Abstract
Nucleotide analog interference mapping (NAIM) is a powerful chemogenetic approach that allows RNA structure and function to be characterized at the atomic level. Random modifications of base or backbone moieties are incorporated into the RNA transcript as nucleotide analog phosphorothioates. The resulting RNA pool is then subjected to a stringent selection step, in which the RNA has to accomplish a specific task, for example, folding. RNA functional groups important for this process can be identified by physical isolation of the functional and the nonfunctional RNA molecules and subsequent mapping of the modified nucleotide positions in both RNA populations by iodine cleavage of the susceptible phosphorothioate linkage. This approach has been used to analyze a variety of aspects of RNA biochemistry, including RNA structure, catalysis and ligand interaction. Here, I describe how to set up a NAIM assay for studying RNA folding. This protocol can be readily adapted to study any RNAs and their properties. The time required to complete the experiment is dependent on the length of the RNA and the number of atomic modifications tested. In general, a single NAIM experiment can be completed in 1–2 weeks, but expect a time frame of several weeks to obtain reliable and statistically meaningful results.
INTRODUCTION
Despite recent advances in X-ray crystallography and NMR techniques for studying the architecture of RNA molecules, classical biochemical tools remain essential complementary approaches in RNA structure/function analyses. These methods are used, for instance, to investigate RNA structure and to identify ligand-binding sites and solvent-inaccessible regions within the folded RNA (chemical and enzymatic probing assays)1-3; to obtain structural information on the spatial proximity of various functionalities within an RNA or an RNA-protein complex (UV cross-linking)4-6 and to identify metal-binding sites within an RNA (metal-induced cleavage assays)7-9. Nucleotide analog interference mapping (NAIM10-12) is a very elegant combinatorial technique that allows the chemical groups within the RNA essential for its function to be rapidly and efficiently pinpointed. This method provides information at the atomic level in contrast to conventional mutational analyses, which work at the nucleotide level. Using nucleotide analogs carrying different modifications of base or backbone moieties (Fig. 1), it is possible to simultaneously, yet individually, assess the contribution of a particular functional group to RNA function at every position within the molecule. It is also noteworthy that, compared to NAIM, classical chemical probing or interference assays require the use of very harmful reagents such as DMS (dimethyl sulfate), DEPC (diethyl pyrocarbonate) or CMCT (1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate). These chemicals can also assess the importance of nucleobase atoms, but the detection of these positions commonly requires reverse transcription of the modified RNA1. In contrast, NAIM allows direct mapping of the sites of interference due to the phosphorothioate tag of the nucleotide analog, which is susceptible to iodine cleavage.
Figure 1. Commercially available analogs.
(a) Adenosine analogs, (b) guanosine analogs, (c) uridine analogs and (d) cytidine analogs. The parent phosphorothioate analogs (AαS, GαS, UαS and CαS) are boxed. The nucleoside modifications are indicated with a gray box.
Overview of NAIM
NAIM can be used to determine the functional groups critical for RNA structure, folding, catalysis and ligand (metal ions, proteins or metabolites) interactions13-36. The cornerstones for developing this assay were set in the laboratories of F. Eckstein12,37-39 and S.A. Strobel11,40-42 and it has since been applied to several RNA model systems such as RNase P14-16, group I (refs. 17-19) and group II introns20-22, HDV24, hammerhead25,26,31, hairpin27-29 and VS ribozymes30, tRNA32, glmS riboswitch33 and the signal recognition particle34.
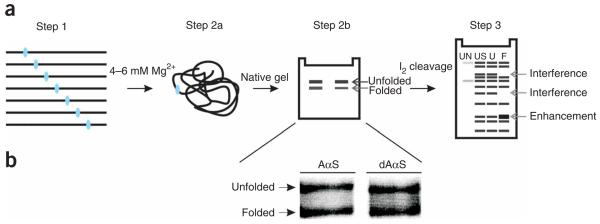
NAIM is based on using nucleotide analogs carrying a base and/or backbone modification in addition to the phosphorothioate tag (Fig. 1), and consists of three individual steps (Fig. 2).
Figure 2. Experimental design.
(a) Scheme of the individual steps involved in identifying the functional groups essential for the tertiary collapse of the Sc. ai5γ D135 ribozyme. Blue ellipses indicate the site of incorporation of an analog. In Step 3 lane 1 (untreated (UN)) shows background degradation of the labeled RNA. Lane 2 (unselected (US)) shows the relative incorporation level for an analog at a specific position (RNA, which did not undergo the selection step, was iodine cleaved). Lane 3 (unfolded (U)) shows iodine cleavage products for RNA, which derived from the unfolded population purified from the native gel. Lane 4 (folded (F)) shows iodine cleavage products for RNA, which was obtained from the folded population eluted from the native gel. (b) Representative preparative native gels are shown for different analogs. The unfolded and folded populations indicated here were physically isolated by cutting them out of the gel.
Step 1: Creating a pool of randomly modified RNA molecules
In this step, a pool of randomly modified RNA is obtained by in vitro transcription in the presence of a nucleotide analog phosphorothioate followed by 5′- or 3′-end labeling.
Step 2: Selection for critical functional groups
The chemogenetically modified RNAs are subjected to a stringent selection step, to select for the chemical groups essential for RNA function. This is followed by physical separation of the functional and nonfunctional RNA populations, which is most commonly achieved through gel electrophoresis. The selection step can be readily adapted to study RNA folding, catalysis or ligand interaction. For example, if RNA folding, metal ion interactions or protein binding is investigated, native gel electrophoresis can also be used to separate the folded from the unfolded or the bound from the unbound RNA populations, respectively. Alternatively, a filter-binding assay can be used to separate RNA–ligand complexes from the free RNA. For studying atoms involved in RNA catalysis, such as ribozyme cleavage or ligation reactions, the precursor molecules are typically separated from the product by standard denaturing PAGE. In any case, reaction conditions (pH, metal ion concentration, RNA concentration, temperature and incubation time) need to be optimized to achieve the required stringency of the selection step (not > 20–40% of the functional RNA formed).
Step 3: Mapping NAIM effects
After the selection step, the location of the nucleotide analog moieties can be visualized due to the susceptibility of the phosphorothioates to iodine cleavage. The cleavage products are analyzed on standard denaturing polyacrylamide gels. In my case study, I compared the band intensities of cleavage products stemming from folded and unfolded RNA species to detect which atoms are essential for compaction of the Sc. ai5γ group II ribozyme D135 (Figs. 2 and 3). For example, if the N6 atom at a specific adenosine residue within D135 is important for folding, then the incorporation of an m6AαS analog at this site will interfere with D135 folding and cause these modified RNA molecules to be under-represented in the folded relative to the unfolded species, thereby resulting in an interference (Fig. 3). On the other hand, if the atomic modification provides an advantage to RNA folding, a NAIM enhancement is observed (Fig. 3). In order to calculate respective interferences and enhancements, band intensities are quantified using appropriate software and NAIM effects are then determined using various approaches described in the literature10,11,22. To this end, individual NAIM effects as well as their clustering is anticipated to provide novel insights into the biochemical behavior of the RNA and process in question.
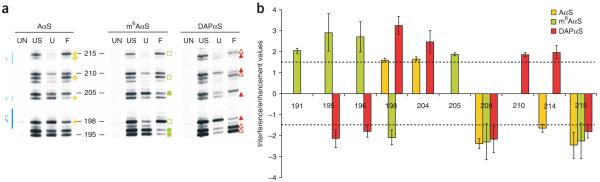
Figure 3. Functional groups critical for the tertiary collapse of the D135 ribozyme.
(a) Representative nucleotide analog interference mapping (NAIM) gels show interferences and enhancements within the folding control element comprising the κ-ζ element. For each gel, lane 1 (untreated (UN)) shows background degradation of the labeled RNA. Lane 2 (unselected (US)) shows the relative incorporation level for an analog at a specific position (RNA, which did not undergo the selection step, was iodine cleaved). Lane 3 (unfolded (U)) shows iodine cleavage products for RNA, which derived from the unfolded population purified from the native gel. Lane 4 (folded (F)) shows iodine cleavage products for RNA, which was obtained from the folded population eluted from the native gel. Yellow star indicates adenosine phosphorothioate analog; green square indicates m6AαS; red triangle indicates DAPαS. Closed symbols indicate interferences and open symbols represent enhancements. (b) Bar graph shows the average value and s.d. for each raw interference or enhancement value found within the 5′ region of the folding control element. The dotted line indicates the cutoff below which interferences were considered insignificant. Only interferences/enhancements that exceed the cutoff are shown. Color code is same as in panel a. For representation purposes, interference and enhancement values are shown as positve and negative numbers, respectively.
Applications of NAIM
As the selection step involves either folding, binding to a ligand or a catalytic reaction, this technique can be modified to explore a number of biochemical features of RNA. For example, this technique allows functional groups important for catalysis versus overall folding (e.g., ref. 36) to be distinguished. In addition, the NAIM data can be correlated with other pieces of information, such as existing structural data43. By choosing appropriate analogs, it is also possible to determine whether specific structural features (e.g., local helical stability or tertiary contacts) are essential for the formation of a particular conformational state22. Moreover, NAIM signatures can be established for specific RNA motifs (e.g., the GNRA tetraloop–receptor interaction20,44). Finally, NAIM results set the foundation for performing nucleotide analog interference suppression (NAIS45-47), a method enabling the identification of novel tertiary interactions within an RNA molecule. Taken together, the variety of potential applications and anticipated results emphasizes that techniques using nucleotide analogs are powerful tools and important complementary approaches to the structural characterization by conventional chemical modification assays, X-ray crystallography and NMR.
I recently used NAIM to provide insight into the structural requirements and the driving forces of the Mg2+-induced compaction of a group II intron ribozyme22,23. By employing the three-step NAIM approach (Fig. 2), I was able to identify a folding control element, which dictates Mg2+-dependent stability22 as well as the time scale23 of folding of this group II ribozyme (named D135). The experimental procedure involved in this study22 is described in the protocol below to provide guidelines on how to set up a NAIM assay to dissect RNA folding. In brief, after in vitro transcribing a pool of randomly modified D135 RNA, these chemogenetically modified RNAs were stringently folded. Subsequently, the unfolded and folded RNA populations were physically separated through native gel electrophoresis followed by mapping the sites of NAIM interferences and enhancements.
Limitations of NAIM
The length of the RNA plays a critical role in successfully applying NAIM to investigate RNA biochemical properties. First, the transcription yield might be reduced for very large RNAs (>1,000 nt). Specifically, some analogs are very poor substrates for the RNA polymerase and, accordingly, for these analogs the incorporation efficiency is very low (e.g., bulky modifications of the 2′-hydroxyl group), thereby also affecting the transcription yield. This adverse effect increases if there are >3 nt in a row to be substituted with such analogs. Second, mapping NAIM effects throughout very long RNAs is also significantly more elaborate in terms of time frame and required material. Another limitation is the availability of analogs, as many of them have to be synthesized and are not commercially available. Chemical synthesis of phosphorothioates is a difficult procedure for nonspecialists. In addition, RNA transcripts that were randomly modified with certain analogs might be difficult to handle, if the incorporation of the analog results in a severe inhibition of RNA function, thereby precluding the performance of a selection step. In my case, the 2APαS analog perturbed RNA folding severely and could therefore not be used in my NAIM study22. Along this line, a serious drawback of the method is the possibility of a strong interference by a phosphorothioate tag itself that sometimes makes it impossible to identify any additional interference caused by nucleoside modification of an analog. Finally, the selection step has to be carefully optimized for each novel NAIM application to detect the chemical groups essential for RNA function. At the same time, this aspect also makes NAIM a versatile technique that is adaptable to a vast repertoire of applications. Using NAIM, only the critical atoms but not their interaction partner are identified; however, this can be accomplished by performing NAIS.
Experimental design
Before starting a NAIM experiment, there are a number of important considerations to bear in mind, as discussed below. These guidelines should help to avoid any potential pitfalls of this elaborate three-step approach and to assist in designing a successful NAIM strategy for your favorite RNA and its function.
Choice of nucleotide analog
There are a growing number of nucleotide analogs, many of which are commercially available (Fig. 1). However, there is also a growing number of noncommercially available analogs (3-deaza-AαS, 8-aza-AαS, 6-thio-GαS, N2-methyl-GαS, 6-aza-CαS, 5-fluoro-CαS, pseudo-iso-CαS, etc.), which can be synthesized using published protocols14,17,19,35,37,40,41,44,45,48-50. Nucleotide analogs can be approximately classified into three groups: those deleting a functional group, those adding a steric bulk to the functional group and those altering the chemical nature of the functional group. In order to narrow down the number of analogs tested in a NAIM study, it is recommended to define which particular RNA biochemical property to investigate and to take into account the available information (such as phylogenetic conservation of residues, known RNA motifs or data from mutational analyses). For example, analogs with shifted pKa values (e.g., 3-deaza-AαS, 8-aza-AαS, 6-aza-CαS, 5-fluoro-CαS) have been successfully applied to identify nucleobases involved in ribozyme catalysis in a pH-dependent NAIM assay24,28,30. Furthermore, RNA motifs can be identified by their distinct NAIM signature: A-minor interactions are an ubiquitous motif important for helix packing in RNA tertiary architectures51 and are detectable by 3-deaza-AαS interference mapping45, as the N3 imino group of adenosine contributes significantly to the formation of this motif. Similarly, using 6-thio-GαS in conjunction with selective metal ion interference rescue with Tl+ allows for the identification of monovalent metal ion-binding sites in RNA35,44.
To study RNA folding chemogenetically, I decided to use nucleotide analogs that address the importance of tertiary contact formation and of local duplex stability within the Sc. ai5γ group II intron-derived D135 ribozyme22. As D135 follows a folding pathway, in which the Mg2+-dependent collapse is slow52,53, I aimed at identifying the functional groups important for its compaction. Given that the Sc. ai5γ intron has a low GC content and adenosines are major players in RNA tertiary structure formation54, I chose several adenosine phosphorothioates with modifications on either major or minor groove edges (AαS, 7dAαS, m6AαS, DAPαS and 2dAαS). In addition, as group II introns contain some conserved guanosine residues, I also probed the minor groove of guanosine using appropriate phosphorothioate analogs (GαS, IαS and dGαS).
Choice of T7 RNA polymerase
A nucleotide analog is different from its parental NTPαS, as it harbors base or ribose modifications in addition to the substitution of a nonbridging oxygen atom with sulfur (Fig. 1). T7 RNA polymerase is capable of incorporating these phosphorothioate analogs efficiently during in vitro transcription, whereby the ratio of analog and unmodified NTP (Table 1) determines the incorporation level (generally 5%)10,40, which has to be established for each analog39,55. Importantly, for commercially available analogs, this optimization has already been performed and these analogs are accordingly sold at the required concentration (as 10× stocks) to yield the 5% incorporation efficiency. Note that some phosphorothioates, mostly analogs with 2′-hydroxyl modifications, require a mutant T7 RNA polymerase (Y639F (ref. 56)) to be incorporated into the transcript (see Table 1).
TABLE 1.
Guidelines for nucleotide analog incorporation during in vitro transcription.
| Incorporation | NTPαS (mM) | NTP (mM) | T7 RNA polymerase | Reference(s) |
|---|---|---|---|---|
| AαS, GαS, UαS, CαS | 0.05 | 1 | Wt | 61 |
| dAαS, dCαS | 1.5 | 1 | Y639F | 17,40 |
| dGαS, dUαS | 0.5 | 1 | Y639F | 40 |
| IαS, m6AαS | 0.4 | 1 | Wt | 17,19 |
| 7dAαS | 0.1 | 1 | Wt | 17 |
| 7dGαS | 0.05 | 1 | Wt | 14 |
| PurαS, 2APαS | 2 | 0.5 | Wt | 17 |
| DAPαS | 0.025 | 1 | Wt | 17 |
| m2 GαS | 1.5 | 0.5 | Y639F | 50 |
Abbreviations: wt, wild type. Concentrations of commercially available analogs and their unmodified NTP required to yield a 5% NαS incorporation efficiency are indicated.
Optimizing the selection step
Selection is the most fundamental step of the entire approach and can be adapted to investigate a variety of aspects in RNA biochemistry (for review, see refs. 10,11,41). Consequently, this selection step defines the outcome of the NAIM study and requires considerable optimization to ensure a successful outcome. For NAIM assays with folding as a selection step, it is important to establish the optimal: temperature, pH, concentration of monovalent and/or divalent ions and incubation time required to yield ~20–40% of folded RNA relative to the unfolded species—this ensures the necessary stringency of the selection process. For the D135 ribozyme, this was accomplished by folding it at a [Mg2+] below the KMg for the tertiary collapse (13 mM (ref. 53)), thereby allowing D135 to sample the collapsed intermediate state without proceeding to the native state (for which KMg is 20–40 mM (ref. 57)).
Importantly, optimizing the ionic and temperature requirements for RNA folding is critical for investigating any RNA function whether this is RNA structure formation per se, catalytic activity or ligand interaction. If these experimental conditions are not defined carefully, it will be difficult to set up a stringent selection step. Similarly, basic characterization of an RNA's ability to interact with a ligand (e.g., KD) or an RNA's catalytic properties and strategies (e.g., reaction rate-constants) is a prerequisite for successfully exploring the chemical basis of these processes by NAIM.
The selection step is equally dependent on stringency and on proper physical separation of the functional and nonfunctional RNA species, for example, through native gel electrophoresis in the case of a folding-based NAIM assay. Thus, it is also critical to find optimal native gel electrophoresis conditions for the RNA of interest: acrylamide concentration, magnesium or other metal ion concentration and power settings. Notably, incorporating different analogs in your favorite RNA might require minor adjustments of the selection procedure (such as small changes in metal ion concentration, reaction time for folding or power settings, ionic strength during PAGE).
Analytical gel
In order to map NAIM effects throughout an RNA molecule, the iodine-cleaved samples and the control for background degradation typically have to be resolved on several denaturing polyacrylamide gels (Fig. 3). For large RNAs (>250 nt), it is advisable to run long denaturing gels (90 cm) for an increased read-out in addition to the standard sequencing gels (45 cm). Finally, by varying the percentage of the gels distinct sequence regions of the RNA can be mapped (see Table 2).
TABLE 2.
Dye migration in denaturing polyacrylamide gels.
| Acrylamide solution (%) |
Bromophenol blue (BB) (nt) |
Xylene cyanol (XC) (nt) |
|---|---|---|
| 5 | 35 | 130 |
| 6 | 26 | 106 |
| 8 | 19 | 75 |
| 10 | 12 | 55 |
| 20 | 8 | 28 |
The dyes (BB and XC) co-migrate with RNA or DNA fragments of a specific length, as shown above (number of nucleotides). Importantly, this correlation varies with the percentage of the gel.
Controls
Given that all nucleotide analogs have a nonbridging oxygen replaced by sulfur (Fig. 1), it is essential to always study the parent phosphorothioate as a control. In other words if interested in the exocyclic amine of guanosine, both analogs GαS and IαS have to be analyzed to distinguish the effect of the phosphorothioate and that of the nucleoside modification. For each analog tested, it is appropriate to assess the relative incorporation efficiency by iodine cleaving unselected end-labeled RNA and resolving it on the analytical denaturing polyacrylamide gels together with an untreated sample to control for background degradation and the RNA which underwent the selection step and was subsequently iodine cleaved (Figs. 2 and 3). Information from all these samples is required to evaluate NAIM effects.
Here, I provide a protocol for applying NAIM to characterize RNA folding, based on our previously published methods22,23. Where appropriate, I indicate where the Procedure can be adapted to study other aspects of RNA function, according to the considerations listed above.
MATERIALS
REAGENTS
Restriction enzyme (20 U μl−1; NEB) producing 5′ overhang ▲ CRITICAL T7 RNA polymerase has a higher efficiency transcribing from such templates resulting in an increased yield.
10× Restriction buffer (NEB)
Wild-type (wt)T7 RNA polymerase (e.g., EpiCentre or Roche) can be used for incorporating most nucleotide analogs into RNA transcripts (see Table 1)
Mutant (Y639F)T7 RNA polymerase (50 U μl−1; EpiCentre) has to be used to incorporate certain nucleotide analogs (e.g., 2′ deoxy analogs) into RNA transcripts (see Table 1). It is supplied with a 5× transcription buffer and 0.1 M DTT stock solution
5× Mutant (Y639F) T7 RNA polymerase transcription buffer (EpiCentre)
RNase inhibitor (40 U μl−1; Roche)
Alkaline phosphatase (20 U μl−1; Roche)
10× Dephosphorylation buffer (Roche)
T4-polynucleotide kinase (PNK, 10 U μl−1; NEB)
10× T4-PNK buffer (NEB)
DNA polymerase I Klenow fragment exo− (2 U μl−1; Roche)
10× Nucleotide analog solution (Glen Research)
NTPs (100 mM stock; Roche)
γ-P32-ATP (6,000 Ci mmol−1; Perkin Elmer) ! CAUTION Radiation can cause cell damage; wear protective gloves, goggles and lab coat and use a protective shield.
α-P32-dCTP (6,000 Ci mmol−1; Perkin Elmer) ! CAUTION Radiation can cause cell damage; wear protective gloves, goggles and lab coat and use a protective shield.
EDTA (0.5 M stock, pH 8.0; Sigma)
MOPS (1 M stocks, pH 6.0 and pH 7.0; Sigma)
HEPES (Fisher)
Tris base (Fisher)
Urea (J.T. Baker)
Boric acid (Fisher)
40% Acrylamide solution (29:1; Fisher) ! CAUTION Toxic, carcinogenic; prevent skin or eye contact by wearing protective gloves, goggles and lab coat.
50% Long-Ranger acrylamide solution (Cambrex) ! CAUTION Toxic, carcinogenic; prevent skin or eye contact by wearing protective gloves, goggles and lab coat.
Tetramethylethylenediamine (TEMED; Sigma) ! CAUTION Flammable, toxic by inhalation, irritating to eyes.
Ammonium persulfate (10% (wt/vol) APS stock; Sigma) ▲ CRITICAL Store at 4 °C for up to 1 month.
Agarose (Fisher)
Ethidium bromide (10 mg ml−1; Sigma) ! CAUTION Intercalates with DNA, mutagenic; prevent skin contact by wearing protective gloves and lab coat and work in a fume hood.
Sucrose (50% stock (wt/vol); Sigma-Aldrich)
Bromophenol blue (BB; Sigma)
Xylene cyanol (XC; Sigma)
Glycerol (Sigma)
Isoamylalcohol (Sigma)
Chloroform (Sigma)
Phenol (Sigma) ! CAUTION Toxic, corrosive; prevent skin or eye contact by wearing protective gloves, goggles and lab coat and work in a fume hood to prevent inhalation.
Ethanol (Fisher)
NaOAc (3 M stock pH 5.0; Sigma)
KCl (3 M stock; Sigma)
MgCl2 (1 M stock; Sigma)
Iodine (10 mM stock in ethanol; Sigma) ! CAUTION Toxic by inhalation; skin irritant (wear protective gloves and lab coat).
Glycogen (20 mg ml−1 stock, dilute with ddH2O to a 10 mg ml−1 stock; Roche)
Spermidine (1 M stock; Sigma) ▲ CRITICAL Store frozen at −20 °C for up to 12 months.
DTT (1 M stock; Sigma) ▲ CRITICAL Store frozen at −20 °C for up to 6 months.
10× Transcription buffer (see REAGENT SETUP)
Elution buffer (see REAGENT SETUP)
ME (see REAGENT SETUP)
TE (see REAGENT SETUP)
10× Annealing buffer (see REAGENT SETUP)
20× Mg2+/DTT solution (see REAGENT SETUP)
10× TBE (for denaturing polyacrylamide gels and agarose gels) (see REAGENT SETUP)
20× Running buffer without Mg2+ (for native polyacrylamide gels) (see REAGENT SETUP)
1× Running buffer with Mg2+ (for native polyacrylamide gels) (see REAGENT SETUP)
1× Loading buffer (for denaturing polyacrylamide gels) (see REAGENT SETUP)
4× Loading buffer (for native polyacrylamide gels) (see REAGENT SETUP)
Stop solution for iodine cleavage reaction (see REAGENT SETUP)
Denaturing acrylamide solutions (15%) (see REAGENT SETUP)
Denaturing acrylamide solutions (5% or 8%) (see REAGENT SETUP)
Nondenaturing acrylamide solution (4%-4 mM MgCl2) (see REAGENT SETUP)
0.7% Agarose gel (see REAGENT SETUP)
CI (see REAGENT SETUP)
PCI (see REAGENT SETUP)
Ethanol/0.3 M NaOAc mix (see REAGENT SETUP)
RNase Zap (Ambion)
EQUIPMENT
RNase/DNase-free 1.5-ml Eppendorf tubes (VWR)
RNase/DNase-free 14-ml polystyrene tubes (VWR)
Filter units (0.2 μm; Nalgene)
Acrodisc syringe filters (0.2 μm; VWR)
Syringes (5 or 10 ml; Fisher)
Thermomixer (Eppendorf) and water bath (VWR)
Refrigerating (4 °C) centrifuges for 1.5-ml tubes (e.g., Eppendorf centrifuge) and 14-ml tubes (e.g., Sorvall centrifuge, SA 600 rotor)
Vertical gel electrophoresis apparatus (adjustable) without special cooling systems; gel plate sets [90 cm (L) or 45 cm (L) × 29 cm (W); 25 cm (L) × 16 cm (W)], combs (0.4 mm × 25 cm with 7 or 31 wells or 1.2 mm × 12 cm with 4–8 wells) and spacers (0.4 mm or 1.2 mm). I used custom-made gel chambers, plates, combs and metal plates, but most commercially available systems (e.g., C.B.S. Scientific) are also suitable
High-voltage power supply (Thermo EC) for PAGE and low-voltage power supplies (Thermo EC) for agarose gels
UV lamp (254 nm; Fisher) and fluor-coated thin layer chromatography plate for detecting RNA transcripts in gel matrix
Gel documentation system to visualize agarose gels and PAGE (cold RNA transcripts)
UV spectrophotometer (Agilent Technologies) and quartz cuvettes for determining RNA or DNA concentration
Instant Imager (GE Healthcare; formerly Packard); alternatively X-ray film and related equipment
Storm Phosphorimager and adequate screens (Molecular Dynamics)
ImageQuant Software (Molecular Dynamics)
Semi-automated footprinting analysis (SAFA) software
REAGENT SETUP
10× Transcription buffer (for wt T7 RNA polymerase) 400 mM Tris–HCl pH 7.5, 260 mM MgCl2,30 mM spermidine. ▲ CRITICAL Store at −20 °C for up to 6 months.
Elution buffer 40 mM MOPS pH 6.0, 10 mM EDTA. ▲ CRITICAL Store at 4 °C in the dark for up to 6 months.
ME 10 mM MOPS pH 6.0, 1 mM EDTA. ▲ CRITICAL Store at room temperature (24 °C) in the dark for up to 12 months.
TE 10 mM Tris–HCl pH 8.0, 1 mM EDTA. ▲ CRITICAL Store at room temperature for up to 12 months.
10× Annealing buffer 140 mM Tris–HCl pH 7.5, 400 mM NaCl, 2 mM EDTA. ▲ CRITICAL Store at −20 °C for up to 6 months.
20× Mg2+/DTT solution 140 mM MgCl2, 20 mM DTT. ▲ CRITICAL Store at −20 °C for up to 6 months.
10× TBE (for denaturing polyacrylamide gels and agarose gels) 0.89 M Tris base, 0.89 M boric acid, 20 mM EDTA. ▲ CRITICAL Store at room temperature for up to 12 months.
20× Running buffer without Mg2+ (for native polyacrylamide gels) 2 mM EDTA, 1.32 M HEPES, 680 mM Tris base. ▲ CRITICAL Store at 4 °C for up to 12 months.
1× Running buffer with Mg2+ (for native polyacrylamide gels) 4 mM MgCl2 in 1× running buffer (prepare before use).
1× Loading buffer (for denaturing polyacrylamide gels) 7 M Urea, 0.25% (wt/vol) BB, 0.25% (wt/vol) XC in 1× TBE. ▲ CRITICAL Store at room temperature for up to 12 months.
4× Loading buffer (for native polyacrylamide gels) 40% (vol/vol) Glycerol, 0.25% (wt/vol) BB, 0.25% (wt/vol) XC. ▲ CRITICAL Store at 4 °C for up to 12 months.
Stop solution for iodine cleavage reaction 13.6 g Urea, 0.8 ml of 2% (wt/vol) XC, 0.8 ml of 2% (wt/vol) BB, 0.16 ml of 0.1 M EDTA, 3.2 ml of 50% (wt/vol) sucrose, 4.64 ml ddH2O. ▲ CRITICAL Store at room temperature for up to 12 months.
Denaturing acrylamide solutions (15%) 15% (vol/vol) Acrylamide solution (29:1), 7 M urea in 1× TBE. ▲ CRITICAL Store at 4 °C for up to 3 months.
Denaturing acrylamide solutions (5 or 8%) 5–8% (vol/vol) Long-Ranger acrylamide solution, 7 M urea in 1× TBE. ▲ CRITICAL Store at 4 °C for up to 3 d.
Nondenaturing acrylamide solution (4%-4 mM MgCl2) 4% (vol/vol) Acrylamide solution (29:1), 4 mM MgCl2 in 1× running buffer (prepare before use).
0.7% Agarose gel 0.7% (wt/vol) Agarose, 0.5 μg ml−1 ethidium bromide in 1× TBE. ▲ CRITICAL Store at room temperature for up to 3 months.
CI Chloroform and isoamylalcohol mixed at a ratio of 24:1. ▲ CRITICAL Store at room temperature in the dark for up to 6 months.
PCI Phenol and CI mixed at a ratio of 1:1. ▲ CRITICAL Store at 4 °C in the dark for up to 6 months.
Ethanol/0.3 M NaOAc mix Absolute ethanol and 3 M NaOAc pH 5.0 mixed at a ratio of 9:1. ▲ CRITICAL Store at room temperature for up to 3 months.
PROCEDURE
Synthesis of nucleotide analog containing RNA: template preparation
1| Prepare the template for in vitro transcription by digesting 250 μg plasmid DNA containing the gene of interest as outlined in the table below. Incubate the sample at 37 °C for at least 2 h.
| Component | Amount per reaction | Final |
|---|---|---|
| Plasmid DNA | Variable | 250 μg |
| 10× Restriction enzyme buffer | 50 μl | 1× |
| 100× BSA (if necessary) | 5 μl | 1× |
| Restriction enzyme (20 U μl−1) | 20 μl | 400 U |
| Reaction volume | 500 μl |
▲ CRITICAL STEP Choose a restriction enzyme that produces a 5′-overhang (e.g., EcoRI or HindIII) to linearize the plasmid to increase the yield of the transcription; the T7 RNA polymerase has a preference for templates with a 5′-overhang.
■ PAUSE POINT Alternatively, the restriction digest can be left to incubate at 37 °C overnight.
2| Phenol extract the DNA by adding an equal volume of PCI, vortex strongly and centrifuge for 5 min at room temperature (24 °C) at 18,000g.
3| Take the aqueous phase and mix it with an equal volume of CI. Vortex and centrifuge the sample as outlined in Step 2.
4| Transfer the aqueous phase into a clean tube and precipitate with 2.5× volume of ethanol/0.3 M NaOAc mix. Place the sample at −20 °C for a minimum of 30 min.
■ PAUSE POINT Alternatively, the DNA can be left to precipitate at −20 °C overnight.
5| Centrifuge the sample for 30 min at 4 °C at 18,000g. Remove and discard the supernatant.
6| Dry the pellet (air dry or use a SpeedVac) and resuspend in an appropriate volume (~100 μl) of ddH2O or TE. Determine the concentration of the linearized vector by UV-spectroscopy (260 nm).
■ PAUSE POINT The sample can be stored at −20 °C for up to a year.
7| To confirm that the plasmid is properly linearized, load and run an aliquot (~1 μg) of the digested plasmid and an aliquot of the undigested plasmid (~1 μg) next to a size marker on a 0.7% agarose gel. In contrast to supercoiled or relaxed plasmid species, the linearized vector migrates in the gel according to its length (compare with size marker). If the restriction digest is incomplete, additional bands corresponding to uncut plasmid species will be visible (compare with undigested sample).
▲ CRITICAL STEP It is important that all plasmid is fully linearized, as the remaining uncut plasmid will produce an incorrect transcript, which may not be easy to separate from the correct one in the case of long RNAs.
Synthesis of nucleotide analog containing RNA: in vitro transcription
8| Set up the in vitro transcription reaction as outlined in the table below. Incubate the sample at 37 °C for 3 h. Note that some nucleotide analogs (see Table 1) can only be incorporated by the mutant (Y639F) T7 RNA polymerase56, in which case the manufacturer's instruction should be followed, scaling the reaction volume to 1 ml.
| Component | Amount per reaction | Final |
|---|---|---|
| Template DNA (linearized plasmid) | Variable | 25 μg |
| 10× Transcription buffer | 100 μl | 1× |
| 100 mM NTP (each)* | 10 μl | 1 mM |
| 10× Nucleotide analog mix | 100 μl | 1× |
| 1 M DTT | 10 μl | 10 mM |
| RNase inhibitor (40 U μl−1) | 10 μl | 400 U |
| T7 RNA polymerase (home made) | 50 μl | ~500 U |
| Reaction volume | 1,000 μl |
For some analogs, the final concentration of the parent NTP should be 0.5 mM (instead of 1 mM; see Table 1).
9| Precipitate the in vitro transcripts by adding 50 μl EDTA pH 8.0 and 3 ml ethanol/0.3 M NaOAc mix in 14-ml polystyrene tubes. Freeze the sample at −80 °C for 60 min.
■ PAUSE POINT Alternatively, the RNA can be left to precipitate at −80 °C overnight.
10| Centrifuge the sample for 30 min at 4 °C at 12,000g (Sorvall centrifuge, SA 600 rotor). Remove and discard the supernatant.
11| Dry and resuspend the pellet in 50 μl ME and 100 μl loading buffer (7 M urea).
12| Purify the transcripts on a denaturing polyacrylamide gel (use 1.2-mm spacers, 2 cm well size). Choose the percentage of the gel according to the size of the RNA (see Table 2). For example, a 600-nt long RNA is typically purified on a 5% gel, which is run until the XC dye has reached the bottom of the gel.
13| After the gel run, detect the RNA transcripts by UV shadowing.
? TROUBLESHOOTING
14| Cut out the major band corresponding to full-length RNA using a sterile blade, put the gel piece in a sterile syringe and squeeze it into a 14-ml polystyrene tube. Add 3 ml elution buffer, vortex and freeze the sample at −80 °C for 15 min.
■ PAUSE POINT Alternatively, the RNA can be left to precipitate at −80 °C overnight.
15| Thaw the sample and shake it at 4 °C for 2 h to elute the RNA from the gel.
16| Filter the eluate through an acrodisc filter, according to the manufacturer's instructions.
17| Precipitate the filtrate with 2.5× volume of ethanol/0.3 M NaOAc mix. Freeze the sample at −80 °C overnight.
18| Next day, centrifuge the sample (12,000g,30 min, 4 °C) and discard the supernatant.
19| Dry the pellet carefully and then resuspend it in 30–50 μl ME. Determine the concentration by UV spectroscopy (260 nm).
? TROUBLESHOOTING
Synthesis of nucleotide analog containing RNA: end-labeling of RNA
20| 5′-End-label (option A) or 3′-end-label (using the splint method58, option B) the RNA. Depending on the size of the RNA, it might be sufficient to use either 5′-or 3′-labeled RNA to map the entire molecule. Mostly, 5′-end-labeling is preferred, as it is faster and typically has a higher yield of labeled RNA. In contrast, for larger RNA molecules (>250 nt), both end-labeling techniques need to be used to map the entire RNA.
- (A) 5′-End-labeling of RNA
- (i) To 5′-label in vitro-transcribed RNA, remove its 5′-triphosphate group before the labeling reaction. For this purpose, mix 100 pmol RNA with 10 μl of 10× dephosphorylation buffer and ddH2O to a final volume of 100 μl. Add 5 μl alkaline phosphatase and incubate the sample at 37 °C for 30 min.
- (ii) Add 30 μl of 3 M NaOAc pH 5.0 and 70 μl ddH2O and phenol extract to the sample. Use 200 μl phenol (acidic pH of < 6.0), vortex strongly and centrifuge the sample at room temperature for 5 min (18,000g).
- (iii) Take the aqueous phase, mix it with 200 μl PCI and repeat the vortexing and centrifugation steps described in Step 20A(ii).
- (iv) Mix the aqueous phase with 200 μl CI and repeat the vortexing and centrifugation steps described in Step 20A(ii).
- (v) Precipitate the RNA with 1 μl glycogen and 2.5× volume of ethanol/0.3 M NaOAc mix. Freeze the sample at −80 °C for 60 min.
- ■ PAUSE POINT Alternatively, the RNA can be left to precipitate at −80 °C overnight.
- (vi) Centrifuge the sample at 4 °C at 18,000g for 30 min and resuspend the dried pellet in 20 μl ME. Determine the concentration by UV spectroscopy (at 260 nm).
- (vii) Set up the labeling reaction by mixing 30 pmol dephosphorylated RNA, 50 pmol γ-P32-ATP, 2 μl of 10× PNK buffer and add ddH2O to a final volume of 20 μl. Add 2 μl T4-PNK and incubate the sample at 37 °C for 30 min.
- (B) 3′-End-labeling of RNA
- (i) Mix 20 pmol RNA with 1 μl of 200 μM splint oligo, 0.5 μl of 10× annealing buffer and ddH2O to a final volume of 5 μl.
- (ii) Incubate the annealing reaction at 95 °C for 1 min, take out the heat block and allow the sample to cool to 42 °C slowly. Afterward, place the reaction on ice for 25 min.
- (iii) After annealing of the splint to the 3′-end of the RNA, add 15 μl extension mix consisting of 1 μl of 20× DTT/MgCl2, 23 pmol α-P32-dCTP, ddH2O to a final volume of 15 μl and 2 μl Klenow polymerase exo−. Incubate the reaction at 37 °C for 2h.
21| Stop the reaction (from Step 20A(vii) or 20B(iii)) by adding 1 μl of 0.5 M EDTA pH 8.0.
22| Heat the sample to 95 °C for 1 min, then immediately place the tube on ice for 2 min. (Optional: To remove the unused excess radioactive nucleotides before gel purification, precipitate the stopped labeling reaction by adding 1 μl glycogen and 2.5× volume ethanol/0.3 M NaOAc mix. Freeze at −20 °C for 30 min; centrifuge at 4 °C for 30 min (18,000g). Remove the supernatant and air dry the pellet.)
■ PAUSE POINT Samples can be stored overnight at −80 °C, before proceeding with gel purification.
23| Add 30 μl loading buffer (7 M urea) to the sample (or pellet) from Step 22. Gel purify the end-labeled RNA by denaturing PAGE (use 0.4-mm spacers, 2 cm well size; for choosing the proper percentage of gel, see Table 2).
▲ CRITICAL STEP Gel purification of RNA after the labeling reaction is essential to remove degradation products and unused excess radioactivity.
24| After the gel run, visualize the radioactively labeled RNA by autoradiography (use an instant imager; alternatively, use a phosphorimager or film exposure).
? TROUBLESHOOTING
25| Cut out the major band corresponding to full-length RNA using a sterile blade, put the gel piece in a 1.5-ml RNase-free Eppendorf tube, crush it and then add 800 μl elution buffer. Freeze the sample at −80 °C for 15 min.
■ PAUSE POINT Alternatively, the RNA can be left at −80 °C overnight.
26| Thaw the sample and shake the tube at 4 °C for 1.5 h to elute the RNA from the gel.
27| Filter the eluate through an acrodisc filter.
28| Precipitate the filtrate with 1 μl glycogen and 2.5× volume of ethanol/0.3 M NaOAc mix. Freeze the sample at −80 °C overnight.
29| Centrifuge the sample (30 min, 4 °C, 18,000g), dry the pellet carefully and then resuspend it in 20–30 μl ME yielding a concentration of ~0.5 μM. Retain an aliquot of the end-labeled RNA (~10–15 μl) to prepare the untreated and unselected controls in Steps 42 and 40, respectively.
Selection step: functional assay
30| Perform the optimized functional (folding, catalysis or ligand interaction) assay relevant to your experiment. Here, I provide an example of a folding assay optimized for the Sc.ai5γ D135 ribozyme, which requires unusually high salt concentrations. Denature the end-labeled D135 RNA (~1.5 pmol) in the presence of 0.5 M KCl and 80 mM MOPS pH 7.0 by heating the sample at 95 °C for 1 min and cooling the sample for 2 min at room temperature.
▲ CRITICAL STEP This step can be modified to investigate other RNAs of interest by adapting the monovalent ion and its concentration as well as the cooling procedure after the heating step.
31| Refold the RNA by adding 4 mM MgCl2 and 5 μl 4× glycerol loading buffer to the sample yielding a final volume of 20 μl. Incubate the sample at 42 °C for 10 min. While a much higher [Mg2+] is required to reach the native, catalytically active state of D135, I chose an Mg2+ concentration that allows the RNA to reach the compact intermediate state, as I aimed to monitor which functional groups are critical for the tertiary collapse of the D135 ribozyme22.
▲ CRITICAL STEP The ionic conditions, temperature and time for folding have to be optimized for the RNA of interest.
Selection step: native gel electrophoresis
32| To separate the unfolded from the compact population, load the RNA sample immediately on a 4%-4 mM Mg2+ native (nondenaturing) polyacrylamide gel (0.4-mm spacer and 2-cm wide wells). Native gel electrophoresis can also be used to separate products of ligand interaction assays, while denaturing gel electrophoresis is used for catalytic assays.
▲CRITICAL STEP The percentage of the gel as well as the amount of MgCl2 in the gel and in the running buffer have to be optimized before the NAIM selection.
33| Run the gel at 25 W for ~3 h in the cold room (4 °C) until the XC dye has reached the last third of the gel (size of the gel is 29 cm (W) × 25 cm (L)).
34| Perform autoradiography to detect folded and unfolded species, as indicated in Step 24.
? TROUBLESHOOTING
35| Cut out both bands (Fig. 2b) using a sterile blade and treat each RNA species (unfolded and folded) separately: crush each gel piece through a syringe in a 14-ml tube and add 2 ml elution buffer each. Freeze the samples at −80 °C for 15 min.
■ PAUSE POINT Alternatively, the RNA can be left at −80 °C overnight.
36| Thaw the samples and shake the tubes at 4 °C for 1.5 h to elute the RNA.
37| Filter the eluate through an acrodisc filter.
38| Split the filtrate into four 1.5-ml tubes and precipitate it with 1 μl glycogen and 1 ml ethanol/0.3 M NaOAc mix. Freeze the samples at −80 °C overnight.
▲ CRITICAL STEP I found it preferable to precipitate the samples as described rather than in a 14-ml tube as the pellet and the resuspension volume are both small.
39| Centrifuge the samples (30 min, 4 °C, 18,000g), dry the pellets carefully and then resuspend all four pellets in a total of 6 μl ME (keep tubes on ice).
■ PAUSE POINT Store the sample at −80 °C (up to a week) until ready to perform iodine cleavage (Step 40).
Mapping of NAIM effects: iodine sequencing
40| As all nucleotide analogs carry a phosphorothioate tag, it is possible to detect the sites of analog incorporations by performing iodine sequencing. Set up separate iodine cleavage reactions for both RNA samples (unfolded and folded species) purified from the native gel (Steps 35–39) and half the RNA sample that has not undergone the selection step (from Step 29). Mix 6 μl of each RNA sample with 1 μl of 10 mM iodine and incubate at 37 °C for 3 min.
▲ CRITICAL STEP The nonselected sample is required (see lanes US in Fig. 3) to determine the relative incorporation efficiency of a nucleotide analog at a given position. Comparing band intensities between the unfolded and folded samples (see lanes U and F in Fig. 3) reveals any sites of NAIM interferences or enhancements.
41| Quench each reaction by adding 8 μl preheated stop solution.
▲ CRITICAL STEP The stop solution must be heated to 95 °C ahead of time to ensure it is liquefied.
■ PAUSE POINT Store the samples at −80 °C.
Mapping of NAIM effects: denaturing gel electrophoresis
42| To map NAIM effects throughout the RNA, run the required number of denaturing polyacrylamide gels of the appropriate lengths and concentrations (see Table 2 and Experimental design for guidelines). In addition to the three samples from Step 41 (US, U and F; Fig. 3), load a control (UN lane in Figs. 2 and 3) to determine background degradation of the RNA (use the remaining RNA aliquot from Step 29 and mix it with stop solution). For example, for the ~600-nt long D135 RNA, I used 90-cm long 5 and 8% long-ranger acrylamide gels. These were run at 90 and 80 W, respectively, for 6–11 h. I also used regularly sized (45 cm) standard 15% gels (35 W, ~2–3 h) to map the very 5′-or 3′-end of the RNA.
43| Dry the gels and expose them in phosphorimager cassettes overnight. The next day, scan the screen using the phosphorimager.
? TROUBLESHOOTING
Quantification
44| Quantify the band intensities using ImageQuant Software (Molecular Dynamics). Alternatively, SAFA59 software could be used, which is a semiautomated program and therefore reduces the time required for data analysis. Notably, this software was specifically designed for footprinting analysis (e.g., ref. 60) and therefore calculates counts for every position (all nucleotides) in the RNA. In the case of NAIM, this is somewhat disadvantageous, as each tested analog only refers to a specific nucleotide type (e.g., for studying 7dAαS one only needs data for all adenosine residues).
45| Process the output numbers in Excel (Microsoft) or in a comparable program. Correct for background by subtracting the counts observed for the individual bands in the UN lane from the respective bands in the U and F lanes (Supplementary Table 1 online).
46| Correct for loading differences by calculating the ratio of counts for each band in the U and F lanes and subsequently the mean of these ratios. Then normalize the F lane by this mean factor (Supplementary Table 1 online).
47| Calculate NAIM effects from the ratio of normalized counts in the folded and unfolded lanes for a given position (Supplementary Table 1 online).
48| Set a threshold value, below which interferences and enhancements are considered insignificant; the cutoff is typically between 1.5 and 2 (refs. 21,22,40). Additionally, a λ discrimination factor and limits (from the normalized data) can be calculated to determine which effects are significant10.
49| Determine whether an observed NAIM effect is due to the phosphorothioate tag or the specific nucleotide modification, by comparing the NAIM effects for the modified nucleoside (e.g., in case of m6AαS) to those for the corresponding phosphorothioate (AαS). This step can be achieved by following either option A or B:
- (A) ‘A progressive approach’
- (i) Divide the modified nucleoside value (e.g., m6AαS) by that of the parent phosphorothioate (e.g., AαS); this is the approach used in most NAIM studies10,40. This way, if both AαS and m6AαS interferences are observed for a particular adenosine residue in the RNA, it is possible to infer the individual effects resulting from the phosphorothioate tag or from the N6-methylation. This is based on the assumption that the influence of the nonbridging sulfur modification and of the secondary nucleoside modification on RNA folding is essentially independent from one another.
- (B) ‘A conservative approach’
- (i) Present the raw interference values for each modification and display them next to that of the corresponding phosphorothioate22. This more conservative approach has the following advantages. First, it might not be a fair assumption that the effects from the nonbridging sulfur modification and the additional nucleoside modification are entirely independent of one another. Second, it is possible that phosphorothioate enhancements are observed, indicating that the RNA folds better when oxygen is replaced by sulfur at certain positions. Similarly, modified nucleoside enhancements may also be detected. In these cases, any correction for the parent phosphorothioate (as described in Step 49A) would definitively bias the interpretation of the data, and therefore it can be preferable to provide the raw, uncorrected data for interference or enhancement at any given position22.
● TIMING
Steps 1–7, synthesis of nucleotide analog containing RNA (template preparation): ~1 d
Steps 8–19, synthesis of nucleotide analog containing RNA (in vitro transcription): ~2 d
Steps 20–29, synthesis of nucleotide analog containing RNA (end-labeling of RNA): ~2 d
Steps 30 and 31, selection step (functional assay): ~30 min
Steps 32–39, selection step (native gel electrophoresis): ~1–2 d
Steps 40 and 41, mapping of NAIM effects (iodine sequencing): 15 min
Steps 42 and 43, mapping of NAIM effects (denaturing gel electrophoresis): 5 d
Steps 44–49, quantification: ~1 month
? TROUBLESHOOTING
Steps 13 and 24
Problem: RNA degradation.
Solution: RNA (in particular radioactively labeled RNA) is very sensitive to degradation. Therefore, it is of utmost importance to follow some general considerations when working with RNA: (i) always wear gloves; (ii) keep the RNA on ice unless otherwise indicated; (iii) avoid freezing and thawing the RNA stock too often (aliquot, if necessary); (iv) prepare solutions in RNA grade using RNase-free or DEPC-treated water; chemicals should be of high quality and reserved for RNA; (v) reagents and consumables should be maintained free of RNases or bought as RNase-free grade. If you encounter serious RNase contamination, it is often helpful to clean the equipment with ‘RNase Zap’ (Ambion) or a comparable product. Also, clean the bench thoroughly including the water bath. In addition, it is highly recommended to aliquot the reagents (e.g., ddH2O, MgCl2 solution, buffers); if a problem is encountered, simply take a new aliquot without having to remake all the solutions.
Step 19
Problem: Low in vitro transcription yield and incorporation of analogs.
Solution: For optimal transcription efficiency, it is preferable to optimize the transcription reaction conditions for each novel construct. Parameters, such as incubation time, concentration of nucleotides and Mg2+ as well as of the DNA template can significantly influence the yield of transcripts. Furthermore, adding an increased amount of DTT or 0.1% (final concentration) Triton X-100 can improve the yield. At last, adjusting the ratio of NTPαS and unmodified NTP (Table 1) is important for optimizing the incorporation level of the analog during transcription, especially if noncommercially available nucleotide analogs are used39,40,48,49,55.
Step 24
Problem: Low efficiency of labeling reaction.
Solution: For 5′ labeling, it is noteworthy that the T4-PNK is an enzyme highly sensitive to changes in temperature (e.g., when taking the stock vial out of the freezer). Also, the enzyme buffer is sensitive to multiple rounds of freeze–thaw. Therefore, I suggest repeating the experiment using fresh T4-PNK and aliquot the provided buffer. Moreover, it is possible that the RNA was insufficiently dephosphorylated, in which case Step 20A (i–vi) should be repeated. In the case of 3′ labeling, check whether the melting temperature of the splint oligo is >42 °C. The TM of the splint should be at least 5 °C higher than the temperature during the extension reaction (37 °C), to ensure that the splint remains annealed to the RNA during this procedure. Also, it is advantageous to use a splint with a 2′,3′ dideoxynucleotide at its 3′-end; this prevents the Klenow enzyme from extending thesplintatits 3′-end, especially if the next 5′-residue in the RNA is complementary to the radioactive nucleotide, such as α-P32-dCTP.
Step 34
Problem: Low resolution of RNA populations in the native gel.
Solution: The native gel assay has to be optimized for the RNA of interest (nonanalog-containing transcript) before applying it to a NAIM procedure. However, it is possible that despite the initial optimization, the incorporation of analogs then results in a decreased resolution of the different RNA species, such as the unfolded and folded population. Some analogs can cause a severe smear between the bands, which may no longer be visible as sharp bands. This problem can be overcome for most analogs by (i) adjusting the native gel setup (e.g., reducing the applied power, increasing Mg2+ concentration in the gel and running buffer, subtly changing the percentage of the native gel, adjusting the loaded sample volume with respect to well size) and (ii) reducing the nucleotide analog incorporation level. Nevertheless, it remains possible that some analogs may not be suitable for your folding-based NAIM approach. In my case, the 2APαS analog perturbed folding of the Sc. ai5γ D135 ribozyme severely and could therefore not be used in our NAIM study22.
Step 43
Problem: Poor or excessive iodine cleavage.
Solution: If this is observed, vary the concentration of iodine from 0.1 to 2 mM (final concentration) and alter the incubation time (1–5 min) or temperature (37 °C or room temperature). It is also recommended to prepare fresh iodine solution every 2 weeks.
Step 43
Problem: Uneven loading.
Solution: Although small differences in loading can be accounted for by normalization during the quantification of the data, substantially different band intensities usually render the data analyses impossible requiring a re-run of the gel. Thus, it is generally recommended to determine the counts of each sample (control, unselected, unfolded and folded) and balance them before loading on the denaturing polyacrylamide gel.
Step 43
Problem: Salt front.
Solution: If RNA samples have a high salt content, it typically results in the narrowing of lanes during denaturing gel electrophoresis. In order to avoid this problem, it is generally recommended to wash RNA/DNA pellets with 70% ethanol before drying and resuspending them. If this problem is encountered with the iodine-cleaved samples, it is advisable to precipitate the RNA after iodine cleavage (Step 40) instead of direct loading onto the denaturing polyacrylamide gel. In addition, by increasing the percentage of the denaturing gel and the time of the gel run, this problem is usually overcome, as the salt front runs out of the gel. For example, to read more or less the same stretch of RNA (e.g., nucleotides 70–200), one can run a 8% gel until the XC is close to the bottom (dye migrates with ~75 nt; see Table 2) instead of a 5% gel until the BB is at the bottom (dye migrates with ~35 nt; see Table 2), thereby running the salt front out of the gel and avoiding narrowing of lanes due to the salt.
Step 43
Problem: Low signal-to-noise ratio.
Solution: This makes quantification difficult and less reliable. In order to improve the signal-to-noise ratio, load more sample per lane. Of course, this requires an adjustment of the amount of starting material. I recommend optimizing the amount of RNA used in the selection step (and accordingly the amount of RNA used before in the labeling reaction) with respect to the counts required per lane. Also, it should be taken into consideration that it is of course preferable to map NAIM effects throughout the entire RNA (or half of it in case of large RNAs, such as the D135 ribozyme) from the same selection batch (of either 5′-or 3′-end-labeled RNA).
ANTICIPATED RESULTS
The NAIM experiment usually consists of four lanes (per nucleotide analog) resolved on a sequencing gel (Fig. 3a). Figure 3 shows the results of a representative experiment performed using the D135 ribozyme having AαS, m6AαS or DAPαS incorporated to identify whether these functional groups are essential for folding of this RNA molecule. The first lane (UN) always indicates background degradation of the labeled RNA, which has to be taken into account during quantification. The second lane US depicts the relative incorporation level of a particular analog at a given position. For example, in this experiment, analogs are less efficiently incorporated at position 214 than 215. This information is important to evaluate band intensities better in the subsequent lanes (U and F), in which faint bands are observed at position 214 in both lanes. Finally, NAIM effects are detected by comparing band intensities between lanes U and F and quantifying their ratio (Fig. 3): interferences are detected as gaps in the RNA sequence ladder among the folded RNA population, while enhancements are seen as under-representation in the unfolded species. As all nucleotide analogs harbor a phosphorothioate linkage, it is essential to differentiate whether a NAIM effect is caused by the phosphorothioate tag or by the nucleotide modification, whereby I chose the more conservative quantification approach (Step 49B), as a number of phosphorothioate enhancements had been identified22. For example, incorporating AαS at position 209 results in a pronounced phosphorothioate enhancement, indicating that this modification is beneficial for D135 folding. Note that this enhancement remains unchanged when m6AαS or DAPαS are incorporated, implying this effect is entirely due to the phosphorothioate tag and there is no additive effect from the nucleoside modification (Fig. 3b). On the other hand, introducing AαS at positions 198 and 204 causes interferences, suggesting that this modification is detrimental to folding and therefore these molecules are under-represented in the folded population. Notably, incorporating DAPαS at these positions (A198 and A204) results in increased interference values than those observed for the AαS analog (Fig. 3b), implying an additional folding perturbation is caused by the nucleobase modification. As Mg2+ ions preferentially coordinate with oxygen, these phosphorothioate interferences suggest that metal ions play an important role in organizing the κ-ζ element (Fig. 3). In addition to phosphorothioate effects, numerous nucleoside-specific NAIM effects were observed: incorporation of m6AαS resulted in interferences at positions 195 and 196, while DAPαS substitution of the very same positions offers an advantage to D135 folding resulting in enhancements. Intriguingly, I often observed m6AαS interferences and DAPαS enhancements for the same adenosines in helical segments of D135, implying that local duplex stability in the D1 core is essential for folding of this RNA22.In contrast to m6AαS, which weakens the A–U base pair by one hydrogen bond, DAPαS is able to base pair with uridine forming an additional H-bond. At last, the NAIM study also provided information on the D135 architecture, as the observed NAIM effects (Fig. 3) are mostly inconsistent with the κ region forming a canonical tetraloop and ζ element adopting a receptor structure in the collapsed D135 molecule22,23. In summary, plotting the individual NAIM effects onto the secondary structure map of the D135 ribozyme provided novel insights into the structural requirements of D135 compaction and led to the identification of a folding control element22,23.
NAIM results are strongly dependent on the design and stringency of the selection step, during which the pool of molecules is analyzed for their performance of a specific RNA function. It is therefore recommended to optimize carefully this most critical step of the entire NAIM procedure, as this ensures proper selection for crucial atoms and functional groups. The absolute number of NAIM effects significantly varies between individual studies due to a difference in stringency, length of RNA, process investigated, number of analogs tested and choice of the threshold value among other reasons. Importantly, although in most studies only NAIM interferences have been reported, this does not preclude the possibility of observing NAIM enhancements. In my NAIM study, albeit surprisingly, multiple enhancements were indeed identified, suggesting that altering certain atoms at specific residues is beneficial for the formation of the collapsed RNA folding intermediate22,23. Independent of the actual number of NAIM effects, individual interferences and enhancements as well as their clustering are expected to provide an enormous amount of novel insights into the atomic requirements of the process under investigation: RNA structure, folding, catalysis and ligand interaction.
Supplementary Material
ACKNOWLEDGMENTS
Anna Marie Pyle is acknowledged for her support and helpful suggestions. I thank Olga Fedorova for many invaluable discussions and for critically reading the manuscript. Funding was in part provided by National Institutes of Health (NIH) grant GM50313 to A.M. Pyle when I was a postdoctoral associate in her lab and by the Austrian Science Foundation (FWF; J2332) to C.W.
Footnotes
Note: Supplementary information is available via the HTML version of this article.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Brunel C, Romby P. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- 2.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 2006;34:e48. doi: 10.1093/nar/gkl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampel KJ, Burke JM. Time-resolved hydroxyl-radical footprinting of RNA using Fe(II)-EDTA. Methods. 2001;23:233–239. doi: 10.1006/meth.2000.1134. [DOI] [PubMed] [Google Scholar]
- 4.Urlaub H, Hartmuth K, Luhrmann R. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods. 2002;26:170–181. doi: 10.1016/S1046-2023(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 5.Reed R, Chiara MD. Identification of RNA-protein contacts within functional ribonucleoprotein complexes by RNA site-specific labeling and UV crosslinking. Methods. 1999;18:3–12. doi: 10.1006/meth.1999.0751. [DOI] [PubMed] [Google Scholar]
- 6.Noah JW, Lambowitz AM. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003;42:12466–12480. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- 7.Sigel RK, Pyle AM. Lanthanide ions as probes for metal ions in the structure and catalytic mechanism of ribozymes. Met. Ions Biol. Syst. 2003;40:477–512. [PubMed] [Google Scholar]
- 8.Streicher B, Westhof E, Schroeder R. The environment of two metals ions surrounding the splice site of a group I intron. EMBO J. 1996;15:2556–2564. [PMC free article] [PubMed] [Google Scholar]
- 9.Polacek N, Barta A. Metal ion probing of rRNAs: evidence for evolutionarily conserved divalent cation binding pockets. RNA. 1998;4:1282–1294. doi: 10.1017/s1355838298980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedorova O, Boudvillain M, Kawaoka J, Pyle AM. Nucleotide analog interference mapping and suppression: specific applications in studies of RNA tertiary structure, dynamic helicase mechanism and RNA–protein interactions. In: Hartmann RK, Bindereif A, Schön A, Westhof E, editors. Handbook of RNA Biochemistry. Wiley-VCH; Weinheim: 2005. pp. 259–293. [Google Scholar]
- 11.Ryder SP, Ortoleva-Donnelly L, Kosek AB, Strobel SA. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 2000;317:92–109. doi: 10.1016/s0076-6879(00)17008-9. [DOI] [PubMed] [Google Scholar]
- 12.Vortler LC, Eckstein F. Phosphorothioate modification of RNA for stereochemical and interference analyses. Methods Enzymol. 2000;317:74–91. doi: 10.1016/s0076-6879(00)17007-7. [DOI] [PubMed] [Google Scholar]
- 13.Christian EL, Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 14.Kazantsev AV, Pace NR. Identification by modification-interference of purine N-7 and ribose 2′- OH groups critical for catalysis by bacterial ribonuclease P. RNA. 1998;4:937–947. doi: 10.1017/s1355838298980384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rox C, Feltens R, Pfeiffer T, Hartmann RK. Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme. J. Mol. Biol. 2002;315:551–560. doi: 10.1006/jmbi.2001.5261. [DOI] [PubMed] [Google Scholar]
- 16.Siew D, Zahler NH, Cassano AG, Strobel SA, Harris ME. Identification of adenosine functional groups involved in substrate binding by the ribonuclease P ribozyme. Biochemistry. 1999;38:1873–1883. doi: 10.1021/bi982329r. [DOI] [PubMed] [Google Scholar]
- 17.Ortoleva-Donnelly L, Szewczak AA, Gutell RR, Strobel SA. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauss-Soukup JK, Strobel SA. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J. Mol. Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- 19.Strobel SA, Shetty K. Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. Proc. Natl. Acad. Sci. USA. 1997;94:2903–2908. doi: 10.1073/pnas.94.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorova O, Pyle AM. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. EMBO J. 2005;24:3906–3916. doi: 10.1038/sj.emboj.7600852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat. Struct. Mol. Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 23.Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oyelere AK, Kardon JR, Strobel SA. pK(a) perturbation in genomic Hepatitis Delta Virus ribozyme catalysis evidenced by nucleotide analogue interference mapping. Biochemistry. 2002;41:3667–3675. doi: 10.1021/bi011816v. [DOI] [PubMed] [Google Scholar]
- 25.Tuschl T, Ng MM, Pieken W, Benseler F, Eckstein F. Importance of exocyclic base functional groups of central core guanosines for hammerhead ribozyme activity (published erratum appears in Biochemistry 33, 848 (1994)) Biochemistry. 1993;32:11658–11668. doi: 10.1021/bi00094a023. [DOI] [PubMed] [Google Scholar]
- 26.Williams DM, Pieken WA, Eckstein F. Function of specific 2′-hydroxyl groups of guanosines in a hammerhead ribozyme probed by 2′ modifications. Proc. Natl. Acad. Sci. USA. 1992;89:918–921. doi: 10.1073/pnas.89.3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowrira BM, Burke JM. Extensive phosphorothioate substitution yields highly active and nuclease-resistant hairpin ribozymes. Nucleic Acids Res. 1992;20:2835–2840. doi: 10.1093/nar/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder SP, et al. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 29.Ryder SP, Strobel SA. Nucleotide analog interference mapping of the hairpin ribozyme: implications for secondary and tertiary structure formation. J. Mol. Biol. 1999;291:295–311. doi: 10.1006/jmbi.1999.2959. [DOI] [PubMed] [Google Scholar]
- 30.Jones FD, Strobel SA. Ionization of a critical adenosine residue in the Neurospora Varkud Satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- 31.Ruffner DE, Uhlenbeck OC. Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res. 1990;18:6025–6029. doi: 10.1093/nar/18.20.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vortler CS, Fedorova O, Persson T, Kutzke U, Eckstein F. Determination of 2′-hydroxyl and phosphate groups important for aminoacylation of Escherichia coli tRNAAsp: a nucleotide analogue interference study. RNA. 1998;4:1444–1454. doi: 10.1017/s1355838298980967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen JA, McCarthy TJ, Soukup GA, Soukup JK. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat. Struct. Mol. Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- 34.Cochrane JC, Batey RT, Strobel SA. Quantitation of free energy profiles in RNA-ligand interactions by nucleotide analog interference mapping. RNA. 2003;9:1282–1289. doi: 10.1261/rna.5102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu S, et al. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- 36.Luptak A, Doudna JA. Distinct sites of phosphorothioate substitution interfere with folding and splicing of the Anabaena group I intron. Nucleic Acids Res. 2004;32:2272–2280. doi: 10.1093/nar/gkh548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckstein F. Nucleoside phosphorothioates. Annu. Rev. Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 38.Eckstein F, Gish G. Phosphorothioates in molecular biology. Trends Biochem. Sci. 1989;14:97–100. doi: 10.1016/0968-0004(89)90130-8. [DOI] [PubMed] [Google Scholar]
- 39.Gish G, Eckstein F. DNA and RNA sequence determination based on phosphorothioate chemistry. Science. 1988;240:1520–1522. doi: 10.1126/science.2453926. [DOI] [PubMed] [Google Scholar]
- 40.Ryder SP, Strobel SA. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 41.Strobel SA. A chemogenetic approach to RNA function/structure analysis. Curr. Opin. Struct. Biol. 1999;9:346–352. doi: 10.1016/S0959-440X(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 42.Cochrane JC, Strobel SA. Probing RNA structure and function by nucleotide analog interference mapping. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Current Protocols in Nucleic Acid Chemistry. John Wiley & Sons; Hoboken, New Jersey: 2004. pp. 6.9.1–6.9.21. [DOI] [PubMed] [Google Scholar]
- 43.Ryder SP, Strobel SA. Comparative analysis of hairpin ribozyme structures and interference data. Nucleic Acids Res. 2002;30:1287–1291. doi: 10.1093/nar/30.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu S, Strobel SA. Biochemical detection of monovalent metal ion binding sites within RNA. Methods. 2001;23:264–275. doi: 10.1006/meth.2000.1137. [DOI] [PubMed] [Google Scholar]
- 45.Soukup JK, Minakawa N, Matsuda A, Strobel SA. Identification of A-minor tertiary interactions within a bacterial group I intron active site by 3-deazaadenosine interference mapping. Biochemistry. 2002;41:10426–10438. doi: 10.1021/bi020265l. [DOI] [PubMed] [Google Scholar]
- 46.Boudvillain M, Delencastre A, Pyle AM. A new RNA tertiary interaction that links active-site domains of a group II intron and anchors them at the site of catalysis. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- 47.Szewczak AA, Ortoleva-Donnelly L, Ryder SP, Moncoeur E, Strobel SA. A minor groove RNA triple helix within the catalytic core of a group I intron. Nat. Struct. Biol. 1998;5:1037–1042. doi: 10.1038/4146. [DOI] [PubMed] [Google Scholar]
- 48.Arabshahi A, Frey PA. A simplified procedure for synthesizing nucleoside 1-thiotriphosphates: dATP alpha S, dGTP alpha S, UTP alpha S, and dTTP alpha S. Biochem. Biophys. Res. Commun. 1994;204:150–155. doi: 10.1006/bbrc.1994.2438. [DOI] [PubMed] [Google Scholar]
- 49.Oyelere AK, Strobel SA. Biochemical detection of cytidine protonation within RNA. J. Am. Chem. Soc. 2000;122:10259–10267. [Google Scholar]
- 50.Ortoleva-Donnelly L, Kronman M, Strobel SA. Identifying RNA minor groove tertiary contacts by nucleotide analogue interference mapping with N2-methylguanosine. Biochemistry. 1998;37:12933–12942. doi: 10.1021/bi980723j. [DOI] [PubMed] [Google Scholar]
- 51.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 52.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem. Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutell RR, Cannone JJ, Shang Z, Du Y, Serra MJ. A story: unpaired adenosine bases in ribosomal RNAs. J. Mol. Biol. 2000;304:335–354. doi: 10.1006/jmbi.2000.4172. [DOI] [PubMed] [Google Scholar]
- 55.Basu S, Pazsint C, Chowdhury G. Analysis of ribozyme structure and function by nucleotide analog interference mapping. Methods Mol. Biol. 2004;252:57–75. doi: 10.1385/1-59259-746-7:057. [DOI] [PubMed] [Google Scholar]
- 56.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swisher J, Su L, Brenowitz M, Anderson V, Pyle A. Productive folding to the native state by a group II intron ribozyme. J. Mol. Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 58.Huang Z, Szostak JW. A simple method for 3′-labeling of RNA. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shcherbakova I, Brenowitz M. Perturbation of the hierarchical folding of a large RNA by the destabilization of its Scaffold's tertiary structure. J. Mol. Biol. 2005;354:483–496. doi: 10.1016/j.jmb.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 61.Christian EL, Yarus M. Analysis of the role of phosphate oxygens in the group I intron from Tetrahymena. J. Mol. Biol. 1992;228:743–758. doi: 10.1016/0022-2836(92)90861-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.