Abstract
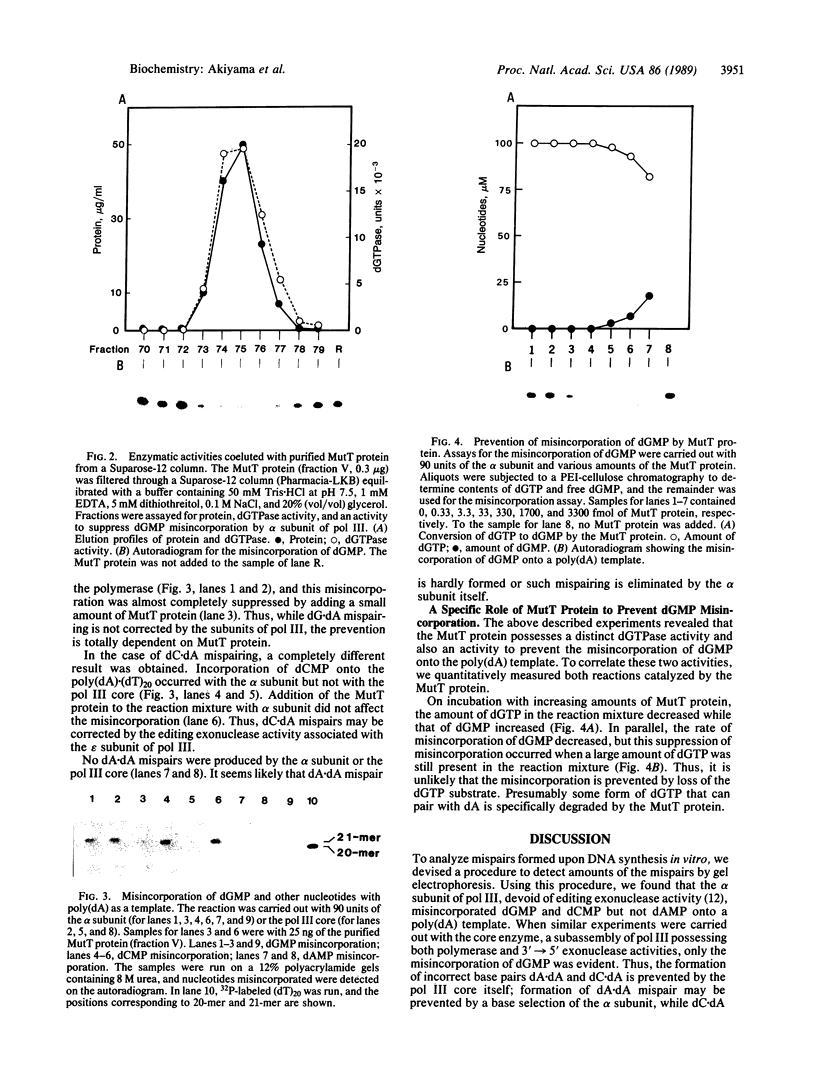
Occurrence of the transversion mutation A.T to C.G is specifically enhanced in Escherichia coli mutT mutants. With the aid of the cloned mutT gene, the MutT protein, which has a molecular mass of 15 kilodaltons, was overproduced and purified to near homogeneity. The protein catalyzes hydrolysis of dGTP to dGMP. dGDP and GTP were also hydrolyzed by the protein, but at a lower rate than seen with dGTP. No other deoxynucleoside triphosphates were hydrolyzed. Using poly(dA).(dT)20 as a template-primer, we investigated the misincorporation of dGMP, dCMP, and dAMP by the alpha subunit and the core of E. coli DNA polymerase III. When the polymerization reaction was performed with the alpha subunit, both dCMP and dGMP were misincorporated. The core, composed of alpha, epsilon, and theta subunits, misincorporated only dGMP. This would imply that the proofreading function of the epsilon subunit of DNA polymerase III may correct the dC.dA mispair but not the dG.dA mispair. Misincorporation of dAMP was not observed in reactions with the alpha subunit or core. The misincorporation of dGMP, but not dCMP, was almost completely suppressed by adding purified MutT protein to the reaction mixture. Under these conditions, only a portion of dGTP present in the reaction mixture was degraded. It is therefore likely that the MutT protein may prevent dGMP misincorporation by degrading a specific form of dGTP, probably the syn form, which can pair with deoxyadenosine.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama M., Horiuchi T., Sekiguchi M. Molecular cloning and nucleotide sequence of the mutT mutator of Escherichia coli that causes A:T to C:G transversion. Mol Gen Genet. 1987 Jan;206(1):9–16. doi: 10.1007/BF00326530. [DOI] [PubMed] [Google Scholar]
- Bhatnagar S. K., Bessman M. J. Studies on the mutator gene, mutT of Escherichia coli. Molecular cloning of the gene, purification of the gene product, and identification of a novel nucleoside triphosphatase. J Biol Chem. 1988 Jun 25;263(18):8953–8957. [PubMed] [Google Scholar]
- Conrad S. E., Dussik K. T., Siegel E. C. Bacteriophage Mu-1-induced mutation to mutT in Escherichia coli. J Bacteriol. 1976 Mar;125(3):1018–1023. doi: 10.1128/jb.125.3.1018-1023.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Cox E. C. Mutator gene studies in Escherichia coli: the mutT gene. Genetics. 1973 Apr;73(Suppl):67–80. [PubMed] [Google Scholar]
- Cox E. C., Yanofsky C. Mutator gene studies in Escherichia coli. J Bacteriol. 1969 Oct;100(1):390–397. doi: 10.1128/jb.100.1.390-397.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. DNA polymerase accuracy and spontaneous mutation rates: frequencies of purine.purine, purine.pyrimidine, and pyrimidine.pyrimidine mismatches during DNA replication. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4251–4255. doi: 10.1073/pnas.78.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler R. G., Schaaper R. M., Glickman B. W. Characterization of mutational specificity within the lacI gene for a mutD5 mutator strain of Escherichia coli defective in 3'----5' exonuclease (proofreading) activity. J Bacteriol. 1986 Jul;167(1):130–137. doi: 10.1128/jb.167.1.130-137.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Kunkel T. A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988 Sep 9;54(6):805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- Maki H., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985 Oct 25;260(24):12987–12992. [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Nghiem Y., Cabrera M., Cupples C. G., Miller J. H. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Escherichia coli mutT mutator effect during in vitro DNA synthesis. Enhanced A.G replicational errors. J Biol Chem. 1987 Dec 5;262(34):16267–16270. [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane D. L., Goodman M. F., Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988 Jul 25;16(14A):6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- Treffers H. P., Spinelli V., Belser N. O. A Factor (or Mutator Gene) Influencing Mutation Rates in Escherichia Coli. Proc Natl Acad Sci U S A. 1954 Nov;40(11):1064–1071. doi: 10.1073/pnas.40.11.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Cox E. C., Horn V. The unusual mutagenic specificity of an E. Coli mutator gene. Proc Natl Acad Sci U S A. 1966 Feb;55(2):274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]