Abstract
A central component of our ability to navigate an environment is the formation of a memory representation that is allocentric and thus independent of our starting point within that environment. Computational models and rodent electrophysiological recordings suggest a critical role for the CA1 subregion of the hippocampus in this type of coding; however, the hippocampal neural basis of spatial learning in humans remains unclear. We studied subjects learning virtual environments using high-resolution functional magnetic resonance imaging (1.6 mm × 1.6 mm in-plane) and computational unfolding to better visualize substructural changes in neural activity in the hippocampus. We show that the right posterior CA1 subregion is active and positively correlated with performance when subjects learn a spatial environment independent of starting point and direction. Altogether, our results demonstrate that the CA1 subregion is involved in our ability to learn a map-like representation of an environment.
Introduction
Forming an internal representation of an environment underlies our ability to form novel routes and therefore find our way within that environment. The process of linking landmarks and locations within their spatial context is thought to involve the formation of a cognitive map of space (O'Keefe and Nadel, 1978; Nadel and MacDonald, 1980; Kumaran and Maguire, 2005; Tolman, 1948). Electrophysiological recordings during navigation identified place cells in the rodent and human hippocampus that fire preferentially in response to specific locations within an environment (O'Keefe and Dostrovsky, 1971; Wilson and McNaughton, 1993; Ekstrom et al., 2003), supporting the idea that the hippocampus plays an important role in forming spatial representations. The hippocampus comprises CA fields 1, 2, and 3, dentate gyrus, and subiculum. Medial temporal lobe cortices adjacent to the hippocampus include the parahippocampal, entorhinal, and perirhinal cortices. These cortical regions, in addition to the hippocampus, support the general formation of declarative memories (Squire et al., 2004).
Due to the small and convoluted nature of the hippocampus, visualization and localization of activity within human hippocampal subregions is challenging. Previous high-resolution imaging studies have shown CA2, CA3, and dentate gyrus activity increases during encoding of novel face–name and object–object associations, while subiculum activity increases during retrieval of these learned associations (Zeineh et al., 2003; Eldridge et al., 2005). No studies to date, however, in humans have demonstrated activity specific to CA1, a region strongly implicated in memory formation, although bilateral loss of this region does result in memory deficits (Zola-Morgan et al., 1986). Furthermore, these high-resolution functional magnetic resonance imaging (fMRI) methods have yet to be applied to a spatial learning paradigm involving virtual reality.
Computational theories of hippocampal involvement in learning and memory suggest that different cortical regions and hippocampal CA fields subserve distinct roles. Specifically, CA3 may be involved in pattern separation processes necessary for encoding and CA1 in updating or integrating these memories with novel information entering via the entorhinal cortex (Levy, 1989; Vinogradova, 2001; Lee et al., 2004; Bakker et al., 2008; Goodrich-Hunsaker et al., 2008). The CA1 region of the hippocampus receives input from both the CA3 pyramidal cells and the entorhinal cortex (Witter et al., 2000). Encoding spatial information into a cognitive map would involve the integration of information about novel routes with that of previously learned routes to a particular learned location in space. We hypothesized a central role for CA1 in this type of spatial learning in humans. In this study, we use high-resolution (1.6 × 1.6 mm in-plane) fMRI to measure activation in the medial temporal lobe while subjects learned to locate landmarks in a virtual environment from multiple start points. In a second task, subjects learned to locate landmarks using a repeated start point. We hypothesized that learning to navigate from novel starting points would engage CA1 in that it requires the integration of novel and previously learned information. In contrast, we hypothesized that learning locations relative to a repeated start point would activate medial temporal lobe structures but would result in less activation in CA1 than learning locations from multiple starting points.
Materials and Methods
Subjects.
Eighteen right-handed, healthy subjects (nine male, nine female) between the ages of 20 and 31 (24.89 ± 0.72 years) provided informed consent and participated in the experiment. We recruited 19 subjects but dropped one who was unable to complete the study. Studies were performed under University of California, Los Angeles Institutional Review Board testing protocols.
Experimental design.
Subjects were navigated through novel spatial environments using single starting points (SSP task) and multiple starting points (MSP task) in which they learned various store locations (Fig. 1). Before each task, subjects were given an alternate version of each task while in the scanner (different stores and locations) to familiarize them with the 4 × 3 layout of the city. Both tasks involved passive viewing of previously recorded navigation videos through continuously refreshed (60 Hz) virtual reality environments each in a 4 × 3 grid design (Fig. 1), surrounded by a wall and containing stores (which subjects were driven to), buildings (which served to maintain the “block” structure of the cities), and roads (on which driving occurred). To create the 4 × 3 grid design of the virtual cities, the same building was used repeatedly across the grid where no stores are located; subjects were never taken to any of these buildings.
Figure 1.
Virtual city snapshots. A, Snapshot of virtual city from a sample starting point. B, Sample store stimulus used. C, Subjects' learned store locations within 4 × 3 grid cities from a varying initial starting point within a city for the MSP encoding condition (blue arrows) and from a single initial starting point within the city for the SSP encoding condition (red arrows). D, Layout of city without stores used in the direction-pressing control baseline condition. Both tasks used this control condition. E, Both the SSP and MSP tasks consisted of alternating blocks of encoding (Learn) and retrieval (Recall) interspersed with blocks of control (Ctl).
During both spatial learning tasks, subjects passively viewed navigation to stores in a blocked design of alternating encoding, control, and retrieval blocks (Fig. 1E). Encoding blocks consisted of passively viewing navigation to novel stores (Fig. 1B); store locations were repeated across encoding blocks. Retrieval blocks consisted of passively viewing navigation to old previously learned (target) or novel (lure) store locations (same store stimuli were used), where subjects were instructed to determine whether the stores were in old or new locations within the city and respond by pressing one of two assigned buttons. Subjects learned and recalled 12 store locations (12 trials) in each task during a scan time of 477 s for each task. In the direction-pressing control condition for both tasks (Fig. 1D), subjects passively viewed navigation through identical cities but without stores (just buildings and roads), and were instructed to press the corresponding button on the keypad every time the direction was changed (left and right). This baseline task was chosen over typical rest tasks for its higher demand on non-mnemonic cognitive processes. It has previously been shown that using a simple rest task activates the hippocampus and thus provides a nonoptimal baseline level of activity (Stark and Squire, 2001). Because we were solely interested in the mnemonic processes associated with our tasks, we chose a baseline task that differed in only this respect. SSP and MSP spatial learning tasks were matched in aspects including experimental design (Fig. 1E), baseline condition (Fig. 1D), virtual city layout (Fig. 1C), length and timing of navigation videos, and number of stores driven to. Alternate stores were used in each task and task order was counterbalanced across subjects.
What differed between spatial learning tasks were the instructions and starting points used within and between encoding and retrieval. For encoding (study) during the SSP task, subjects were instructed to learn store locations from an initial starting point within the city; the starting point was repeated across learning trials (Fig. 1C, red arrows). Instructions during retrieval (test) were the same for both tasks (“Is the store in the same location? Press ‘1’ for yes and ‘2’ for no.”). During SSP retrieval, subjects' starting point and route were also repeated across retrieval trials. The same stores were used in encoding and retrieval, although locations were changed for lure trials. In the SSP version, study and test both relied on the same starting location and route used in the city. For the MSP task, subjects were instructed to learn store locations relative to other stores; the starting point within the city varied across trials during encoding blocks as well as between encoding and retrieval blocks (Fig. 1C, blue arrows). Therefore, subjects could not learn locations relative to the starting point. Study and test both rely on learning locations from multiple starting points; starting points and routes presented during retrieval differed with respect to encoding, and therefore subjects could not depend on previously presented routes for learning. All subjects completed both the SSP and MSP experimental tasks; order was counterbalanced across subjects.
Navigation videos were displayed and recorded using pyepl (http://pyepl.sourceforge.net/), Snapz Pro X (Ambrosia software), and an adapted version of yellowcab2 (with buildings, city layout changed, and passengers removed; original download http://memory.psych.upenn.edu/Software). Stimuli were edited using iMovie (Apple) and presented using MacStim 3.2.1 software (WhiteAnt Occasional Publishing).
Imaging procedure.
Subjects were scanned with a Siemens Allegra head-only 3 tesla scanner at the Ahmanson-Lovelace Brain Mapping Center at the University of California, Los Angeles. High in-plane resolution structural images with a matrix size of 512 × 512 [spin echo, repetition time (TR) = 5200 ms, echo time (TE) = 105 ms, 19 slices, contiguous; voxel size: 0.391 × 0.391 × 3 mm] and echo-planar images (TR = 3000 ms, TE = 39 ms, 128 × 128, 19 slices, contiguous; voxel size = 1.6 × 1.6 × 3 mm) were acquired in the same oblique coronal plane and registered using a matched-bandwidth sequence (TR = 5000 ms, TE = 66 ms, 19 slices, contiguous; voxel size = 1.6 × 1.6 × 3 mm). Two volumes were introduced at the beginning of each functional scan to allow equilibration to steady state and were subsequently excluded from the analysis. The coronal plane was chosen because the structures are relatively homogeneous along the long axis but differ in-plane; thus, we maximized in-plane resolution. For all scans, 19 slices were acquired perpendicular to the long axis of the hippocampus during acquisition. See Zeineh et al. (2000) for further details on method. Visual stimuli were presented to the subject using 512 × 512 resolution magnet-compatible three-dimensional (3-D) goggles and headphones under computer control (Resonance Technologies). The stimuli were presented using a Macintosh G4 Powerbook computer, and key presses were recorded for behavioral analysis.
Imaging and statistical analysis.
fMRI analysis was conducted using FEAT (fMRI Expert Analysis Tool), part of FMRIB Software Analysis (FSL version 3.3, www.fmrib.ox.ac.uk/fsl), to investigate differences in overall activity in contrasts of interest. Functional volumes were motion corrected to the median volume using MCFLIRT (Jenkinson et al., 2002) (FMRIB's motion correction linear image registration tool) using a normalized correlation ratio cost function and linear interpolation. Brains were skull stripped using BET (brain extraction tool) (Smith, 2002). Images were spatially smoothed using a Gaussian kernel of full-width at half-maximum 3 mm and intensity normalized, and a high-pass temporal filter (Gaussian-weighted least-squares straight line fitting, with σ = 100.0 s) was applied. Time-series statistical analysis was performed using FILM (FMRIB's improved linear model) with local autocorrelation correction (Woolrich et al., 2001). Regressors of interest were created by convolving a delta function representing trial onset times with a canonical (gamma) hemodynamic response function, along with their temporal derivatives. Contrasts were cluster corrected at Z > 2.3 and p < 0.05. Functional images were aligned using FMRIB's Linear Image Registration Tool to high-resolution coplanar images via an affine transformation with 6 degrees of freedom. The high-resolution coplanar images were then aligned to the subject's high-resolution structural images using an affine transformation with 6 degrees of freedom.
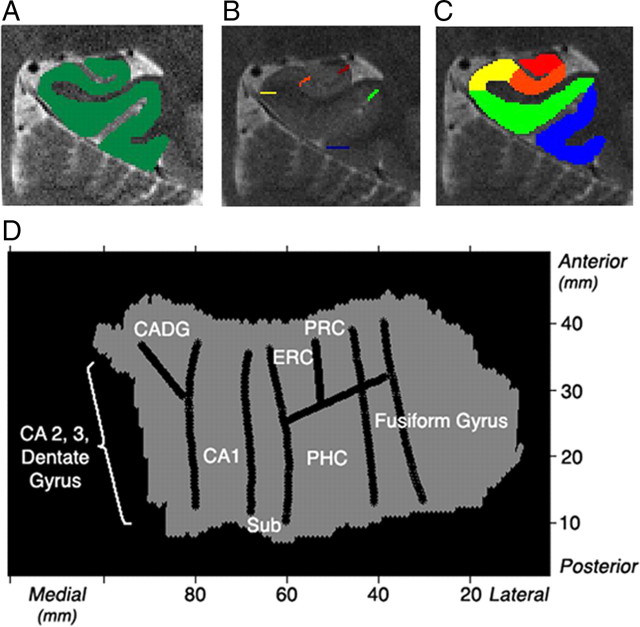
For those slices selected for functional scanning, the 3-D gray matter of the MTL subregions were created (Fig. 2A) by manually segmenting the high-resolution structural images into white matter and CSF using mrGray segmentation software (Teo et al., 1997). The gray matter was then computationally unfolded using an iterative algorithm based on metric multidimensional scaling using mrUnfold software and interpolated by a factor of 7 to improve segmentation along the long axis of the hippocampus (Engel et al., 1997) (mrGray and mrUnfold download: http://white.stanford.edu/∼brian/mri/segmentUnfold.htm), yielding a final voxel size of 0.391 × 0.391 × 0.429 mm (Fig. 2D). The position of the various CA fields, subiculum, entorhinal cortex (ERC), perirhinal cortex (PRC), parahippocampal cortex (PHC), and fusiform gyrus on the unfolded map were found by mapping pixels from points demarcated (Fig. 2B) in the structural images based on the atlases by Amaral and Insausti (1990) and Duvernoy (1998). The raw functional images were aligned with the structural images using matched-bandwidth images and then onto the unfolded hippocampus using the same transformation parameters (Fig. 2D). The activation maps for the functional images were superimposed onto the structural images for precise localization of effects. In this way, we were able to clearly differentiate activation in the substructures of the hippocampus and the parahippocampal gyrus. Anatomical regions of interest (ROIs) were created a priori by defining voxels in 2-D space and then projected into 3-D space (Fig. 2C). These ROIs included anterior and posterior CA2,3 and dentate gyrus (CA23DG), anterior and posterior CA1, anterior and posterior subiculum, ERC, PRC, PHC, and fusiform gyrus. With our current human imaging methods, the dentate gyrus is not distinguishable from adjacent CA fields and therefore grouped in an encompassing region labeled CA23DG (but see Ekstrom et al., 2009). The average percentage signal change was computed in FSL for each ROI from average parameter estimates using the height of an isolated event as the scaling factor, and is relative to the voxel mean (see http://mumford.bol.ucla.edu/perchange_guide.pdf). Correlationanalysis was done by calculating the Spearman rank coefficient (p < 0.05, corrected) between percentage signal change and subjects' average behavioral performance (percentage correct) on the task. We chose the Spearman rank correlation coefficient, rather than the Pearson ρ correlation, because we did not assume a linear correlation between metrics but rather a monotonically increasing relationship between the variables. Conducting the analysis using the Pearson ρ correlation, however, did not change our overall results.
Figure 2.
Unfolding method. A, B, Each subject's gray matter (green) is created by segmenting white matter and CSF. The gray matter is then computationally unfolded (A) and boundaries between regions are projected onto the unfolded flat map (B). C, Voxels in 2-D space are projected into 3-D space to create anatomical regions of interest showing posterior regions (left): CA23DG (red), CA1 (orange), subiculum (yellow), PHC (green), and fusiform gyrus (blue). D, An averaged group flat map (shown is the left) is created showing regions CA2, 3, and dentate gyrus, CA1, subiculum (Sub), ERC, PRC, PHC, and fusiform gyrus.
A group 2-D hippocampal template shown in Figure 3 was created based on the 18 individual subject anatomical images and boundaries. Each subject's individual anatomical and time functional activations were warped into the template. Group activation maps were produced by averaging individual subregional boundaries and then transforming each subject's flat map to this template. The degree of fit between each individual subject's behavior and fMRI signal (e.g., β values), based on model contrasts discussed earlier, were then compared across subjects for each voxel using a mixed-effects t test (t ≥ 2.4; p < 0.05, corrected). See Thompson et al. (2000) and Zeineh et al. (2001) for details on these methods. Correction for multiple comparisons for single-subject contrasts were cluster corrected at Z > 2.3 and p < 0.05. Group activation maps were corrected using a Bonferroni correction for 10 ROIs (p < 0.005). Spearman rank correlation analysis was corrected for multiple comparisons using a Bonferroni correction (p < 0.025).
Figure 3.
A, Group voxel-based mixed-effects unfolded t test maps (n = 18, statistical maps of significantly activated and deactivated regions; −2.4 ≥ t ≥ 2.4; p < 0.05 corrected) for the left and right MTL regions during the MSP and SSP tasks separately compared with baseline. B, Group voxel-based mixed-effects unfolded t test maps (n = 18, statistical maps of significantly difference in activity between MSP and SSP conditions; −2.4 ≥ t ≥ 2.4; p < 0.05 corrected) for the left and right MTL regions. Regions shown include CA2, 3, and dentate gyrus, CA1, subiculum (Sub), ERC, PRC, PHC, and fusiform gyrus.
Results
Subjects' average performance and learning rates on the SSP and MSP spatial tasks did not differ significantly (supplemental Fig. S1, available at www.jneurosci.org as supplemental material) (n = 18; percentage correct: mean ± SEM, SSP, 73.33 ± 3.89; MSP, 68.33 ± 5.18; t(17) = 0.85; p = 0.41).
The focus of this paper was to determine hippocampal subregion activity during the processing of spatial information, specifically during encoding. To investigate changes during encoding of store locations, activity during encoding stimuli from both tasks were analyzed separately during periods of navigation without stores (where subjects were navigated through a city layout that lacked store landmarks) (Fig. 1A) and periods of navigation with stores (where stores were present in the city and subjects could make store–place associations) (Fig. 1B). As expected, no significant increases were found during navigation without stores during encoding blocks compared with navigation during the direction-pressing control blocks (baseline). However, in both spatial tasks activity during navigation with stores showed significant increases compared with the control condition across various MTL subregions bilaterally, including the parahippocampal gyrus and adjacent fusiform gyrus (Fig. 3A). To determine any significant differences between the MSP and SSP conditions, we directly contrasted encoding blocks from these two conditions. Learning locations from varying compared with a single start point yielded a cluster of significant increase within the right CA1 subregion of the hippocampus (Fig. 3B). No significant clusters were found in any extrahippocampal regions or hippocampal subregions CA23DG and subiculum. Furthermore, the reverse contrast yielded no areas of significant difference in activity within the MTL (for example, activation data superimposed on an oblique-coronal high-resolution anatomical image) (see supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
In addition to the voxelwise analysis performed above, we completed an independent analysis based solely on anatomical ROIs. ROIs were determined independent of functional activity, and average percentage signal change was then calculated for each region. These ROIs were based on the anatomical definitions of CA23DG, anterior and posterior CA1, anterior and posterior subiculum, ERC, PRC, PHC, and fusiform gyrus. Average percentage signal change compared with baseline within hippocampal regions is shown in Figure 4. For the CA1 and subiculum, the posterior anatomical ROIs are shown; anterior regions yielded no significant differences between conditions or hemispheres. Based on the a priori hypothesis that CA1 activity would differ between learning conditions, we performed a 2 × 2 hemisphere (right vs left) × condition (MSP vs SSP) ANOVA in posterior CA1, which yielded a significant main effect of hemisphere (F(2,34) = 7.93; p = 0.006) and significant interaction between hemisphere and condition (F(2,34) = 4.90; p = 0.029). Post hoc comparisons using paired t tests for anatomical regions of interest found that activity within the right posterior CA1 subregion was significantly greater during the MSP compared with SSP encoding (t(17) = 2.36; p = 0.026). Furthermore, activity in the right posterior CA1 during MSP encoding correlated significantly with behavioral performance across subjects (Fig. 5A) (Spearman's ρ = 0.53; p = 0.02; n = 18). Activity in this region during SSP encoding was not significantly correlated with performance (Fig. 5B) (ρ = −0.488; n.s.; n = 18). No significant differences between SSP and MSP encoding conditions were found in the left CA1 subregion (t(17) = −0.93; n.s.), although overall activity was significantly greater in the right hemisphere compared with the left (right > left, t(17) = 3.29; p = 0.004). In contrast to this pattern, activity within the right and left CA23DG region did not differ between learning conditions (SSP > MSP, right, t(17) = 0.81; n.s.; left, t(17) = 0.17; n.s.), nor was there a significant right versus left difference (right > left, t(17) = 1.75; n.s.). Similar to CA23DG, the anterior and posterior subiculum showed no significant differences in condition or hemisphere (right, t(17) = −0.41; n.s.; left, t(17) = −0.33; p = 0.74; right > left, t(17) = 0.98; n.s.).
Figure 4.

Results from hippocampal anatomical regions of interest. Average percentage signal change (n = 18, error bars correspond to the SE across subjects) for the SSP and MSP tasks separately compared with baseline within left and right hippocampal subregions CA23DG (A) and subiculum (C) show no significant differences between MSP and SSP conditions or between right and left hemispheric regions. B, Average percentage signal change from baseline within left and right posterior CA1 for MSP and SSP encoding indicate a significant difference between hemisphere (right > left, t(17) = 3.29; p < 0.05) and conditions (MSP > SSP, t(17) = 2.26; p < 0.05).
Figure 5.
Behavioral performance (percentage correct) versus percentage signal change. A, Subjects' performance on the MSP task significantly correlated with percentage signal change in the right CA1 subregion during encoding (n = 18, Spearman's ρ = 0.53; p < 0.05). B, Subjects' performance on the SSP task did not significantly correlate with percentage signal change in the right CA1 subregion during encoding (n = 18, Spearman's ρ = −0.488; n.s.).
For the adjacent PHC, activation increased bilaterally during both encoding conditions compared with baseline (MSP right, t(17) = 4.83; MSP left, t(17) = 5.27; SSP right, t(17) = 4.00; SSP left, t(17) = 4.80; p < 0.0001) (supplemental Fig. 2C, available at www.jneurosci.org as supplemental material). Similarly we found increases in activity within the fusiform gyrus (MSP right, t(17) = 6.39; MSP left, t(17) = 5.39; SSP left, t(17) = 6.19; SSP right, t(17) = 5.79; p < 0.0001) (supplemental Fig. 2D, available at www.jneurosci.org as supplemental material). However, no significant differences were found in these regions with respect to condition (supplemental Fig. 2C,D, available at www.jneurosci.org as supplemental material). Activity within the entorhinal cortex showed no significant increase from baseline (Fig. 3A; supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). Within the right PRC, activity increased during both MSP and SSP conditions (MSP, t(17) = 2.28; p = 0.03; SSP, t(17) = 3.37; p = 0.003); however no significant differences were found with respect to condition (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material).
Discussion
The present findings demonstrate that while MTL regions such as the PHC are engaged during learning of spatial information, processing of locations from multiple starting points further recruits the hippocampus. We isolated the increase in hippocampal activation to CA1, supporting computational models suggesting a role for CA1 in updating memories with novel information. Furthermore, CA1 activity positively correlated with behavioral performance when navigating from multiple starting points, suggesting that this region supports performance during this type of learning.
Our results are consistent with the idea that the hippocampus plays a critical role in forming a viewpoint-independent map of an environment. Spatial information can be processed using a view-dependent (egocentric) or view-independent (allocentric) frame of reference (Tolman, 1948; Howard, 1982). A key difference between allocentric and egocentric spatial representations is that allocentric representations can support navigation from novel starting points. Studies in rodents have suggested that an allocentric representation of space is formed in the CA1 subfield of hippocampus (Leutgeb et al., 2005; Brun et al., 2008) and place cells in CA1 are able to abruptly shift map representations during an incremental shift in the environment, suggesting these cells code for distinct, orthogonal map representations for different environments (Wilson and McNaughton, 1993; Wills et al., 2005). Lesions of CA1 impair memory for relative placement of landmarks to each other (Goodrich-Hunsaker et al., 2008). Hippocampal activity increases during a spatial memory task using novel starting points (Parslow et al., 2004), and hippocampal damage impairs recall of object–place associations when tested from shifted viewpoints (King et al., 2002). While several studies have suggested that the hippocampus plays a major role in forming an allocentric representation of an environment (Morris et al., 1982; Spiers et al., 2001; Kumaran and Maguire, 2005), these studies were not able to measure the contribution of subregions. Thus, our results provide novel evidence suggesting that CA1 is involved in allocentric spatial computations necessary for spatial navigation.
Various studies of spatial learning have shown that the PHC is preferentially involved during encoding and retrieval of object–place associations (Köhler et al., 2002; Malkova and Mishkin, 2003; Sommer et al., 2005). In these studies the objects are presented in front of the observer on a screen and are likely to be encoded egocentrically. Therefore, we would not expect this condition to necessitate formation of a viewpoint-independent spatial memory representation. In contrast, navigation in a virtual environment from different starting points involves movement of the observer relative to objects and thus requires allocentric encoding. Consistent with prior studies, we show increases in activity within the PHC during SSP and MSP learning conditions. However, using virtual reality to manipulate starting point may further recruit allocentric processing and thus hippocampal involvement. Interestingly, learning a map from an aerial perspective does not result in hippocampal activation, while learning a map through ground-level exploration results in significant hippocampal activation (Shelton and Gabrieli, 2002). It may be that the hippocampus plays an important role in building a flexible map-like memory representation of space from route-level information rather than learning spatial maps per se.
Although we did not find increases in activity during SSP compared with MSP learning, there are likely to be regions outside the MTL, which show increased activity. Previous studies have shown caudate nucleus involvement during route learning from single starting points (Hartley et al., 2003; Iaria et al., 2003). Thus, it is possible the caudate nucleus would show increased activity in SSP compared with MSP learning. In the present study, we restricted our field of view to MTL structures to increase resolution necessary to differentiate hippocampal subregions. Using whole-brain fMRI methods could describe the role of regions outside the MTL in spatial learning from single and multiple start points.
A possible difference between allocentric and egocentric spatial learning is that forming an allocentric map requires a higher memory load resulting from multiple viewpoints (Shrager et al., 2007). Increased CA1 activation may therefore be due to a higher memory load in our MSP encoding condition. However, although subjects learned locations from only one starting point in the SSP task, we used routes that were sufficiently complex to ensure that performance was at the same level as in the MSP condition. Thus, the amount of information in memory needed to support performance was approximately equivalent in the two tasks, at least as assessed by learning rate. During MSP encoding and retrieval subjects approached stores from different directions and locations were learned and recalled relative to other stores rather than relative to a repeated starting location. Integration of previously learned routes in memory with novel routes necessitates the formation of a viewpoint-independent memory representation of the environment. We therefore suggest increase in CA1 activity in the MSP condition reflects the nature of spatial representations learned rather than memory load.
CA1 regions active during the MSP learning condition were in the posterior hippocampus. These findings are consistent with other studies indicating that the posterior hippocampus is activated during spatial learning (Gabrieli et al., 1997; Colombo et al., 1998; Fernández et al., 1998; Maguire et al., 2006). In a recent study, patterns of activity in the posterior hippocampus predicted the location of an individual in a virtual environment (Hassabis et al., 2009). These data suggest that the posterior hippocampus may be particularly involved in mnemonic functions involving spatial representations.
Previous neuroimaging studies using virtual reality navigation found PHC activity in absence of hippocampal activation (Aguirre et al., 1996; Mellet et al., 2000; Rosenbaum et al., 2004). One explanation for hippocampal activity detected in our study may be that subregions within the hippocampus are likely to serve functionally distinct roles and thus averaging across these areas would result in reduced signal. In this study, we separately evaluate individual subregions without losing information during group averaging. Individual hippocampi are segmented and warped to an averaged group template constrained by individual subregional boundaries. In addition to unfolding we used an ROI-based method to calculate percentage signal change in anatomical regions of individual subjects. This analysis independently confirmed our unfolding results and allowed for an additional method of high-resolution hippocampal imaging analysis. However, current in vivo imaging methods are unable to reach the resolution necessary to visualize cell histology to determine exact subregional boundaries. Furthermore, the hippocampus is small in structure and variable across subjects. We attempted to deal with this challenge by demarcating each individual subject's boundaries using atlases by Amaral and Insausti (1990) and Duvernoy (1998), which identify landmarks on the MR images using postmortem histological images.
Surprisingly, we did not see CA3 activity differences between MSP and SSP conditions, although this region has been shown to be involved in encoding associative information (Zeineh et al., 2003; Eldridge et al., 2005). While there was activity within voxels of the region encompassing CA23DG in both learning conditions, there was no differential activity when learning from multiple start points. Region CA3 has been associated with pattern separation to form distinct memory traces during learning (Leutgeb et al., 2007; Bakker et al., 2008). It may be that learning a route using a single start point and learning a cognitive map from multiple start points depends to the same degree on pattern separation processes.
In the MSP condition, subjects have to integrate different paths stored in memory to infer where they are located in relation to the goal. Thus, this encoding condition may rely more heavily on retrieval. Navigation depending on an allocentric spatial representation is thought to depend on memory retrieval (Knowlton and Fanselow, 1998). Stronger demands on retrieval to integrate previously learned information with novel information might cause an increase in activation in CA1. One possibility is that the CA1 activation reflects retrieval of other routes during the task. Previous work has shown an increase in CA1 activity during memory retrieval in a nonspatial memory task, although this activation was not extremely robust (Eldridge et al., 2005).
Last, our findings are consistent with computational models proposing that CA1 integrates previously learned information (encoded in CA3) with incoming novel information from the neocortex (via ERC) (Levy, 1989; Vinogradova, 2001). Navigating through a novel spatial environment requires integrating multiple spatial relationships into a flexible memory representation. Forming an allocentric cognitive map requires integration of previously learned viewpoints with newly encountered viewpoints to update the memory representation into one that is independent of starting point. This process would thus presumably recruit more cells within CA1, leading to a detectable fMRI response. In the rodent hippocampus, place cells are context specific (Muller and Ranck, 1987; Wilson and McNaughton, 1993) and remap when a rat is placed in a different spatial environment. CA1 neurons have been shown to be sensitive to the intended destination and trajectory of the animal (Frank et al., 2000; Ainge et al., 2007). These lines of evidence support a role for this structure in acquiring a flexible memory representation of a spatial environment. Overall, the present results suggest that the CA1 region may also play a critical role in allocentric spatial memory formation in humans.
Footnotes
This work was supported by National Institute of Mental Health Grant 5T32 MH015795, National Institute of Neurological Disorders and Stroke Grant F32 NS50067-03, National Institutes of Health Grant T90 431587-BH-29793, National Science Foundation Grant GK-12 0742410, and National Institute on Aging Grants 2R01 AG013308 and 5P01 AG025831. For generous support, we thank the Brain Mapping Medical Research Organization, Brain Mapping Support Foundation, Pierson-Lovelace Foundation, The Ahmanson Foundation, William M. and Linda R. Dietel Philanthropic Fund at the Northern Piedmont Community Foundation, Tamkin Foundation, Jennifer Jones-Simon Foundation, Capital Group Companies Charitable Foundation, Robson Family, and Northstar Fund. We thank Michael J. Kahana for sharing and Aaron Geller and Josh Jacobs for support of the virtual navigation task “yellowcab” with financial support from Dr. Kahana's National Institutes of Health Grant MH61975. We also thank Michael Zeineh and Paul Thompson for assistance with group unfolding scripts, and Michael Jones for technical assistance. Finally, we also thank all of the subjects for their participation in this study.
References
- Aguirre GK, Detre JA, Alsop DC, D'Esposito M. The parahippocampus subserves topographical learning in man. Cereb Cortex. 1996;6:823–829. doi: 10.1093/cercor/6.6.823. [DOI] [PubMed] [Google Scholar]
- Ainge JA, Tamosiunaite M, Woergoetter F, Dudchenko PA. Hippocampal CA1 place cells encode intended destination on a maze with multiple choice points. J Neurosci. 2007;27:9769–9779. doi: 10.1523/JNEUROSCI.2011-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DG, Insausti R. The hippocampal formation. In: Paxinos G, editor. The human nervous system. San Diego: Academic; 1990. pp. 711–755. [Google Scholar]
- Bakker A, Kirwan CB, Miller M, Stark CE. Pattern separation in the human hippocampal CA3 and dentate gyrus. Science. 2008;319:1640–1642. doi: 10.1126/science.1152882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun VH, Leutgeb S, Wu HQ, Schwarcz R, Witter MP, Moser EI, Moser MB. Impaired spatial representation in CA1 after lesion of direct input form entorhinal cortex. Neuron. 2008;57:290–302. doi: 10.1016/j.neuron.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Colombo M, Fernandez T, Nakamura K, Gross CG. Functional differentiation along the anterior-posterior axis of the hippocampus in monkeys. J Neurophysiol. 1998;80:1002–1005. doi: 10.1152/jn.1998.80.2.1002. [DOI] [PubMed] [Google Scholar]
- Duvernoy HM. Berlin: Springer; 1998. The human hippocampus: functional anatomy, vascularization, and serial sections with MRI. [Google Scholar]
- Ekstrom AD, Kahana MJ, Caplan JB, Fields TA, Isham EA, Newman EL, Fried I. Cellular networks underlying human spatial navigation. Nature. 2003;425:184–188. doi: 10.1038/nature01964. [DOI] [PubMed] [Google Scholar]
- Ekstrom AD, Bazih AJ, Suthana NA, Al-Hakim R, Ogura K, Zeineh M, Burggren AC, Bookheimer SY. Advances in high-resolution imaging and computational unfolding of the human hippocampus. Neuroimage. 2009;47:42–49. doi: 10.1016/j.neuroimage.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge LL, Engel SA, Zeineh MM, Bookheimer SY, Knowlton BJ. A dissociation of encoding and retrieval processes in the human hippocampus. J Neurosci. 2005;25:3280–3286. doi: 10.1523/JNEUROSCI.3420-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SA, Glover GH, Wandell BA. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cereb Cortex. 1997;7:181–192. doi: 10.1093/cercor/7.2.181. [DOI] [PubMed] [Google Scholar]
- Fernández G, Weyerts H, Schrader-Bölsche M, Tendolkar I, Smid HG, Tempelmann C, Hinrichs H, Scheich H, Elger CE, Mangun GR, Heinze HJ. Successful verbal encoding into episodic memory engages the posterior hippocampus: a parametrically analyzed functional magnetic resonance imaging study. J Neurosci. 1998;18:1841–1847. doi: 10.1523/JNEUROSCI.18-05-01841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LM, Brown EN, Wilson M. Trajectory encoding in the hippocampus and entorhinal cortex. Neuron. 2000;27:169–178. doi: 10.1016/s0896-6273(00)00018-0. [DOI] [PubMed] [Google Scholar]
- Gabrieli JD, Brewer JB, Desmond JE, Glover GH. Separate neural bases of two fundamental memory processes in the human medial temporal lobe. Science. 1997;276:264–266. doi: 10.1126/science.276.5310.264. [DOI] [PubMed] [Google Scholar]
- Goodrich-Hunsaker NJ, Hunsaker MR, Kesner RP. The interactions and dissociations of the dorsal hippocampus subregions: how the dentate gyrus, CA3, and CA1 process spatial information. Behav Neurosci. 2008;122:16–26. doi: 10.1037/0735-7044.122.1.16. [DOI] [PubMed] [Google Scholar]
- Hartley T, Maguire EA, Spiers HJ, Burgess N. The well-worn route and the path less traveled: distinct neural bases of route following and wayfinding in humans. Neuron. 2003;37:877–888. doi: 10.1016/s0896-6273(03)00095-3. [DOI] [PubMed] [Google Scholar]
- Hassabis D, Chu C, Rees G, Weiskopf N, Molyneux PD, Maguire EA. Decoding neuronal ensembles in the human hippocampus. Curr Biol. 2009;19:546–554. doi: 10.1016/j.cub.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard IP. Human visual orientation. Chichester, UK: Wiley; 1982. [Google Scholar]
- Iaria G, Petrides M, Dagher A, Pike B, Bohbot VD. Cognitive strategies dependent on the hippocampus and caudate nucleus in human navigation: variability and change with practice. J Neurosci. 2003;23:5945–5952. doi: 10.1523/JNEUROSCI.23-13-05945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- King JA, Burgess N, Hartley T, Vargha-Khadem F, O'Keefe J. The human hippocampus and viewpoint dependence in spatial memory. Hippocampus. 2002;12:811–820. doi: 10.1002/hipo.10070. [DOI] [PubMed] [Google Scholar]
- Knowlton BJ, Fanselow MS. Hippocampus, consolidation, and on-line memory. Curr Opin Neurobiol. 1998;8:293–296. doi: 10.1016/s0959-4388(98)80154-2. [DOI] [PubMed] [Google Scholar]
- Köhler S, Crane J, Milner B. Networks of domain-specific and general regions involved in episodic memory for spatial location and object identity. Hippocampus. 2002;12:718–723. doi: 10.1016/s0028-3932(97)00098-5. [DOI] [PubMed] [Google Scholar]
- Kumaran D, Maguire EA. The human hippocampus: cognitive maps or relational memory? J Neurosci. 2005;25:7254–7259. doi: 10.1523/JNEUROSCI.1103-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Yoganarasimha D, Rao G, Knierim JJ. Comparison of population coherence of place cells in hippocampal subfields CA1 and CA3. Nature. 2004;430:456–459. doi: 10.1038/nature02739. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Treves A, Meyer R, Barnes CA, McNaughton BL, Moser MB, Moser EI. Pregressive transformation of hippocampal neuronal representations in “morphed” environments. Neuron. 2005;48:345–358. doi: 10.1016/j.neuron.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser MB, Moser EI. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science. 2007;315:961–966. doi: 10.1126/science.1135801. [DOI] [PubMed] [Google Scholar]
- Levy WB. A computational approach to the hippocampal function. In: Hawkins RD, Gower GH, editors. Computational models of learning in simple neural systems. Orlando, FL: Academic; 1989. pp. 243–305. [Google Scholar]
- Maguire EA, Woollett K, Spiers HJ. London taxi drivers and bus drivers: a structural MRI and neuropsychological analysis. Hippocampus. 2006;12:1091–1101. doi: 10.1002/hipo.20233. [DOI] [PubMed] [Google Scholar]
- Malkova L, Mishkin M. One-trial memory for object-place associations after separate lesions of hippocampus and posterior parahippocampal region in the monkey. J Neurosci. 2003;23:1956–1965. doi: 10.1523/JNEUROSCI.23-05-01956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara TP, Shelton AL. Cognitive maps and the hippocampus. Trends Cogn Sci. 2003;7:333–335. doi: 10.1016/s1364-6613(03)00167-0. [DOI] [PubMed] [Google Scholar]
- Mellet E, Briscogne S, Tzourio-Mazoyer N, Ghaëm O, Petit L, Zago L, Etard O, Berthoz A, Mazoyer B, Denis M. Neural correlates of topographic mental exploration: the impact of route versus survey perspective learning. Neuroimage. 2000;12:588–600. doi: 10.1006/nimg.2000.0648. [DOI] [PubMed] [Google Scholar]
- Morris RG, Garrud P, Rawlins JN, O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Muller RU, Kubie JL, Ranck JB., Jr Spatial firing patterns of hippocampal complex-spike cells in a fixed environment. J Neurosci. 1987;7:1935–1950. doi: 10.1523/JNEUROSCI.07-07-01935.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel L, MacDonald L. Hippocampus: cognitive map or working memory? Behav Neural Biol. 1980;29:405–409. doi: 10.1016/s0163-1047(80)90430-6. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L. The hippocampus as a cognitive map. Oxford: Clarendon; 1978. pp. 190–230. [Google Scholar]
- Parslow DM, Rose D, Brooks B, Fleminger S, Gray JA, Giampietro V, Brammer MJ, Williams S, Gasston D, Andrew C, Vythelingum GN, Loannou G, Simmons A, Morris RG. Allocentric spatial memory activation of the hippocampal formation measured with fMRI. Neuropsychology. 2004;18:450–461. doi: 10.1037/0894-4105.18.3.450. [DOI] [PubMed] [Google Scholar]
- Rosenbaum RS, Ziegler M, Winocur G, Grady CL, Moscovitch M. I have often walked down this street before: fMRI studies of hippocampus and other structures during mental navigation of an old environment. Hippocampus. 2004;14:826–835. doi: 10.1002/hipo.10218. [DOI] [PubMed] [Google Scholar]
- Shelton AL, Gabrieli JD. Neural correlates of encoding space from route and survey perspectives. J Neurosci. 2002;22:2711–2717. doi: 10.1523/JNEUROSCI.22-07-02711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager Y, Bayley PJ, Bontempi B, Hopkins RO, Squire LR. Spatial memory and the human hippocampus. Proc Natl Acad Sci U S A. 2007;104:2961–2966. doi: 10.1073/pnas.0611233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Rose M, Gläscher J, Wolbers T, Büchel C. Dissociable contributions within the medial temporal lobe to encoding of object-location associations. Learn Mem. 2005;12:343–351. doi: 10.1101/lm.90405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers HJ, Burgess N, Maguire EA, Baxendale SA, Hartley T, Thompson PJ, O'Keefe J. Unilateral temporal lobectomy patients show lateralized topographical and episodic memory deficits in a virtual town. Brain. 2001;124:2476–2489. doi: 10.1093/brain/124.12.2476. [DOI] [PubMed] [Google Scholar]
- Squire LR, Stark CEL, Clark RE. The medial temporal lobe. Annu Rev Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- Stark CE, Squire LR. When zero is not zero: the problem of ambiguous baseline conditions in fMRI. Proc Natl Acad Sci U S A. 2001;98:12760–12766. doi: 10.1073/pnas.221462998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo PC, Sapiro G, Wandell BA. Creating connected representations of cortical gray matter for functional MRI visualization. IEEE Trans Med Imaging. 1997;16:852–863. doi: 10.1109/42.650881. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Hum Brain Mapp. 2000;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman EC. Cognitive maps in rats and men. Psychol Rev. 1948;55:189–208. doi: 10.1037/h0061626. [DOI] [PubMed] [Google Scholar]
- Vinogradova OS. Hippocampus as comparator: role of the two input and two output systems of the hippocampus in selection and registration of information. Hippocampus. 2001;11:578–598. doi: 10.1002/hipo.1073. [DOI] [PubMed] [Google Scholar]
- Wills TJ, Lever C, Cacucci F, Burgess N, O'Keefe J. Attractor dynamics in the hippocampal representation of the local environment. Science. 2005;308:873–876. doi: 10.1126/science.1108905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Dynamics of the hippocampal ensemble code for space. Science. 1993;261:1055–1058. doi: 10.1126/science.8351520. [DOI] [PubMed] [Google Scholar]
- Witter MP, Wouterlood FG, Naber PA, Van Haeften T. Anatomical organization of the parahippocampal-hippocampal network. Ann N Y Acad Sci. 2000;911:1–24. doi: 10.1111/j.1749-6632.2000.tb06716.x. [DOI] [PubMed] [Google Scholar]
- Woolrich MW, Ripley BD, Brady M, Smith SM. Temporal autocorrelation in univariate linear modeling of FMRI data. Neuroimage. 2001;14:1370–1386. doi: 10.1006/nimg.2001.0931. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Bookheimer SY. Application of cortical unfolding techniques to functional MRI of the human hippocampal region. Neuroimage. 2000;11:668–683. doi: 10.1006/nimg.2000.0561. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Unfolding the human hippocampus with high resolution structural and functional MRI. Anat Rec. 2001;265:111–120. doi: 10.1002/ar.1061. [DOI] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science. 2003;299:577–580. doi: 10.1126/science.1077775. [DOI] [PubMed] [Google Scholar]
- Zola-Morgan S, Squire LR, Amaral DG. Human amnesia and the medial temporal lobe region: enduring memory impairment following a bilateral lesion limited to field CA1 of the hippocampus. J Neurosci. 1986;6:2950–2967. doi: 10.1523/JNEUROSCI.06-10-02950.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]