Summary
The endoplasmic reticulum (ER) must target potentially toxic misfolded proteins for retrotranslocation and proteasomal degradation while avoiding the destruction of productive folding intermediates. For luminal proteins, this discrimination typically depends not only on the folding status of a polypeptide, but also on its glycosylation state. Two putative sugar binding proteins, Htm1p and Yos9p, are required for degradation of misfolded glycoproteins, but the nature of the glycan degradation signal and how such signals are generated and decoded remains unclear. Here we characterize Yos9p’s oligosaccharide-binding specificity and find that it recognizes glycans containing terminal α1,6-linked mannose residues. We also provide evidence in vivo that a terminal α1,6-linked mannose-containing oligossacharide is required for degradation and that Htm1p acts upstream of Yos9p to mediate the generation of such sugars. This strategy of marking potential substrates by Htm1p and decoding the signal by Yos9p is well suited to provide a proofreading mechanism that enhances substrate specificity.
Introduction
The endoplasmic reticulum (ER) contains sophisticated quality control systems that monitor protein folding to ensure that only properly folded and oligomerized forms are transported forward through the secretory pathway (Anelli and Sitia, 2008; Fewell, 2001). Despite the highly specialized folding environment of the ER, inevitably some fraction of newly made polypeptides misfolds (Casagrande et al., 2000; Friedlander et al., 2000; Helenius and Aebi, 2001; Jensen et al., 1995; Travers et al., 2000; Ward et al., 1995). Such terminally misfolded forms are cleared from the ER through ER-Associated Degradation (ERAD) pathways in which they are first recognized and then retrotranslocated into the cytosol for destruction by the ubiquitin-proteasome degradation system (Nakatsukasa and Brodsky, 2008; Romisch, 2005).
The cell must maintain a balance between overly promiscuous destruction of inherently slow-folding proteins or potentially functional mutants, while at the same time preventing the escape of toxic forms from the ER (Drumm et al., 1991; Sekijima et al., 2005). In order to achieve this balance, the ERAD machinery must accurately distinguish terminally misfolded proteins from abundant folding intermediates (Hebert and Molinari, 2007; Helenius and Aebi, 2001). The ERAD-L pathway, which degrades proteins that contain misfolded domains within the ER lumen (Huyer et al., 2004; Vashist and Ng, 2004), uses a bipartite recognition mechanism that interrogates both the glycosylation and the folding state of potential substrates (Denic et al., 2006; Gauss et al., 2006a; Knop et al., 1996b; Spear and Ng, 2005). This process is carried out by a large multi-protein complex that includes the E3 ubiquitin ligase Hrd1p (Bays et al., 2001; Deak and Wolf, 2001) and several transmembrane and cystosolic factors involved in extraction and ubiquitination of substrates (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006b). The luminal side of the complex contains a putative lectin, Yos9p, (Bhamidipati et al., 2005; Buschhorn et al., 2004; Kim et al., 2005; Szathmary et al., 2005) as well as Hrd3p (Gardner et al., 2000; Plemper et al., 1999), which recruits substrates based on the presence of misfolded domains. It has been recently shown that the disruption of the Yos9p sugar-binding domain or elimination of the sugar modification sites from the model ERAD-L substrate CPY* (CPY*0000) results in substrate stabilization (Finger et al., 1993; Knop et al., 1996b; Kostova and Wolf, 2005; Spear and Ng, 2005), however, CPY*0000 still efficiently interacts with Hrd3p in a manner that is dependent on folding status but not the glycosylation state of the substrate (Denic et al., 2006; Gauss et al., 2006a). These results suggest that recognition occurs as a multi-step process including the recruitment of misfolded proteins to the complex and a distinct commitment step where the presence of the glycans conveys information that is critical for substrate retrotranslocation and degradation (Denic et al., 2006; Gauss et al., 2006a).
While it is clear that the identification of misfolded forms critically depends on the glycosylation status of the misfolded protein, the nature of the glycan species that triggers destruction and how it contributes to specificity remains unclear. Previously, it has been shown that that the processing of the pre-assembled Glc3Man9GlcNac2 oligosaccharide that is initially transferred to proteins at N-X-S/T sites is involved in ERAD recognition. This initial glycan is trimmed through the sequential action of two glucosidases and a mannosidase (Mns1p) to yield a Man8GlcNac2 species (Hebert et al., 2005; Helenius and Aebi, 2004). These trimming steps are required for ERAD-L, and it has been suggested that the period of time it takes to reach a certain glycan structure provides proteins a period of time to fold without risk of being degraded (Helenius, 1994; Hitt and Wolf, 2004; Jakob et al., 1998; Knop et al., 1996b; Wu et al., 2003). However, recent studies in yeast suggest that the above trimming steps occur rapidly on the scale of synthesis and degradation and thus are not well suited to increase specificity (Szathmary et al., 2005). Here we reveal that Yos9p recognizes trimmed glycans that expose a terminal α1,6-linked mannose and provide evidence that suggests that Htm1p, a mannosidase-like protein whose critical role in ERAD has been largely uncharacterized (Jakob et al., 2001; Nakatsukasa et al., 2001), is required to generate such sugars. Our results suggest a model where Htm1p “marks” misfolded glycoproteins by revealing a terminal α1,6-linked mannose that is recognized by Yos9p as the signal for degradation. This dual checking mechanism could provide increased specificity during ERAD.
Results
Yos9p structure and function does not depend on its N-linked glycans
A hallmark of Yos9p and its mammalian homologues is the presence of a mannose 6-phosphate receptor homology (MRH) domain (Munro, 2001). Mutations in this domain (e.g. R200A) that ablate the putative sugar-binding pocket disrupt Yos9p’s ability to support ERAD of glycoproteins suggesting that Yos9p acts as a lectin during substrate recognition (Bhamidipati et al., 2005; Szathmary et al., 2005). In order to explore Yos9p’s role as a lectin, we sought to produce biochemical amounts of Yos9p and directly evaluate its sugar-binding specificity.
Recombinant production of suitable quantities of Yos9p posed a technical challenge as Yos9 is glycosylated and contains many disulfide bonds. First, we mutated all the N-linked glycosylation consensus sites on Yos9p and found that glycosylation of Yos9p is not required for function, as the mutant protein fully supported ERAD of CPY* in vivo (Figure 1A). Attempts to produce native protein in E. coli yielded soluble disulfide cross-linked aggregates (Figure S1). We therefore developed an alternate expression strategy in which Yos9p was isolated under denaturing conditions from E.coli inclusion bodies, refolded under optimized conditions, and purified to apparent homogeneity (Figure 1B, S2). The refolded material appeared well-ordered by circular dichroism spectroscopy (Figure 1D), contained minimal intermolecular disulfide crosslinks (Figure 1B) and migrated as a single peak by gel filtration that is consistent with it being a trimer (Figure 1C). Using the same protocol, we also produced the R200A Yos9p MRH mutant. The R200A mutation did not disturb the folding or structure of Yos9p which is consistent with the proposal that this mutation interferes with ERAD function specifically by preventing sugar-binding rather than causing global unfolding (Figure 1D) (Bhamidipati et al., 2005; Szathmary et al., 2005).
Figure 1. Purification of biochemical amounts of Yos9p from E.coli.
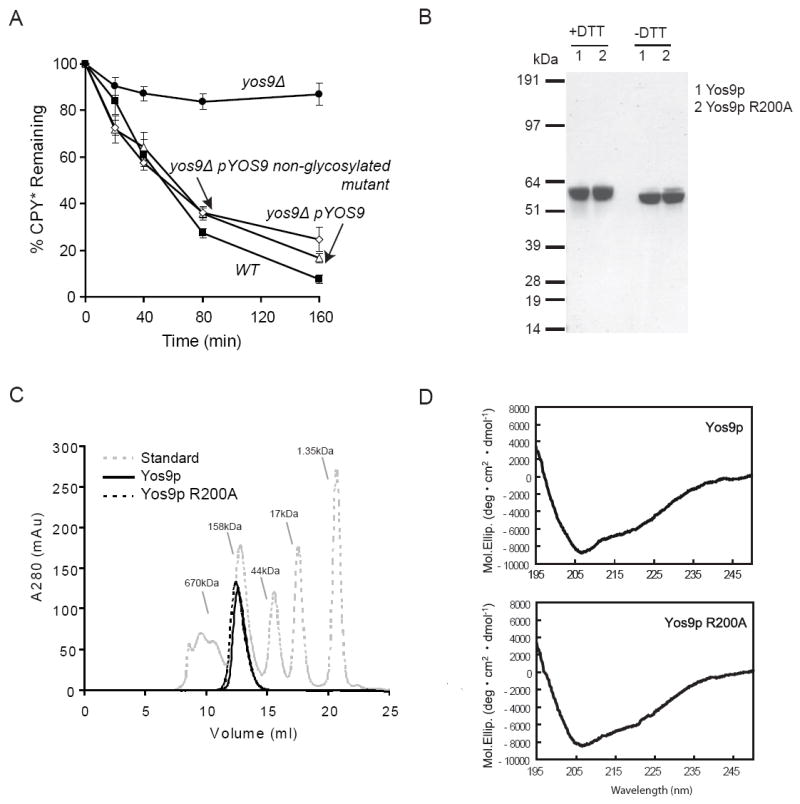
(A) Degradation of CPY* in (■)wild-type (WT), and (●) a yos9Δ strain harboring an empty vector, and yos9Δ strain covered with a plasmid expressing YOS9-flag (△) or a Yos9p variant that is missing glycosylation sites (◇) was monitored by cycloheximide chase. Equal amounts of log phase cells were removed at the indicated times following addition of cycloheximide. Samples were resolved by SDS-PAGE and detected by Western analysis using anti-HA and anti-hexokinase antibodies. Each time point represents the average and +/- standard error of the mean (SEM) of 4 measurements (two independent experiments done in duplicate) and normalized to the hexokinase loading control.
(B) Refolded recombinantly expressed Yos9p (1) and Yos9p R200A (2) was purified and analyzed by SDS-PAGE with sample buffer containing either DTT or N-ethylmaleimide (-DTT) and stained with Coomassie.
(C) Gel Filtration analysis of Yos9p (—), Yos9p R200A (- - -) and molecular size standards (gray).
(D) Circular Dichroism spectra were acquired of Yos9p (top) and Yos9p R200A (bottom) as described in the experimental procedures.
Yos9p recognizes a terminal α1,6-linked mannose
We next determined the sugar-binding specificity of our recombinant Yos9p using frontal affinity chromatography (FAC), which provides a quantitative way to evaluate lectin-oligosaccharide interactions in solution. In this approach, protein is immobilized on a matrix and oligosaccharides, which are fluorescently labeled by pyridylamination (PA), are applied to the column of immobilized protein. The degree of retardation of the sugar, relative to a control sugar that is not recognized by the immobilized protein, provides a quantitative equilibrium measure of the binding affinity, with longer delays corresponding to tighter binding constants (See Figure 2 and Experimental Procedures) (Hirabayashi et al., 2003).
Figure 2. Yos9 recognizes glycans containing a terminal α1,6-linked mannose.
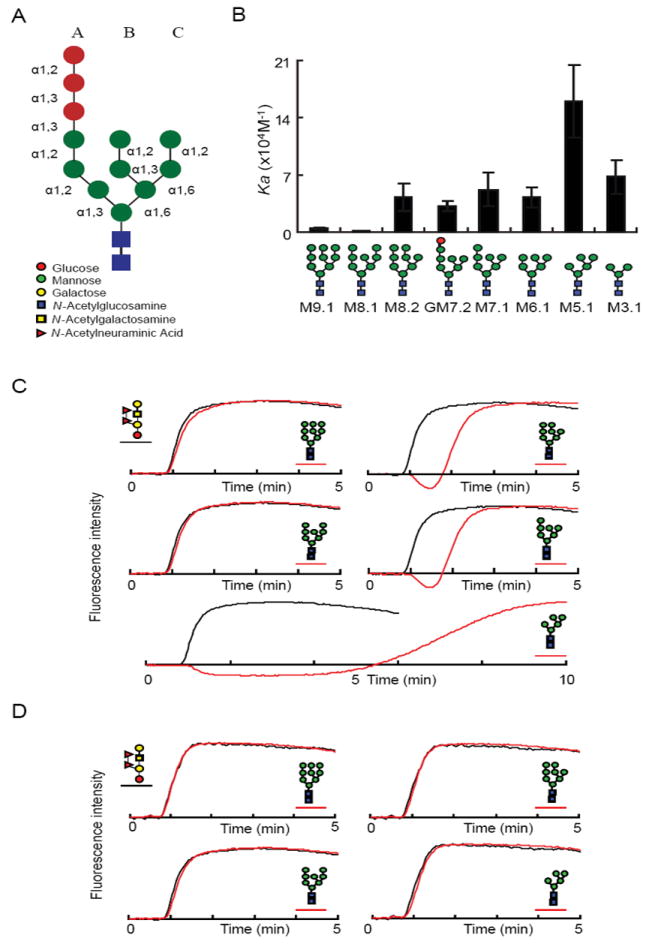
(A) A schematic representation of the initial Glc3Man9GlcNac2 N-linked sugar and the legend for each sugar moiety represented.
(B) FAC analysis of Yos9p sugar binding specificity. Indicated PA-oligasscharides were tested for binding to Yos9p by FAC analysis. Ka values were determined as described in the supplemental experimental procedures and are mean ± S.D. of three independent experiments. Each glycan structure is detailed and given a code name beneath the chart.
(C-D) Elution profiles over time of fluorescently labeled (PA)-oligosaccharides applied over immobilized histidine tagged Yos9p (C, red) or R200A mutant (D, red) in comparison to a negative control sugar (black). PA-glycans are schematically represented next to the corresponding elution profile.
The FAC analysis revealed that Yos9p has a specificity for oligosaccharides containing a terminal α1,6-linked mannose which is not present in the initial N-linked glycan (see Figure 2A, 2B for schematics of high mannose sugars). Importantly, Yos9p’s sugar specificity is dependent on an intact MRH domain, as the R200A lectin mutant shows no affinity for any of the glycans tested (Figure 2D). Yos9p has little affinity for the final trimming product of the two glucosidases (M9.1) or for the trimming product of Mns1p (M8.1) (Figure 2B)(Helenius and Aebi, 2004). In contrast, Yos9p recognizes species with the final mannose on the C branch removed to reveal a terminal α1,6-linked mannose. A comparison between M8.1 and M8.2 highlights the critical role of the α1,6-linked mannose, as both have eight mannoses arranged into three branches but only M8.2, which has the terminal α1,6-linked mannose, interacts with Yos9p. The α1,6-linked mannose seems to be necessary and largely sufficient for recognition by Yos9p because sugars containing this signal show significant affinity for Yos9p. The other residues seem to have relatively small effects on Yos9p binding with one prominent exception being the M5.1 species which has a higher affinity. How and if this species is generated under physiological conditions is unclear (see discussion).
Terminal α1,6-linked mannose containing N-linked glycans serve as the ERAD-L degradation signal
We next tested the functional significance of the terminal α1,6-linked mannose as an ERAD signal in vivo focusing on M7.1 because it is most likely to serve as a degradation signal as it requires minimal modification to the Mns1p produced M8.1 sugar (Herscovics, 2001). To test the role of M7.1, we took advantage of the previous observation by Aebi and coworkers that the M7.1 sugar could be produced in vivo using a series of genetic mutations in the asparagine-linked glycosylation (ALG) biosynthesis pathway (Figure 3A top). Specifically, by simultaneously deleting ALG9, which leads to accumulation of M6.2 sugars, and then artificially bypassing the next step by over-expression of the ALG12 mannosyltransferase, similar levels of M6.2 sugars and M7.1 sugars which contains a terminal α1,6-linked mannose are transferred to proteins (Figure 3A bottom) (Burda et al., 1999).
Figure 3. Production of Man7GlcNac2 sugars in vivo results in ERAD dependent degradation and bypass of HTM1.
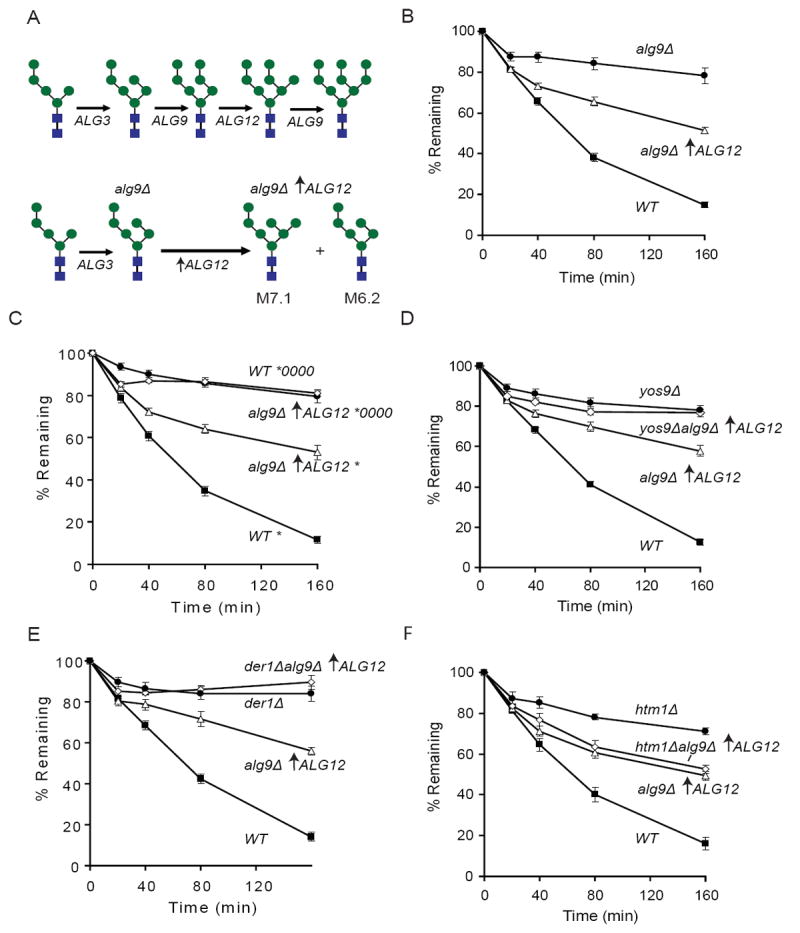
(A) Schematic representation of a portion of the asparagine linked glycosylation (ALG) pathway. Shown are the glycans produced in the wild-type (top) pathway and an alg9Δ over-expressing ALG12 (upward arrow indicates TDH3 driven expression) strain (bottom). Mannose residues are represented as blue circles and N-acetylglucosamine is represented by blue squares.
(B) Degradation of CPY* in a (■) wild-type (WT), (△) alg9↑ALG12Δ, and (●) alg9Δ strains in this and the following panels were monitored as in Figure 1A except that each time point represents the average and +/- SEM of at least 8 measurements (4 independent experiments done in duplicate).
(C) Degradation of CPY* in (■) wild-type (WT) and (△) alg9Δ↑ALG12 (upward arrow represents TDH3 driven expression), or CPY*0000 in (●) wild-type (WT) and (◇) alg9Δ↑ALG12 cells. CPY* is represented as a * and non-glycosylatable CPY* is represented as *0000.
(D) Degradation of CPY* in (■) wild-type (WT), (△) alg9Δ↑ALG12, (◇) yos9Δalg9Δ↑ALG12 and (●) yos9Δ cells.
(E) Degradation of CPY* in (■) wild-type (WT), (△) alg9Δ↑ALG12, (◇) der1Δalg9Δ↑ALG12 and (●) der1Δcells.
(F) Degradation of CPY* in (■) wild-type (WT), (△) alg9Δ↑ALG12, (◇) htm1Δalg9Δ↑ALG12 and (●) htm1Δ cells.
Analysis of ERAD-L in the alg9Δ /ALG12 over-expression strain strongly supports the proposal that Man7.1 is productively recognized by Yos9p. As seen previously, deletion of alg9 which produces M6.2 sugars that lack a terminal α1,6-linked mannose, results in stabilization of CPY* (Figure 3B and S3) (Jakob et al., 1998). By contrast, deletion of alg9 together with overexpression of ALG12 results in degradation of approximately fifty percent of CPY* (Figure 3B and S3). This result is consistent with the ratio of M6.2 and M7.1 sugars produced in the cell strongly suggesting that proteins with M7.1 are being subject to ERAD-L. Importantly, CPY* degradation in the alg9Δ/ALG12 over-expression strain is dependent on presence of substrate glycans (Figure 3C), Yos9p (Figure 3D), and Der1p, another member of the Hrd1p complex (Figure 3E) (Gauss et al., 2006b; Knop et al., 1996a), indicating that degradation in this background goes through the classic ERAD-L pathway (Huyer et al., 2004; Kanehara et al., 2007; Kostova and Wolf, 2005; Spear and Ng, 2005; Vashist and Ng, 2004). Strikingly, in contrast to wild-type cells, we found that in the alg9Δ/ALG12 over-expression background, Htm1p is dispensable for degradation (Figure 3F). An exposed α1,6-linked mannose on a second ERAD-L substrate, KHN, (Vashist et al., 2001; Vashist and Ng, 2004) also bypasses the need for Htm1p (Figure S4). Thus the presence of the M7.1 signal circumvents the requirement for Htm1p without bypassing the need for later-acting components like Yos9p and Der1p that are involved in reading out the signal and the ensuing steps leading to substrate degradation.
Discussion
Here we revealed the sugar-binding specificity of Yos9p, which together with functional studies, supports a model in which two key lectins, Htm1p and Yos9p, cooperate to enhance the specificity of the ERAD-L degradation system (Figure 4). This model builds on the previous observation that degradation by the Hrd1/Hrd3/Yos9 ubiquitin ligase complex requires a bipartite recognition of substrates involving both recognition of misfolded domains and glycans (Denic et al., 2006; Gauss et al., 2006a). Specifically, Hrd3p recruits potential substrates to the Hrd1p ligase complex based on the presence of misfolded domains. Yos9p then queries the glycans for what we have determined to be Yos9p’s preferred oligosaccharide binding specificity, a terminal α1,6-linked mannose, through the action of Htm1p. Upon identification of an appropriate glycan signal, Yos9p commits the substrate for degradation. This model also provides an explanation for the previously enigmatic observation that ERAD-L occurs in alg3Δ strains even though several ALG mutants with less severe defects in the biosynthesis of N-linked glycans abrogate ERAD-L (Jakob et al., 1998), as the Man5GlcNac2 sugar produced in an alg3Δ strain also contains a terminal α1,6-linked mannose. Consistent with this, degradation of CPY* in the alg3Δ background does not require Htm1p (M. Aebi, personal communication and Figure S4).
Figure 4. Model of dual recognition of substrates by ERAD.
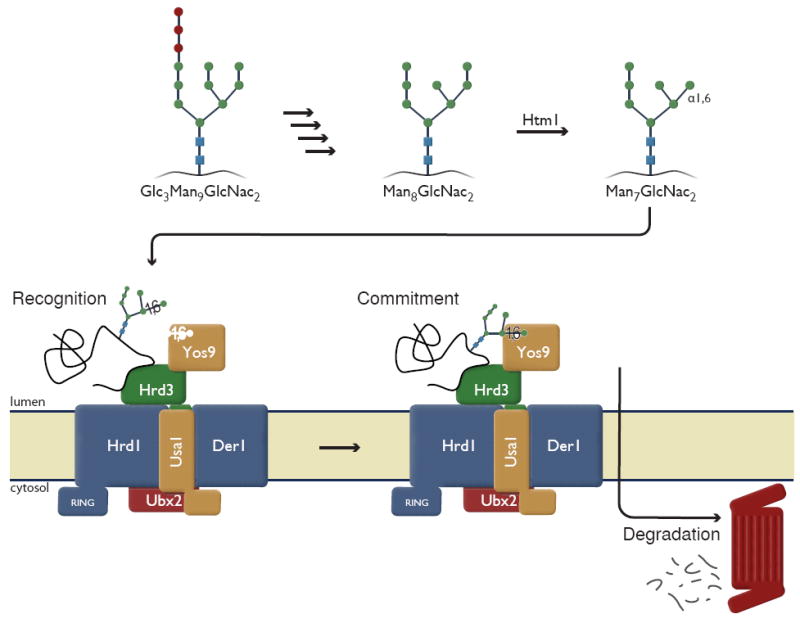
Glycan processing from the initial N-linked Glc3Man9GlcNac2 to Man8GlcNac2 occurs by Glucosidase I, II and Manosidase I, respectively. Htm1p marks potential substrates by playing a role in the generation of Man7GlcNac2 (upper panel). Misfolded proteins are recruited to the Hrd1p complex by recognition of misfolded domains by Hrd3p. Yos9p queries the N-linked glycan and substrates are committed for degradation after Yos9p has identified the presence of a terminal α1,6-linked mannose. Note: Whether Htm1p is an enzyme or cofactor remains to be determined.
While the enzymatic activity of Htm1p has not been directly examined, several observations suggest that Htm1p is required for generating the Man7GlcNac2 signal (either as an enzyme or as a cofactor) that is recognized by Yos9p. Jakob and colleagues reported that deletion of Htm1p results in reduced CPY*-Yos9ps interaction (Szathmary et al., 2005). Htm1p is homologous to α 1,2-mannosidases but lacks conserved cysteine residues that potentially contribute to enzymatic activity (Jakob et al., 2001; Lipari and Herscovics, 1996; Nakatsukasa et al., 2001). However, over-expression of the human Htm1p homologues, EDEM 1 and EDEM 3, which also do not contain the cysteine residues, leads to demannosylation suggesting that they are active mannosidases (Hirao et al., 2006; Olivari et al., 2006). Additionally, Aebi and coworkers (personal communication) find that overexpression of Htm1p results in increased production of protein-bound Man7GlcNac2 oligosaccharide. Finally, we show that a yeast strain engineered to produce Man7GlcNac2 glycans as the starting sugar bypasses the requirement for Htm1p in Yos9p-mediated degradation of CPY*.
How might the requirement for the generation and recognition of a specific degradation glycan increase ERAD-L specificity? One possibility is that Htm1p acts as “timer,” which acts independently of folding status of its substrate thereby providing all polypeptides with a protected window of time in which they can fold without risk of destruction (Helenius, 1994; Jakob et al., 1998; Wu et al., 2003). It is also possible that Htm1p is more sophisticated and that the presence of specific misfolded structures determines whether a substrate is marked by Htm1p. A precedent for such a mechanism is provided by the mammalian UDP-Glc:glycoprotein glucosyltransferase, which adds a glucose only to glycans proximal to a misfolded domain (Ritter and Helenius, 2000; Ritter et al., 2005; Trombetta and Helenius, 2000; Trombetta et al., 1991). Htm1p is also in a complex with Pdi1p whose chaperone activity could confer specificity based on substrate structure (Collins et al., 2007; Krogan et al., 2006). Either way, this modification of sugars adds another level of surveillance to the bipartate recognition of misfolded domains and sugar status by Hrd3p/Yos9p (Denic et al., 2006; Gauss et al., 2006a). The use of multiple query steps separated by irreversible steps would allow for a kinetic proofreading mechanism (Hopfield, 1974) ensuring enhanced specificity in the recruitment of misfolded proteins. In light of this model, it is intriguing that Yos9p shows its highest affinity for a specific Man5GlcNac2 species that is missing the two A- branch mannose residues, as this species has been observed in mammals (Avezov et al., 2008; Frenkel et al., 2003; Hosokawa et al., 2003; Kitzmuller et al., 2003; Lederkremer and Glickman, 2005). While it remains to be seen how or even if this species can be generated through additional sugar trimming in yeast, the enhanced affinity of Yos9p for the Man5GlcNac2 (M5.1) sugar species could provide a mechanism for preferentially degrading a subset of potential substrates.
On a practical note, a more sophisticated understanding of what constitutes a good ERAD substrate should enable a range of biochemical and structural studies to elucidate substrate recognition. In light of the multi-step nature of marking and decoding of ERAD-L substrates, it is perhaps not surprising that in vitro reconstitution of this process has proven so challenging. The discovery that substrate glycans containing a terminal α1,6-linked mannose such as Man7GlcNac2 are recognized by Yos9p should facilitate efforts to create a synthetic substrate with the correct glycan signal attached, thus bypassing the complicated upstream trimming steps. Furthermore, it should now be possible to monitor the specific recognition steps of ERAD-L in vitro. Several immediate questions emerge: How is Htm1p selecting its substrates? Are Yos9p and Hrd3p querying distinct structural features or are they simply double checking Htm1p’s decisions? What is the mechanism of the commitment step once Yos9p confirms that the glycan is correct? Addressing these and related issues will provide a detailed molecular understanding of how and when the cell decides to commit ER proteins for destruction.
Experimental Procedures
Yeast Strains and Plasmids
All yeast strains are derivatives of S288c. Gene deletions, epitope taggings and promoter insertions were done using standard PCR based techniques. Details are available in the Supplemental Data section.
Cycloheximide Degradation Assays
Cycloheximide chase degradation assays were performed as previously described (Denic et al., 2006) with the exception that bands were visualized and quantitated using the LI-COR Odyssey system using an area of the blot with no specific signal as background. Following normalization to the hexokinase loading control, the values were plotted as averages ± standard error of the mean (SEM) with timepoint 0 set to 100%.
Yos9 Purification and Refolding
HIS-tagged Yos9p or Yos9 R200A was purified from Rosetta (DE3) pLysS inclusion bodies, solubilized in 8M urea and purified over a Ni-NTA agarose and a Source Q column. The protein was then refolded into 100mM Tris-HCL (pH 8.5) 150mM NaCl, 1mM CaCl2, 0.5M L-arginine, 5mM GSH, and 0.5mM GSSG at 4°C for 24 hours and subsequently purified over a Resource Q column before being buffer exchanged into 10mM HEPES, 1mM Cacl2, 10% glycerol, and 150mM NaCl (pH 7.4). Further details are given in the Supplemental Data.
Frontal affinity chromatography
FAC analyses were carried out as previously described (Kamiya et al., 2008; Kamiya et al., 2005; Kasai, 1986). Details given in the Supplemental Data.
Circular dichroism spectroscopy
Circular dichroism (CD) measurements were conducted on a Jasco J-725 spectropolarimeter (Jasco Inc., Japan) at room temperature using samples containing 0.05 mg/ml of Yos9p or its R200A mutant in 0.1 M sodium phosphate, at pH 7.4. Each spectrum was recorded as the average of four scans over the range 195 -250 nm with a step size of 0.1 nm and a bandwidth of 1.0 nm.
Supplementary Material
Acknowledgments
We thank V. Van Voorhis for help with the initial observations of the M7.1 glycan CPY* degradation; C. Chu for help with the protein production strategy; M. Bassik, D. Breslow, and N. Ingolia for helpful discussions and critical reading of the manuscript; M. Aebi, D. Ng, and K. Nagata for communicating results prior to publication; D. Ng for reagents; B. Toyama for help with graphics. We are grateful for support from the National Science Foundation (EQ), an EMBO long-term fellowship (1201-2006) (JW), an HFSP long-term fellowship (LT00821/2007- L) (JW), the Howard Hughes Medical Institute (JSW) and Grant-in-aid for Scientific Research from Ministry of Education, Culture, Sports, Science, and Technology of Japan (KK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anelli T, Sitia R. Protein quality control in the early secretory pathway. Embo J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic Reticulum (ER) Mannosidase I Is Compartmentalized and Required for N-Glycan Trimming to Man5 6GlcNAc2 in Glycoprotein ER-associated Degradation. Mol Biol Cell. 2008;19:216–225. doi: 10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Burda P, Jakob CA, Beinhauer J, Hegemann JH, Aebi M. Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology. 1999;9:617–625. doi: 10.1093/glycob/9.6.617. [DOI] [PubMed] [Google Scholar]
- Buschhorn BA, Kostova Z, Medicherla B, Wolf DH. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 2004;577:422–426. doi: 10.1016/j.febslet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zuniga M, Brown PO, Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as an ubiquitin-ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Drumm ML, Wilkinson DJ, Smit LS, Worrell RT, Strong TV, Frizzell RA, Dawson DC, Collins FS. Chloride conductance expressed by delta F508 and other mutant CFTRs in Xenopus oocytes. Science. 1991;254:1797–1799. doi: 10.1126/science.1722350. [DOI] [PubMed] [Google Scholar]
- Fewell SW, Travers KJ, Weissman JS, Brodsky JL. The action of molecular chaperones in the early secretory pathway. Annual Review of Genetics. 2001;35 doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Frenkel Z, Gregory W, Kornfeld S, Lederkremer GZ. Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6-5GlcNAc2. J Biol Chem. 2003;278:34119–34124. doi: 10.1074/jbc.M305929200. [DOI] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006a;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. Embo J. 2006b;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Herscovics A. Structure and function of Class I alpha 1,2-mannosidases involved in glycoprotein synthesis and endoplasmic reticulum quality control. Biochimie. 2001;83:757–762. doi: 10.1016/s0300-9084(01)01319-0. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J, Arata Y, Kasai K. Frontal affinity chromatography as a tool for elucidation of sugar recognition properties of lectins. Methods Enzymol. 2003;362:353–368. doi: 10.1016/s0076-6879(03)01025-5. [DOI] [PubMed] [Google Scholar]
- Hirao K, Natsuka Y, Tamura T, Wada I, Morito D, Natsuka S, Romero P, Sleno B, Tremblay LO, Herscovics A, et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- Hitt R, Wolf DH. DER7, encoding alpha-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res. 2004;4:815–820. doi: 10.1016/j.femsyr.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1- antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Jakob CA, Bodmer D, Spirig U, Battig P, Marcil A, Dignard D, Bergeron JJ, Thomas DY, Aebi M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001;2:423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, Roth J, Aebi M. Degradation of mis-folded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Kamiya Y, Kamiya D, Yamamoto K, Nyfeler B, Hauri HP, Kato K. Molecular basis of sugar recognition by the human L-type lectins ERGIC-53, VIPL, and VIP36. J Biol Chem. 2008;283:1857–1861. doi: 10.1074/jbc.M709384200. [DOI] [PubMed] [Google Scholar]
- Kamiya Y, Yamaguchi Y, Takahashi N, Arata Y, Kasai K, Ihara Y, Matsuo I, Ito Y, Yamamoto K, Kato K. Sugar-binding properties of VIP36, an intracellular animal lectin operating as a cargo receptor. J Biol Chem. 2005;280:37178–37182. doi: 10.1074/jbc.M505757200. [DOI] [PubMed] [Google Scholar]
- Kanehara K, Kawaguchi S, Ng DT. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol. 2007;18:743–750. doi: 10.1016/j.semcdb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Kasai K, Oda Y, Nishikawa M, Ishii S. Frontal affinity chromatoraphy: theory for its application to studies on specofoc interactions of biomolecules. J Chromatogr. 1986;376:33–47. doi: 10.1016/s0378-4347(00)80822-1. [DOI] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Kitzmuller C, Caprini A, Moore SE, Frenoy JP, Schwaiger E, Kellermann O, Ivessa NE, Ermonval M. Processing of N-linked glycans during endoplasmic-reticulumassociated degradation of a short-lived variant of ribophorin I. Biochem J. 2003;376:687–696. doi: 10.1042/BJ20030887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. Embo J. 1996a;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop M, Hauser N, Wolf DH. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast. 1996b;12:1229–1238. doi: 10.1002/(sici)1097-0061(19960930)12:12<1229::aid-yea15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kostova Z, Wolf DH. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci. 2005;118:1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ, Glickman MH. A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Lipari F, Herscovics A. Role of the cysteine residues in the alpha1,2-mannosidase involved in N-glycan biosynthesis in Saccharomyces cerevisiae. The conserved Cys340 and Cys385 residues form an essential disulfide bond. J Biol Chem. 1996;271:27615–27622. doi: 10.1074/jbc.271.44.27615. [DOI] [PubMed] [Google Scholar]
- Munro S. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr Biol. 2001;11:R499–501. doi: 10.1016/s0960-9822(01)00302-5. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Nishikawa S, Hosokawa N, Nagata K, Endo T. Mnl1p, an alpha -mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J Biol Chem. 2001;276:8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- Olivari S, Cali T, Salo KE, Paganetti P, Ruddock LW, Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of foldingdefective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112(Pt 22):4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- Ritter C, Helenius A. Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat Struct Biol. 2000;7:278–280. doi: 10.1038/74035. [DOI] [PubMed] [Google Scholar]
- Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. Embo J. 2005;24:1730–1738. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Spear ED, Ng DT. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Helenius A. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J Cell Biol. 2000;148:1123–1129. doi: 10.1083/jcb.148.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta SE, Ganan SA, Parodi AJ. The UDP-Glc:glycoprotein glucosyltransferase is a soluble protein of the endoplasmic reticulum. Glycobiology. 1991;1:155–161. doi: 10.1093/glycob/1.2.155. [DOI] [PubMed] [Google Scholar]
- Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitinproteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wu Y, Swulius MT, Moremen KW, Sifers RN. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc Natl Acad Sci U S A. 2003;100:8229–8234. doi: 10.1073/pnas.1430537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


