately in thousands of samples. We compared two methods of obtaining CCL3L1 copy number, the paralogue ratio test (PRT)14, and QPCR2. PRT and QPCR rely on comparing the signal from the CNV against that from a reference locus and obtaining the ratio. If the reference has two copies, then a ratio of 1:1 denotes two copies of the CNV (assuming equal PCR efficiencies for both reactions). PRT uses a locus paralogous to the CNV with invariant copy number as the reference14. For the CCL3L1 region three PRT assays were used with CNVs CCL3L1, CCL4L1 and a long terminal repeat (LTR) located between them, and paralogous loci CCL3, CCL4 and a LTR on chromosome 10q22, respectively. These ratios were scored to give integer copy numbers.
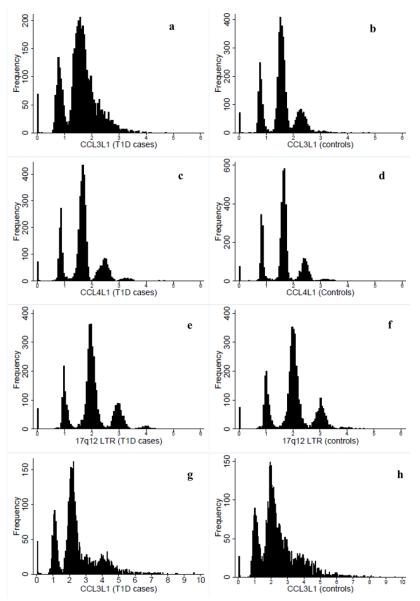
DNA samples from 5,771 British T1D cases and 6,854 geographically matched controls were studied for CCL3L1 variation using QPCR and PRT assays (Supplementary Methods). Owing to small, but potentially crucial, variations in PCR efficiency, the ratios of assay product form a distribution around the whole copy number (Fig. 1). With PRT, discrete clusters were distinguishable for the ratios from both CCL4L1 and the nearby LTR, whereas clusters overlapped for CCL3L1 (Fig. 1a–f). This could be due to sequence specific DNA bound protein interfering with the PCRs for CCL3L1. Differences in the DNA extraction methods for case and control DNA may have left different amounts of DNA bound protein, resulting in differential cluster quality. In contrast to the LTR, both the CCL3L1 and CCL4L1 assay ratios were not centred on integer values, but were shifted towards lower values. In controls, for example, one copy of CCL3L1 was centred on 0.8, two copies on 1.6, three copies on 2.3 (Fig. 1b). Having examined the distributions of the assay ratios, we assigned integer copy numbers using two methods; one was k-means clustering and the other was by rounding the PRT data to the nearest integer (Supplementary Methods, Supplementary Discussion). The original HIV-1/AIDS study used rounding for their QPCR assays2.
Figure 1.
Histograms of the frequency of the chromosome 17q12 CNV measure obtained using PRT and QPCR assays. Ratios of the CNV (CCL3L1, CCL4L1, LTR17) versus the reference (paralogous loci CCL3, CCL4, LTR10 for PRT and the haemoglobin beta gene for QPCR) that are used to assign copy numbers, either by rounding the data to the nearest integer or by using k-means clustering, are presented. (a) Assay ratio (copies) of CCL3L1 obtained using PRT in 3,860 cases and (b) 4,084 controls. (c) Assay ratios (copies) of CCL4L1 obtained using PRT in 4,041 cases and (d) 4,318 controls. (e) Assay ratios (copies) of the LTR on chr17q12 obtained using PRT in 4,044 cases and (f) in 4,266 controls. (g) Assay ratios (copies) of CCL3L1 obtained using QPCR in 3,362 cases and (h) in 3,983 controls.
Except in highly stratified populations, deviation from HWE provides a useful indicator of genotyping error. Therefore, we developed a statistical test for HWE with multi-allelic CNVs (Supplementary Methods). In controls, CCL3L1 was in HWE for the k-means PRT data, but not for the rounded PRT data (Supplementary Table 1), reflecting the inappropriateness of rounding here. Both CCL4L1 and the LTR were in HWE in controls (Table 1, Supplementary Tables 1–2). As the k-means clustered data for each assay individually and averaged across assays were in HWE, they were tested for association with T1D using a logistic regression model, with disease status as outcome variable and copy number as the independent variable (Supplementary methods). No convincing evidence of association was obtained (P > 0.05; Table 1, Supplementary Tables 1–2, Supplementary Results). The CCL3L1 rounded data showed evidence of association with T1D (P =8×10−11), but the deviation from HWE suggests that this was false, and attributable to genotyping error. Evidence of association was also obtained with the rounded data at CCL4L1 (P = 0.0002) due to an excess of two copies at CCL4L1. The majority of the three-copy cluster lay between two and 2.5 and so was incorrectly scored as two copies when rounded (Fig. 1c,d). Therefore, this association was an artefact of the method of assigning whole copy number (which was rounding) and consequently is likely to be false. We concluded that the LTR assay was the most robust of the three assays because, the LTR ratios clustered well around integer copy number and so could be assigned by rounding or k-means, and the assay gave consistent results for the replicated quality control samples. Consequently, we tested the LTR for interaction with the CCR5Δ32 allele in T1D (Supplementary Methods), since combinations of CCR5-CCL3L1 genotypes have been reported to be associated with HIV-1/AIDS risk and progression2,9. No evidence of an interaction in their T1D association was obtained (P = 0.29).
Table 1.
Copy number of the 17q12 CNV
| Copy # | LTR (PRT) | CCL3L1 (QPCR) | ||
|---|---|---|---|---|
| Obs. (Exp.) | Obs. (Exp.) | Obs. (Exp.) | Obs. (Exp.) | |
| Cases | Controls | Cases | Controls | |
| 0 | 75 (62.2) | 78 (73.9) | 53 (56.9) | 29 (68.3) |
| 1 | 726 (752.3) | 829 (837.4) | 598 (590.9) | 750 (668.6) |
| 2 | 2408 (2393.3) | 2510 (2505.0) | 1610 (1610.5) | 1729 (1771.8) |
| 3 | 732 (733.7) | 756 (757.0) | 467 (461.5) | 746 (739.8) |
| 4 | 93 (92.3) | 83 (82.6) | 399 (400.3) | 453 (451.5) |
| 5 | 9 (9.4) | 9 (9.1) | 158 (169.3) | 173 (175.8) |
| 6 | 1 (0.8) | 1 (0.9) | 45 (51.8) | 66 (80.9) |
| 7 | 25 (15.9) | 23 (18.8) | ||
| 8 | 6 (4.2) | 12 (6.0) | ||
| 9 | 1 (0.7) | 1 (1.2) | ||
| 10 | 0 (0.0) | 1 (0.3) | ||
| P HWE | 0.1562 | 0.8242 | 0.1454 | 1×10−8 |
| P T1D | 0.6946 | 0.0002 | ||
Copy numbers were obtained using the LTR PRT assay in 4,044 cases and 4,266 controls, and from the qPCR assay in 3,362 cases and 3,983 controls (2,479 cases and 2,378 controls were successfully genotyped with both assays). Obs=Observed counts; Exp=expected counts; PHWE = Hardy-Weinberg Equilibrium P-value. PT1D = P-value for association with type 1 diabetes.
The distribution of ratios obtained by QPCR for CCL3L1 in cases and controls was right-shifted towards higher than integer copy numbers (Fig. 1g, h). We used k-means clustering (Supplementary Methods), and rounding to assign integer copy number. With both methods of assigning copy number, the CCL3L1 data from QPCR deviated from HWE in controls, due to a lack of “0” copy numbers, and a right-shift in the copy number distribution (Supplementary Table 3). There was evidence for strong observed associations of CCL3L1 with T1D, P = 7×10−34 (k-means) and P = 3×10−7 (rounding) which, in light of the HWE tests, we regard as artefactual.
QPCR used a standard curve on each plate to standardise the concentration of DNA between CNV and reference reactions within a plate, which introduced plate-to-plate variation. Standard curves are not required for PRT, which has reference and CNV reactions in the same well. As cases and controls were dispensed onto different plates, the statistical test for association may have actually detected plate-to-plate variation and not T1D association. Since we had so many plates (105) we were able to estimate the QPCR plate-to-plate variation (6% in controls and 2% in cases) to correct the association test. We reduced the apparent evidence of T1D association, P = 7×10−9(k-means; Table 1) and P = 0.00019 (rounding; Supplementary Table 3). The remaining evidence of association may be attributable to shifts in copy number distribution, caused by unpredictable interactions between the assay and the differential quality and composition of DNA from different sources, leading to HWE deviation in the controls but not the cases15 (Supplementary Discussion). These results not only demonstrate the importance of testing for HWE but also of allowing for plate effects in the analysis. We recommend arraying case and control samples onto the same plates.
5,121 samples were common to the PRT and QPCR experiments. Using the LTR as the most accurately scored measure of CCL3L1 for the PRT method, and the k-means clustered data for the QPCR assay, we found 64% were consistent between the two methods with 25% having one additional copy of CCL3L1 with QPCR than PRT. Nine percent of the data had between two and five additional copies of CCL3L1 obtained using QPCR compared to PRT. Just 2% of the data had one or two copies more of CCL3L1 as measured by PRT than by QPCR. The QPCR assay also exhibited a general trend towards higher copy numbers compared to the other two PRT assays: a shift that may be an artefact of the QPCR primers also binding to a CCL3L1 pseudogene, CCL3L2, as well as to CCL3L12,4,14 (Supplementary Figure 1, Supplementary Discussion).
This is the first report where CCL3L1 copy number has been estimated in such a large sample set using both QPCR and PRT. The PRT LTR assay copy number exhibited good clustering, with no difference in distribution between T1D cases and controls, suggesting that the primers designed for this locus are highly specific, and robust to variations in source of DNA. PRT also avoids potential error in scoring zero copy numbers. We recommend rounding the data if the assay ratios cluster distinctly round integers and using k-means clustering otherwise. QPCR can be used to assay CCL3L1 in large well-powered sample sets, if appropriate quality control measures are implemented. The distribution of copy number, and its dependence on DNA source should be examined statistically and an appropriate method of assigning copy number adopted for each DNA source. Tests for deviation from HWE must be performed and any detected deviations resolved. If they cannot be resolved then an assay such as PRT should be used instead. In small sample-sets (e.g. n ≤ 500), even in the absence of deviations from HWE, any associations should be treated with some scepticism, owing to the limited power to detect deviations from HWE (Supplementary Results).
Finally, we note that there is significant variation in CCL3L1 copy number according to ethnic group, which others have reported2 (Supplementary Table 4). We genotyped 95 African Yoruban samples in duplicate using the LTR PRT assay. Our data was highly reproducible (correlation coefficient > 0.99; Supplementary Figure 2). The CNVs obtained were between two and eight (Supplementary Table 4) with a mean copy number of 4.3. Hence, any inadvertent admixture, as seems possible on detailed evaluation of the original CCL3L1-HIV-1/AIDS study in which European Americans have different CCL3L1 copy numbers than Hispanic Americans2, combined with copy number distribution shifts interacting with DNA source and error prone copy number scoring (i.e. rounding) described here, could lead to apparently highly significant disease associations.
All DNA samples were collected with approval from the Cambridgeshire 2 Research Ethics Committee, and written consent was obtained from all individuals, or parents of individuals who were too young to consent.
Supplementary Material
Acknowledgments
This work was funded by the Juvenile Diabetes Research Foundation (JDRF) International, the Wellcome Trust, and the UK National Institute for Health Research Cambridge Biomedical Research Center. The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). We are grateful for the participation of the type 1 diabetes patients and the control individuals. We would like to thank the Medical Research Council and Wellcome Trust for funding the collection of DNA for the British 1958 Birth Cohort. DNA control samples were prepared and provided by S. Ring, R. Jones, M. Pembrey, W. McArdle of the ALSPAC Laboratory, Department of Social Medicine, University of Bristol; D. Strachan of the Division of Community Health Sciences, St George's, University of London and P. Burton of the Department of Genetics and Health Sciences, University of Leicester. T1D case DNA samples were prepared by K. Bourget, S. Duley, S. Hawkins, G. Coleman, M. Maisuria, S. Hood, E. King, T. Mistry, A. Simpson, S. Wood, S. Clayton, F. Wright and H. Stevens of the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research, University of Cambridge. Double scoring was performed by M. Hardy of the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research, University of Cambridge.
References
- 1.McCarroll SA, et al. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez E, et al. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 3.McCarroll SA. Hum Mol Genet. 2008;17:R135–142. doi: 10.1093/hmg/ddn282. [DOI] [PubMed] [Google Scholar]
- 4.Modi WS. Genomics. 2004;83:735–738. doi: 10.1016/j.ygeno.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman PA, et al. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- 6.Colobran R, et al. J Immunol. 2005;174:5655–5664. doi: 10.4049/jimmunol.174.9.5655. [DOI] [PubMed] [Google Scholar]
- 7.Smyth DJ, et al. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolan MJ, et al. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 9.Ahuja SK, et al. Nat Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulkarni H, et al. PLoS ONE. 2008;3:e3671. doi: 10.1371/journal.pone.0003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mamtani M, et al. Ann Rheum Dis. 2008;67:1076–1083. doi: 10.1136/ard.2007.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao W, et al. Genes Immun. 2007;8:224–231. doi: 10.1038/sj.gene.6364378. [DOI] [PubMed] [Google Scholar]
- 13.Todd JA, et al. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker S, Janyakhantikul S, Armour JA. Genomics. 2009;93:98–103. doi: 10.1016/j.ygeno.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Clayton DG, et al. Nat Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.