Abstract
Mutations of the sequence-specific master regulator p53 that alter transactivation function from promoter response elements (REs) could result in changes in the strength of gene activation or spectra of genes regulated. Such mutations in this tumor suppressor might lead to dramatic phenotypic changes and diversification of cell responses to stress. We have determined “functional fingerprints” of sporadic breast cancer-related p53 mutants many of which are also associated with familial cancer proneness such as the Li-Fraumeni Syndrome and germline BRCA1/2 mutant-associated cancers. The ability of p53, wild type and mutants, to transactivate from 11 human target REs has been assessed at variable expression levels using a cellular, isogenomic yeast model system that allows for the rapid analysis of p53 function using a qualitative and a quantitative reporter. Among 50 missense mutants, 29 were classified as loss-of-function. The remaining 21 retained transactivation towards at least one RE. At high levels of galactose induced p53 expression, 12/21 mutants that retain transactivation appeared similar to WT. When the level of galactose was reduced, transactivation defects could be revealed suggesting that some breast cancer related mutants can have subtle changes in transcription. These findings have been compared with clinical data from an ongoing neoadjuvant chemotherapy treatment trial for locally advanced breast tumors. The functional and nonfunctional missense mutations may distinguish tumors in terms of demographics, appearance and relapse, implying that heterogeneity in the functionality of specific p53 mutations could impact clinical behavior and outcome.
Keywords: p53, breast cancer, altered-function mutations
Introduction
The tumor suppressor p53 is a master regulatory gene that regulates the differential expression of target genes in a sequence-specific manner in response to cellular and environmental insults (1-3). Cell cycle regulation, apoptosis, angiogenesis, replication and repair are processes interconnected within the p53 transcriptional network. p53 exerts itself as a transcription factor by binding as a homotetramer, or dimer of dimers, to a consensus response element (RE) sequence comprised of two decamer half-sites [RRRCWWGYYY]2 (where R= purine; W= A/T; Y=pyrimidine) which vary between target genes. While the canonical consensus sequence allows for spacing between the half-sites of up to 13 nucleotides, we recently showed that transactivation was greatly reduced as the separation between decamers is increased beyond 2 nucleotides (4). Activation of a p53 target gene is dependent on a matrix of factors including cell type, stimuli, post-translational modifications and transcriptional co-factors (5).
The importance of p53 as a tumor suppressor and sequence-specific transcription factor in human cells is highlighted by the occurrence of p53 mutations in the majority of cancers (6). Interestingly, p53 is unique in comparison to other transcription factors in that over 75% of mutations that occur in this tumor suppressor are single amino acid changes which result in missense mutations (7). These missense mutations predominantly occur in the DNA binding domain (DBD) of the protein (>80%) (7). At the molecular level, p53 mutations found in cancers, including breast cancer, are usually associated with loss of the ability to maintain proper cell cycle checkpoints, suppress transformation caused by oncogenes, induce apoptosis and maintain the integrity of the genome (1, 8).
Specific mutations in p53 are known to denature the native protein or abrogate its ability to bind DNA thus completely abolishing its function. Such mutations are thought to provide a selective advantage within cancerous cells by forming a hetero-tetramer with WT p53 and functioning in a dominant-negative fashion (9, 10). Alternatively, gain-of-function mutations can potentiate tumorigenesis through oncogenic mechanisms including aberrant transcriptional regulation of either known or novel target genes--presumably through structure-selective DNA binding or protein-protein interactions (9-11). Many p53 missense mutations have been described that retain function as sequence-specific transcription factors such that the ability to regulate cellular responses is altered, but not completely lost (2, 12).
Functional mutations that alter the transcriptional capacity of the p53 master gene have been identified as super-transactivating, change-in-spectrum or overall downward modulation of transactivation. For example, change-in-spectrum mutants may be capable of regulating genes containing a strong RE, such as p21, but unable to regulate those with a weak RE, such as Bax. This is consistent with the observation of mutant p53s that still induce cell cycle arrest, yet lose the ability to activate apoptosis (13-15). In addition, p53 mutations may alter the active binding sites of potential transcriptional cofactors, thus diminishing the potential maximal level of transcriptional response. Modifications in the transcriptional network due to altered-function mutations may result in cellular responses that impact genome stability, repair, replication, and programmed cell death. Varying patterns of cellular responses, including apoptosis and survival have been elicited in human cells as a consequence of distinct altered-function p53 mutations (16). Furthermore, the aberrant biological consequences of specific altered-function mutations can be influenced by specific cell type and activating stimuli.
Mutations in p53 are associated with approximately 25% of sporadic cases of breast cancer, a frequency lower than that in other sporadic cancers, such as lung and colorectal carcinomas. However, sporadic p53 mutations occur at much higher frequencies in BRCA1/2 germline-associated breast cancers possibly due to a decreased efficiency to repair damage (17, 18). BRCA1/2 and p53 are involved in maintaining genome stability by controlling aspects of homologous recombination and repair, centrosome regulation, cell cycle checkpoints and transcription (19), where loss of either increases the likelihood of cancer (20). Interestingly, BRCA1-associated cancers have an altered spectrum of p53 mutations which may reflect changes in mutagenesis and/or selection for the acquired mutations (17). While BRCA1 mutations are largely absent in somatic breast tumors, silencing of the gene through hypermethylation has been reported in sporadic cases (21). Such epigenetic changes have been reported to associate with estrogen receptor negative (ER-) tumors and occur concomitantly with p53 mutations (21).
At the clinical level, p53 mutations in breast cancer have been associated with poor prognosis, earlier on-set, increased aggressiveness of tumors, aneuploidy, and adverse responses to chemotherapeutic treatments (22). Studies that classify breast cancers based on gene expression profiling have shown p53 mutations are more frequent in the hormone receptor-negative subtypes such as the HER2+/ER− [human epidermal growth factor-2 positive/estrogen receptor negative] and the basal-like subtypes [ER−, PR− (progesterone receptor), HER2−, cytokeratin 5/6+, and/or HER1+] (23-25). Based on a recent population-based study, these subtypes were prevalent among African American and/or premenopausal women and correlated with a more aggressive disease and shortened survival, irrespective of lymph node status (25). Regardless of subtype, p53 status (WT or mutant) also displays a signature expression profile in breast tumors which is a prognostic indicator of patient survival, where WT p53 associates with a more favorable outcome (23, 26, 27).
We have employed a newly developed model system in diploid yeast (4) to analyze the functional consequences of p53 missense mutations found in breast cancers on gene activation endpoints in the p53 transcriptional network at various levels of p53 expression. Transactivation capacities of WT and mutant p53 have been determined using a qualitative and a quantitative reporter, and a “functional fingerprint” was established for each p53 variant towards a subset of human REs that are representative of p53-dependent cellular responses. We have determined that p53 missense mutations found in sporadic and familial breast cancers can retain function and the alterations in transactivation are often subtle where differences can be exaggerated by changes in p53 levels. While patient numbers are limited, the separation of missense p53-associated breast cancer mutations into functional (which include fully functional and altered function) versus nonfunctional classes appears to associate with prognostic factors and outcome in a largely locally-advanced patient population treated with chemotherapy prior to surgery. Functional fingerprinting of cancer-associated p53 mutants may thus be a useful tool for understanding tumor biology and behavior.
Materials and Methods
Isogenomic diploid yeast strains
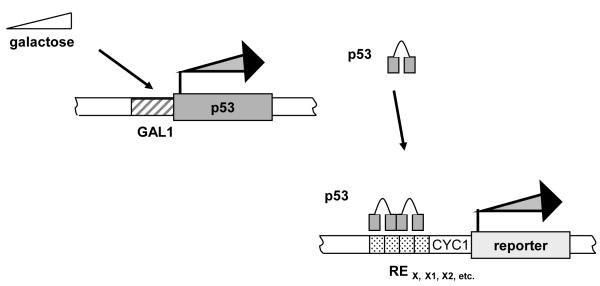
Two panels of isogenic haploid yeast strains, a “p53-host” and “response element (RE) reporter” strains, were developed in the budding yeast, S. cerevisiae with the delitto perfetto site-directed mutagenesis system as previously described (Figure 1) (4, 79). Each “p53-host” strain, yAT-iGAL::p53 (MATa leu2-3,112 trpl-1 his3-11,15 can 1-100 ura3-1, trp5::pGAL1:p53:cyc1-Ter, lys2::HygroR), contains the wild type or mutant p53 cDNA controlled by the inducible, “rheostatable” GAL1 promoter (2) integrated at the TRP5 locus on chromosome VII. p53 mutations were constructed using a derivative of the previously described p53 host strain containing a CORE cassette (CO, counterselectable, KLURA3; RE, reporter, KanMX4 resistance gene) integrated at various nucleotide positions spanning the p53 cDNA (4). Modifications of the p53 cDNA were performed using the delitto perfetto approach so that CORE cassettes were replaced with an oligonucleotide containing the mutation of interest to generate a full-length mutant p53 cDNA. (Oligonucleotide sequences are available upon request.) Replacement of the CORE was confirmed by selection on 5-FOA and kanamycin sensitivity. Specific p53 alterations were confirmed by colony PCR and sequencing (Big dye, Applied Biosystems, Foster City, CA). The second panel of isogenic strains, constructed previously, contain human target p53 REs upstream of the CYC1 minimal promoter and either the ADE2 or firefly luciferase reporter (2, 4). The RE reporter strains are also isogenic with the p53 host strains, but LYS2 and Hygros. Mating of the reporter and p53-host strains followed by selection for diploid cells on Lys− Hygro+ plates, results in isogenomic yeast that enable the assessment of the transactivation potential for WT or mutant p53 proteins towards individual REs in the p53 transcriptional network.
Figure 1. Evaluation of transactivation potential towards REs by WT and mutant p53s at variable levels of expression.
The budding yeast S. cerevisiae is used as an in vivo test tube to assess the consequences of p53 missense mutations associated with breast cancer on transactivation capacity from human RE targets. Isogenomic diploid yeast strains were generated by mating haploid “p53-host” strains with “RE-reporter” strains of opposite mating types. Each haploid strain was constructed with the delitto perfetto in vivo mutagenesis system where p53 and the REs were integrated into the chromosomes as previously reported (4, 79). Importantly, all of the conditions within the diploid yeast are constant except for the p53 mutation of interest or a few nucleotide differences in the RE sequence. The rheostatable GAL1 promoter allows for tight regulation of p53 expression where increasing levels of galactose allow for over 150- fold induction in protein expression (4). Regulation of protein expression allows for ascertainment of functional discrepancies between WT and mutant p53 transactivation at variable expression levels. Since p53 is not endogenous in yeast and human p53 can utilize the yeast transcriptional machinery, placement of the reporter downstream of the minimal CYC1 promoter provides the opportunity to directly monitor p53 interactions with RE sequence.
Qualitative ADE2 color assay
Single colony isolates of the p53-inducible RE-ADE2 reporter strains were streaked onto an YPDA control plate containing glucose and high levels of adenine and grown to equivalent amounts at 30°C. The plates were then replica plated onto a series of 9 plates containing selective media with low levels of adenine [5 mg/L], 2% raffinose and increasing galactose (0, 0.001, 0.002, 0.004, 0.008, 0.016, 0.032, 0.064 and 0.128%). Transactivation capacities for the p53 mutants were determined after three days of growth at 30°C by the ability of the mutant to produce a change in colony pigmentation. Transactivation of the ADE2 gene, which is a direct readout of p53 interaction with the specific RE, results in white colonies where decreased or loss-of-transactivation of the ADE2 results in pink and red colonies, respectively (32). Transactivation capacities for the p53 WT and mutants were determined after three days of growth at 30°C by the ability of the mutant to produce a change in colony pigmentation. Colony pigmentation was manually scored on a scale of 1 to 5, where 1 is no apparent transactivation (red colonies) and 5 is strong transactivation (white colonies) (Supplemental figure 2).
Quantitative luciferase assay
Diploid yeast strains containing GAL1::p53 (WT or mutant) crossed with a specified RE-luciferase reporter were grown overnight in 5ml YPDA plus adenine [200 mg/L] rich media. Overnight cultures were diluted 1:50 in H2O. For each measurement, 1 ml of the diluted culture was spun down, washed of residual glucose with H2O and re-suspended in 2mL synthetic complete -LYS media plus 2% raffinose supplemented with increasing amounts of galactose (0, 0.002, 0.004, 0.008, 0.010, 0.012, 0.016, 0.020, 0.024, 0.028 or 0.032%). These cultures were grown overnight (~ 18 hr) at 30°C to ~2 − 4 × 107 per ml (late log early stationary). The 2 ml cultures were spun down and the supernatant was aspirated. The remaining pellet was resuspended in 100 μl reporter lysis buffer (Promega, Madison, WI) and an equivalent amount of 425-600 micron acid-washed, glass beads was added (Sigma, St. Louis, MO). Samples were homogenized for 30 seconds in the Biospec Products, Inc. mini-bead beater (Bartlesville, OK), briefly incubated on ice and spun for 20 minutes at 16k relative centrifugal force (rcf) in an Eppendorf 5415R centrifuge (Batavia, IL) to separate the soluble protein fraction. The standard protocol recommended by the manufacturer (Promega; Madison, WI) was performed for the luciferase assay system starting with 10 μl of protein extract. Luciferase activity was measured from 96-well, white optiplates (Perkin Elmer, Waltham, MA) in a Wallac Victor2 multilabel counter (Perkin Elmer, Waltham, MA). Light units were standardized per μg protein as determined by a Bio-Rad protein assay (Bio-Rad; Hercules, CA).
Western analysis
Diploid yeast strains containing GAL1::p53 (WT or mutant) crossed with the p21-5′ RE-luciferase reporter were grown as described above. Overnight cultures containing 0.024% galactose were harvested, lysed in 35 μl reporter lysis buffer (Promega, Madison, WI) plus 2% protease inhibitors (cocktail for use with fungal and yeast extracts; Sigma, St. Louis, MO) and processed in the same fashion as those used in the luciferase assay. Protein concentrations were measured with the Bio-Rad protein assay according to the standard protocol (Bio-Rad; Hercules, CA). 50 μg of total protein was run on 4-12% BisTris NuPAGE and transferred as previously described (32). The p53 protein was detected with a mix of DO7 (BD BioSciences Pharmenigen) and pAb1801 (Santa Cruz) antibodies unless otherwise specified according to the manufacturer’s protocol. Bands were detected using horseradish peroxide-conjugated secondary antibodies (Santa Cruz) and the enhanced chemiluminescence (ECL) detection system (Amersham, Cleveland, OH, USA). Membranes were stained with Ponceau S to determine efficiency of protein loading.
Mutation analysis from patients participating in a neoadjuvant trial
p53 mutational analysis was performed on untreated breast cancer tissues obtained from women participating in two clinical-translational trials, a single institution study from the University of North Carolina–Lineberger Comprehensive Cancer Center (UNC-LCCC 9819), and a multiinstitutional cooperative group trial sponsored by the National Cancer Institute (CALGB 150007). In these correlative studies, women with locally advanced breast cancer were treated first with an anthracycline generally followed by taxane-based chemotherapy (with or without trastuzumab, depending on Her2 status). Both trials involved acquisition of breast cancer core biopsy tissue before treatment with any chemotherapy and were designed to examine molecular markers predicting response to cytotoxic chemotherapy (29). These studies were approved by the institutional review boards of the University of North Carolina at Chapel Hill and participating institutions through the Cancer and Leukemia Group B (CALGB). All study subjects gave written informed consent to participate in the clinical trial and the correlative science studies. p53 gene mutations were assessed using a two-tiered screening strategy. A pre-release version of the p53 AmpliChip array (Roche Molecular Systems) was first used to detect point mutations and 1 base pair deletions. Samples identified as potentially positive by the AmpliChip was sequenced to confirm the mutation. Specimens that were mutation-negative by AmpliChip were evaluated by single strand conformational polymorphism (SSCP) analysis in p53 exons 2-11 to detect deletions larger than 1 base pair and insertions (80). Mutations identified as potentially positive by SSCP were sequenced to identify the mutation. To rule out the possibility that mutations occurred due to PCR errors, we re-amplified and re-sequenced all mutation-positive specimens.
Results
Functional fingerprinting of p53 missense mutations associated with breast cancers
Budding yeast lack endogenous p53 and have been used as an in vivo test tube to analyze directly interactions between p53 and REs in a cellular environment (2, 3). Recently, we developed a diploid yeast model system to address the contribution of RE sequence, organization and level of human p53, as well as the consequences of mutations upon p53-mediated transactivation (Figure 1) (4).
We sought to define how specific p53 missense mutations found in breast cancers interfere with p53 function by assessing the ability of mutant p53 to transactivate from a panel of REs associated with p53-dependent downstream target genes (Table 1). Fifty missense mutations were chosen for examination if they were identified in cases of sporadic breast cancers. Importantly, 20 were identified in patients undergoing neoadjuvant treatment for locally advanced breast tumors and participating in clinical trials examining biomarkers predicting response to sequential anthracycline- and taxane-based chemotherapy prior to surgery (28, 29). Furthermore, 18 also associate with familial BRCA1/2 cancers and/or are found as germline mutations in Li-Fraumeni Syndrome (LFS), Li-Fraumeni-like Syndrome (LFL), and/or familial history (FH) cancer patients (Table 1). Of particular interest were mutations present in the L2 loop, L3 loop or zinc binding regions of the protein which have been correlated with breast cancers that are often nonresponsive to chemotherapeutic treatments including doxorubicin, tamoxifen, and/or combined therapies of 5-flurouracil and mitomycin (19, 30, 31).
Table 1.
p53 missense mutations associated with breast cancers: functional status, frequency, and features.
| p53 MUTATION |
FUNCTIONALa STATUS |
IARC DATABASEb | FEATURESc | ||
|---|---|---|---|---|---|
| SOMATIC | GERMLINE FAMILIES |
||||
| TOTAL | BREAST | ||||
| T125R | ALTERED | 2 | 1 | 0 | |
| L130V | ALTERED | 21 | 3 | 0 | Neo. |
| C135F | LOSS | 49 | 3 | 0 | |
| C135Y | LOSS | 70 | 11 | 0 | Neo. |
| A138V | FUNCTIONAL | 48 | 7 | 0 | |
| C141W | ALTERED | 14 | 1 | 0 | Neo. |
| P151A | ALTERED | 17 | 3 | 0 | BRCA1 |
| P151H | FUNCTIONAL | 35 | 4 | 0 | Neo. |
| G154S | ALTERED | 11 | 0 | 0 | Neo. |
| V173L | LOSS | 85 | 10 | 0 | L2; Neo. |
| R174K | ALTERED | 9 | 1 | 0 | L2 |
| R174W | FUNCTIONAL | 14 | 2 | 0 | L2 |
| C176F | LOSS | 181 | 7 | 0 | L2-Zn |
| H179R | LOSS | 139 | 16 | 0 | L2-Zn; Neo. |
| R181P | LOSS | 22 | 2 | 1 | L2; FH |
| S183L | LOSS | 3 | 1 | 0 | L2 |
| P190L | ALTERED | 48 | 4 | 0 | L2; BRCA1 |
| L194P | ALTERED | 14 | 1 | 0 | L2; BRCA |
| L194R | LOSS | 55 | 9 | 0 | L2 |
| H214R | ALTERED | 72 | 5 | 0 | BRCA2 |
| Y220C | ALTERED | 315 | 41 | 4 | LFS; Neo. |
| M237I | ALTERED | 172 | 26 | 1 | L3; LFL |
| C238F | LOSS | 38 | 5 | 0 | L3-Zn; BRCA1 |
| N239D | LOSS | 43 | 5 | 0 | L3; Neo. |
| N239T | LOSS | 9 | 2 | 0 | L3 |
| S241C | LOSS | 33 | 4 | 0 | L3 |
| C242F | LOSS | 86 | 7 | 0 | L3-Zn |
| C242S | LOSS | 32 | 5 | 0 | L3-Zn; BRCA1 |
| C242Y | LOSS | 51 | 5 | 3 | L3-Zn; FH; Neo. |
| M243T | LOSS | 8 | 2 | 0 | L3 |
| G244V | LOSS | 23 | 2 | 1 | L3 |
| G245S | LOSS | 396 | 35 | 18 | L3; LFS, LFL, FH; Neo.; BRCA1 |
| M246A | LOSS | 0 | 0 | 0 | L3; Neo. |
| M246T | LOSS | 12 | 3 | 0 | L3 |
| R248L | LOSS | 112 | 6 | 0 | L3, Neo. |
| R249G | LOSS | 43 | 7 | 0 | L3 |
| R249S | LOSS | 374 | 14 | 0 | L3, Neo. |
| D259V | LOSS | 17 | 2 | 0 | Neo. |
| G266R | LOSS | 65 | 5 | 0 | Neo. |
| R267Q | ALTERED | 12 | 3 | 3 | LFS, FH |
| V272L | FUNCTIONAL | 47 | 1 | 1 | LFS |
| R273C | LOSS | 611 | 28 | 9 | LFS, LFL, FH; Neo. |
| R273H | LOSS | 733 | 76 | 16 | LFS, LFL; Neo.; BRCA1/2 |
| P278A | ALTERED | 24 | 5 | 0 | Neo. |
| D281E | LOSS | 48 | 3 | 0 | |
| R283P | ALTERED | 34 | 3 | 0 | Neo. |
| E285K | FUNCTIONAL | 156 | 20 | 0 | Neo. |
| K305M | LOSS | 2 | 2 | 1 | FH |
| R337C | ALTERED | 17 | 4 | 3 | LFS |
| R337H | ALTERED | 2 | 0 | 69 | LFL, FH; ACC |
Based on transactivation capacity from 11 human p53 target REs assessed with the plate color assay and the luciferase assay. Mutations were classified as loss-of-function if they were unable to transactivate from any RE at any protein concentration. Mutations were categorized as functional if they were equivalent to WT p53 in transactivation capacity or altered function if the allele retained the ability to function from a RE(s), but deviated from WT p53 in transactivation capacity from several REs at any level of p53 expression.
Number of reported allele-specific mutations in the R12 release of the IARC p53 mutation database (7): total somatic mutations reported 24,810; missense mutations occurring in breast tumors, 1984; germline mutations, 399; however, families may have multiple tumors in various tissues with the same mutation. The single mutation not reported, M246A, was identified in a patient participating in a multi-institutional prospective trial. p53 status in the neoadjuvant treatment patients was confirmed by single-stranded conformational polymorphism analysis (SSCP) and validated with direct sequencing (71).
Criteria used to select the specific missense mutation for transcriptional analysis. BRCA, associated with BRCA1/2 cancers; LFS, Li-Fraumeni syndrome; LFL, Li-Fraumeni-like syndrome; FH, familial history; L2, L2 loop; L3, L3 loop; Zn, zinc binding; Neo, identified in patients undergoing neoadjuvant treatment for locally advanced breast tumors.
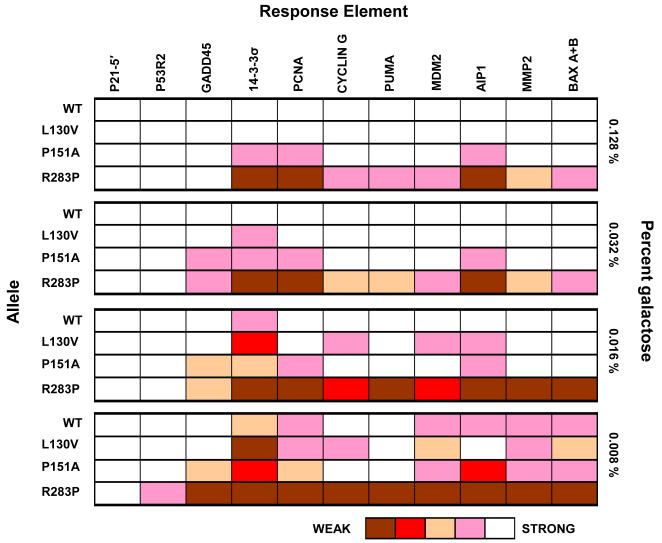
The yeast ADE2 plate color assay (32) was used to determine functional fingerprints for WT and p53 missense mutations based on their ability to transactivate from 11 different human REs at variable levels of protein expression (Figure 2 and Supplemental Figures 1 and 2). Briefly, single colony isolates of yeast strains containing the mutation and RE of interest were replicated onto plates containing increasing concentrations of galactose where the ability of the p53 variant to drive transactivation from a specific sequence could be assessed based on colony pigmentation (see Materials and Methods and Supplemental Figure 2). The REs analyzed are associated with known human p53 target genes involved in cell cycle, DNA repair, apoptosis, angiogenesis, and p53 regulation (Supplementary Table 1). Mutations were categorized as fully functional if they were indistinguishable from WT p53 in transactivation capacity or altered function if the allele retained the ability to function from at least one RE(s), but deviated from WT p53 in transactivation capacity from the REs examined at any of the levels of p53 expression examined.
Figure 2. Functional fingerprints of p53 mutants reveal subtle transactivational differences.
The ADE2 plate color assay was used to assess the transactivation capacity of WT and mutant p53 towards 11 human target REs at various protein concentrations. Transcription of ADE2 is dependent on the ability of p53 (WT or mutant) to interact with and transactivate from the specific RE sequence upstream of the minimal CYC1 promoter and the reporter. The ADE2 color assay scores p53 transactivation capacity from a RE based on colony pigmentation which ranges from red (no transactivation) to pink (weak to moderate) to white (strong) depending on the extent of ADE2 transcription (33). The level of p53 expression was controlled by replica plating strains onto plates containing rich media, raffinose (2%) as the carbon source, plus various amounts of galactose (0 – 0.128%) in the presence of low levels of adenine (5mg/L). Shown are examples of functional fingerprints for WT p53 and several change-in-spectrum mutations at 4 levels of protein expression. At high expression levels (0.128%) several mutations were found to display an altered spectrum of REs regulated in comparison to WT p53, for example, P151A and R283P. However, several mutants, such as L130V, appear indistinguishable from WT p53 in transactivation capacity. Reducing the levels of expression with the rheostatable promoter, exaggerated the subtle transcriptional effects of these mutations and distinguished them from WT p53 and other mutations in transactivation capacity.
Among the 50 missense mutations, 29 were classified as loss-of-function due to their inability to transactivate from any RE (Table 1). The remaining 21 (42%) mutations were able to function from at least one RE, where the transcriptional capacities varied from different levels of functionality to fully functional. Among the 21 functional mutations, 9 were clearly altered in transactivation capacity at high levels of galactose (0.128% galactose) (Supplemental Figure 1) and displayed a change-in-spectrum for REs transactivated, as exemplified by reduced or complete lack of transactivation from the 14-3-3σ and PCNA REs by P151A and R283P, respectively, in Figure 2. Of these 9 mutations, 3 (Y220C, M237I and P278A) retained the ability to transactivate from only the strongest REs, p21-5′ and P53R2 when p53 expression was induced with high levels of galactose (and, in the case of P278A, MDM2 which contains two full-site REs) (Supplemental Figure 1).
At high levels of galactose, the remaining 12 of the 21 mutants (L130V, A138V, C141W, P151H, R174K, R174W, P190L, H214R, R267Q, V272L, E285K, and R337H) looked similar to WT in their transactivation capacity (i.e., L130V in Figure 2, Supplemental Figure 1). Transactivation from three biological replicates was either indistinguishable from WT p53 or nearly identical with the exception of 1 or 2 REs at 0.128% galactose. However, when the levels of galactose were reduced, subtle transactivation defects were revealed that further differentiated 6 of the 12 mutants (underlined) from WT p53 at 3 or more REs. Similar to previous studies (2), the deviation from WT p53 at lower levels of induction consisted of both reduced and enhanced transactivation capacities from specific REs, such as the ability of L130V to transactivate from the Cyclin G and AIP1 REs at 0.008% galactose, respectively (Figure 2). The remaining 6 mutants remained similar to WT p53 in transactivation capacity even at low levels of galactose with the exception of subtle variation at 1 or 2 REs. For example, A138V was only observed to be slightly reduced in transactivation compared to WT p53 towards PCNA at 0.008% galactose (Supplemental Figure 1).
Luciferase assays confirm transcriptional anomalies
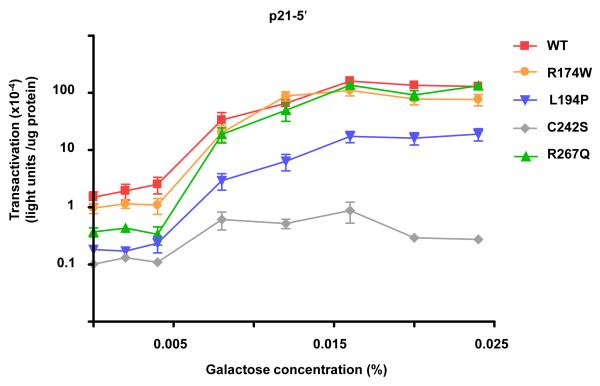
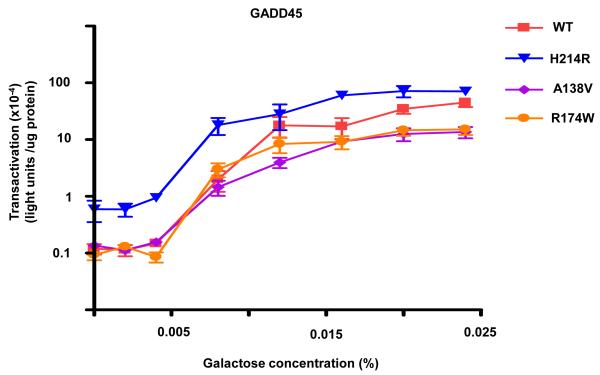
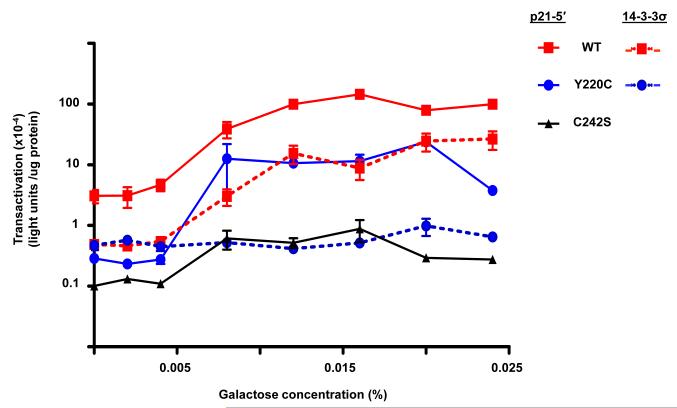
The functional status of the 21 missense mutants that retained function and several loss-of-function mutants was examined with a luciferase reporter assay that provides the opportunity to quantitate transactivation from REs in late log phase growing cells. The transactivation capacities from the p21-5′, GADD45 and 14-3-3σ REs were comparable to those with the color plate assay in terms of classifying functional status; however, the assay provides greater discrimination between mutant and WT p53 transactivation. Assessment of the altered-function mutants with the luciferase assay showed varying degrees of functionality from the p21-5′ RE, where the maximal level of transactivation was dependent on the specific mutation (Figure 3). Similar to the results with the plate assay, several mutants (i.e., R267Q) differed from WT p53 in their ability to transactivate from the p21-5′ RE only at low levels of galactose (low p53 expression), whereas other mutations (i.e., L194P) displayed a decreased ability to transactivate from the p21-5′ RE at all levels of induction and corresponding p53 expression. Of the 6 fully functional mutations examined that were similar to WT p53 in terms of transactivation from the strong p21-5′ RE in both the plate and luciferase assays, one (H214R) showed an altered transactivation potential when assessed for transactivation from the weaker GADD45 RE in the luciferase assay (Figure 4). H214R had an increased ability to transactivate from the GADD45 RE in comparison to WT p53, whereas the remaining fully functional mutations, including A138V and R174W remained indistinguishable from WT p53. Change-in-spectrum mutations which retained transactivation function from some REs, but were devoid of function from others, were also verified with the luciferase assays. As shown in Figure 5, Y220C was able to transactivate from the strong p21-5′ RE, but to reduced levels; the maximal level of transactivation was comparable to that for WT p53 transactivating from the weaker 14-3-3σ RE. The Y220C mutant was actually unable to transactivate from the 14-3-3σ RE.
Figure 3. Assessment of WT and mutant p53 transactivation towards the p21-5′ RE using a luciferase assay.
Diploid yeast strains containing GAL1::p53 (WT or mutant) and the p21-5′ RE-luciferase reporter strain were grown overnight in complete medium, diluted, washed and inoculated into selective medium containing either raffinose (2%) or raffinose (2%) plus increasing concentrations of galactose (0 – 0.024%) for an additional night at which point the cultures in late logarithmic growth. Protein lysates were obtained and a quantitative luciferase assay was used to determine the transactivation capacity for the p53 variants from the p21-5 RE’. The strength of transactivation was calculated as relative light units/ug protein. Depicted are the mean and standard error of measurement (SEM) for 7 independent experiments. The transactivation responses to increasing galactose can be described as basal, linear-increase, and plateau. Maximal transactivation is dependent on the p53 variant. Many p53 missense mutations associated with breast cancers, i.e. R174W and R267Q, do not affect the maximal level of transactivation towards the strong p21-5′ RE in comparison to WT p53. However, transactivation can be altered at low levels of galactose which correspond to low levels of p53 expression. Several mutants, such as L194P, were shown to modulate the levels of transactivation at all concentrations of expression. C242S is a loss-of-function mutation.
Figure 4. Transactivation from the GADD45 RE distinguishes altered-function mutants from WT p53.
The ability of p53 (WT and mutant) to transactivate from the GADD45 RE was measured 24 hours after inoculation into increasing concentrations of inducing media with a quantitative luciferase assay (see Figure 4.2). The strength of transactivation was calculated as relative light units/μg protein. Transactivation from the GADD45 RE can differentiate mutant p53 alleles that looked similar to WT p53 in the ADE2 plate assay. H214R has an increased ability to transactivate from the GADD45 RE in comparison to WT p53 at low and high levels of expression, whereas A138V and R174W had similar transactivation potentials. Presented are the mean and standard error of measurement (SEM) for 6 independent experiments.
Figure 5. Change-in-spectrum p53 missense mutations can eliminate REs from the p53 transcriptional network.
To establish that change-in-spectrum mutations are capable of altering the transcriptional network as a result of eliminating transcription from several REs, transactivation was assessed for WT and Y220C p53 from the p21-5′ and GADD45 REs. Diploid yeast strains were grown overnight in complete medium, diluted, washed and inoculated into induction media. Protein lysates were obtained from overnight cultures and quantitative luciferase assays were used to determine the transactivation capacity of the p53 variants from the p21-5′ RE. Y220C is capable of transactivating from the p21-5′ RE to levels comparable to WT p53 transactivation from the weaker 14-3-3σ RE. However, Y220C was not capable of transactivating from the 14-3-3σ RE suggesting this target gene may be eliminated from the p53 transcriptional network for this particular mutant. The strength of transactivation was calculated as relative light units/μg protein. Depicted are the mean and standard error of measurement (SEM) of 6 independent experiments.
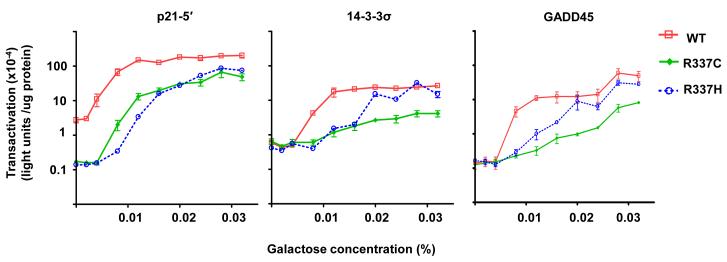
The transactivation profiles with increasing levels of galactose (i.e., increased p53 expression) were similar between the DBD mutants and WT p53 where initial induction occurred between 0.004 – 0.008% galactose and maximal levels of transactivation were between 0.016% - 0.024% galactose (Figures 3-5). This was not observed for the tetramerization domain mutant R337H (Figure 6). Although maximal levels of transactivation appeared similar to WT p53, the R337H mutation clearly altered transactivation from the p21-5′, 14-3-3σ and GADD45 REs, requiring higher levels of p53 expression than WT to initiate transactivation (Figure 6). The requirement for increased p53 levels necessary for initial transactivation by the tetrameric mutants appeared dependent on the strength of the RE. The R337C mutation resulted in overall reduction in transactivation.
Figure 6. Altered-function mutations at the same codon in the tetramerization domain display different transactivation capacities.
Transactivation capacities were obtained for the R337C and R337H mutants towards the p21-5′, 14-3-3σ, and GADD45 REs at increasing levels of expression with the quantitative luciferase assay. The levels of transactivation were severely diminished with R337C. R337H required an increased amount of p53 expression to stimulate transactivation. The magnitude of effect for R337C and R337H was dependent on the RE examined.
Altered-function mutants can maintain protein levels comparable to WT p53
Protein levels were analyzed at 0.024% galactose (within the range of expression where transactivation was shown to plateau in the luciferase assays) by Western analysis for the 21 mutations that retained function and a representative loss-of-function mutant (Supplemental Figure 3). Of the functional mutants, 13 displayed similar levels of protein to WT p53, as did the loss-of-function missense mutation. However, seven mutants (C141W, L194P, Y220C, M237I, P278A, E285K and R337C) had reduced expression compared to WT p53. Surprisingly, several of these mutants were shown to efficiently function from multiple REs in the plate and luciferase assays. Detection of the p53 protein with additional antibodies that recognize different epitopes was consistent with most of these mutants having reduced levels of protein in comparison to WT p53 (Supplemental Figure 4).
Functional status and clinical response
To address how different p53 mutations might influence response to chemotherapeutics, we examined transcriptional functional status of p53 missense mutations found in breast cancers in relation to clinical manifestations. Twenty-nine unique p53 missense mutations analyzed for functionality in this study (20 mutations) or in a related haploid yeast system (14, 33) have been identified in 46 patients with locally advanced breast tumors (primarily ductal carcinomas). The patients were participants in clinical trials that monitor clinical and pathologic responses to neoadjuvant treatment prior to surgery (Supplemental Table 3). As summarized in Table 2 and Supplemental Table 3, among the 46 patients, 10 of the missense mutations (11 patients) resulted in p53s that retained function (9 altered and 1 fully functional mutant) and 19 were loss-of-function (35 patients). This corresponded to ~24% of patients (11 among 46) having functional mutations, suggesting a significant group that might be approached differently regarding treatments.
Table 2.
Clinical characteristics and response to therapy in patients with p53 functional or nonfunctional missense mutants. There were 29 unique breast cancer-associated p53 mutations identified among 46 patients.
| Clinical factor | Functional mutants (10 mutations among 11 patients) |
Nonfunctional mutants (19 mutations among 35 patients) |
|---|---|---|
| Median age | 49 | 49 |
| Race/ethnicity | ||
| Caucasian | 7/11 (64%) | 22/35 (63%) |
| African-Americans | 2/11 (18%) | 12/35 (34%) |
| Asian | 2/11 (18%) | 1/35 (3%) |
| Node-positive | 2/11 (18%) | 17/34 (50%) |
| Stage | ||
| II | 7/11 (64%) | 12/34 (35%) |
| III | 3/11 (27%) | 18/34 (53%) |
| Inflammatory | 1/11 (9%) | 4/34 (12%) |
| Overall grade | ||
| 1 | 1/11 (9%) | 0 |
| 2 | 3/11 (27%) | 8/34 (23%) |
| 3 | 4/11 (36%) | 20/34 (59%) |
| Indeterminate | 3/11 (27%) | 6/34 (18%) |
| Immunohistochemical subtype | ||
| HER2 negative | 7/9 (78%) | 18/31 (58%) |
| Triple negative (ER, PR, HER2) | 5/9 (56%) | 11/23 (48%) |
| Pathologic complete response to therapy | 3/11 (27%) | 7/34 (21%) |
| Clinical response to chemotherapy | 8/10 (80%) | 29/33 (88%) |
| Recurrence (at approximately 3 y follow-up) | ||
| Local | 0/10 | 4/34 (12%) |
| Distant metastasis | 1/10 (10%) | 12/35 (34%) |
While the number of p53 missense mutations examined is limited, there are emerging trends that may differentiate patients with functional (altered or fully functional) versus nonfunctional p53 missense mutants (Table 2 and Supplemental Table 3). Among the tumors with somatic p53 missense mutations where HER2, ER and PR status could be assessed, functional missense mutations were more common in HER2-negative tumors (7 of 25, 28%) compared with HER2-positive (2 of 15, 13%). Among triple negative (ER, PR, and HER2) tumors with p53 missense mutations, 5 of 16 (31%) carried functional mutations. There appeared to be a higher frequency of functional/total missense mutations among Caucasians and Asians (7/29 and 2/3, respectively) than among African American patients (2/14, 14%). Functional mutations appear to be associated with good prognostic factors such as low incidence of nodal involvement (2 of 11, 18%). Conversely, compared with functional mutations, patients carrying nonfunctional mutations were more likely to be stage III at diagnosis (53 vs 27%), have high grade tumors (59 vs 36%) and to relapse in distant sites (34 vs 10%), as well as local sites (12 vs 0%) although the numbers are small. In terms of responsiveness to chemotherapy, the pathologic complete response (pCR; i.e., eradication of tumor) was similar between patients with functional p53 mutations and nonfunctional missense mutations (21% and 27%, respectively). However, women with nonfunctional mutations were more likely to die (36% vs 20%) at 3 years post-treatment than those with functional mutations. While suggestive of trends, none of these differences reached statistical significance.
Discussion
Single amino acid changes in the p53 master regulator protein that differentially impact transactivation may result in the selective advantage of specific mutations in certain tissue types or stages of neoplastic transformation, as well as alter the responsiveness to or the efficacy of chemotherapeutic agents. We have used a diploid yeast system to analyze the potential transactivation capacity for a set of p53 missense mutations associated with breast cancers. The 50 missense mutants examined represent approximately 18% of all somatic p53 mutations reported in the International Agency for Research on Cancer (IARC) TP53 mutation database and 20% of all p53 missense mutations documented in breast tumors (7). Among the mutations analyzed, one (K305M) occurs at an acetylation site in a non-structured portion of the protein, 2 (R337H and R337C) are in the tetramerization domain and the remaining 47 are distributed across the sequence-specific DNA binding domain.
Functional fingerprinting established that 21 out of the 50 missense mutants associated with breast cancer can retain p53 function. The effect on transactivation appears dependent upon the specific amino acid alteration such that even different missense mutations at the same residue can vary in the impact on p53 functionality. This is exemplified at codon 194 where changing the leucine residue to an arginine (L194R) results in loss-of-function, whereas a proline (L194P) results in altered function.
The majority of the altered-function mutations analyzed in this study do not seem to be the result of a general reduction in transactivation from all REs. Rather, they appear to be change-in-spectrum mutants that impact the REs differentially. Since each functional p53 missense mutation had a unique functional fingerprint (Supplemental Figure 1) ranging from severely altered to fully functional, there may be diverse cellular effects. The functional consequences of mutations appear not to be predictable simply by assessments of conservation, topology, or structural models, emphasizing the need to address the function of p53 mutants in vivo (Supplemental Table 2). For example, a recent structure-based analysis of mutations in the DBD does not appear indicative of the in vivo transactivation capacity. Over 45% of the predictions based on computational geometry (which assessed residual score profiles derived from Delaunay tessellations) (7, 34) differed with results from our transactivation assays (Supplemental Table 2).
Importantly, the differences in transactivation for all of the mutations could not be attributed simply to protein stability since most of the p53 mutants were expressed at levels comparable to that for WT protein. Of the mutations that had a reduced level of protein expression, the C141W and E285K mutants were only modestly compromised for transactivation capacity in comparison to WT p53, whereas others such as M237I and Y220C were severely compromised. It is possible that reduced levels of protein may be due to an increased level of degradation within the cell due to conformation changes. This could also explain associated temperature sensitivity of some alleles, specifically Y220C, M237I, and E285K (35-37).
However, regarding these low expressing mutants, the issue arises as to whether cellular p53 protein expression is comparable between yeast and tumor cells. There are examples where expression in yeast is matched by low or undetected levels in breast cancer cell lines (i.e., E285K in BT474 and MDA-MD-134VI cells (38, 39) and Y220C in HCC1419 cells (40, 41)) or alternatively being accumulated in the cells (i.e., M237I in SUM149PT cells (38)). Regardless of the amount of p53 protein expressed in yeast, the results are informative for those cases where the mutant proteins retain transactivation ability as this indicates the potential for mutant protein to function in mammalian cells.
Breast cancer associated mutations in the DBD can modulate p53 transactivation
The DBD of p53 consists of a β-sandwich which provides a scaffold for two large β-loops, L2 and L3 that are stabilized by a zinc ion and a loop-sheet-helix motif (42, 43). Mutations in the DBD have been postulated to affect the binding affinity of p53 towards REs by abolishing DNA contacts, decreasing the thermodynamic stability of the protein, causing local distortions in the DNA binding surface, or enhancing the loss of the Zn ion (44, 45). Mutations in the L2/L3 loops are predicted to be highly destabilizing to the tertiary structure of p53, can cause chemical shifts which alter the DNA binding surface and/or can alter the response to chemotherapeutics (7, 19, 30, 31, 46).
Of the DBD mutations analyzed for transactivation potential, 16 correspond to residues in the L3 loop and 10 are within the L2 loop; among these 26 mutants, 6 are also zinc-binding residues. All 6 mutations (C176F, H179R, C238F, C242F, C242S, C242Y) that interfere with the histidine or cysteine side chains involved in coordination of the Zn ion rendered the protein nonfunctional in terms of transactivation, emphasizing the vital role of Zn in sequence-specific DNA binding and stabilization of the p53 protein (44, 47). Similarly, all the missense mutations analyzed in the L3 loop, which binds the minor groove of DNA and partakes in the dimerization interface between core domains (45), were loss-of-function mutations with the exception of M237I which was very weak for transactivation. However, 4 mutants (R174K, R174W, P190L and L194P) in the L2 loop retained function of which several (underlined) displayed subtle alterations in transactivation capacity. These results suggest that mutations in the L2 loop, which functions as a support for the L3 loop, may be less detrimental to transactivation potential than those of zinc binding or in the L3 loop.
Tetramerization mutations may alter the level of p53 required for transactivation
Although the majority of p53 missense mutations occur in the DBD, several missense mutations have been found in the tetramerization domain that are associated with germline syndromes and are found in sporadic breast tumors. R337C is a partial function mutation associated with LFS (48). R337H has been associated with pediatric cases of adrenocortical carcinoma (ACC); however, it may be a low penetrant LFS or LFL allele as well (49-52). The functional fingerprints varied between these two altered-function mutations in that R337C had a greater impact on p53 transactivation displaying an overall dampening effect from the various REs. This reduced transactivation may reflect a greater instability of the protein as depicted in the protein analysis (Supplemental Figures 1-4). Contrary to previous reports, the p53 missense mutation R337H was not a silent mutation, but displayed altered function when examined in the ADE2 phenotypic assay (Supplemental Figure 1). At high levels of galactose induction (i.e., high levels of p53 expression from the GAL1 promoter), R337H looked identical to WT p53 in the color assay. At low levels of galactose, R337H had a reduced ability to transactivate from REs in comparison to WT p53, presumably due to its reduced ability to form tetramers. Interestingly, the luciferase assay revealed a pattern of transactivation that was unique to R337H where higher levels of protein expression were required to detect transactivation. This altered pattern of transactivation may be a manifestation of the novel features of R337H which include a pH-dependent instability and formation of amyloid-like fibrils (53, 54).
Subtle variation in transactivation capacity of altered-function missense mutations
Especially interesting is the observation that many of the mutants looked similar to WT p53 when examined at high levels of galactose (i.e., high p53 expression) yet were altered function at lower levels of galactose, consistent with our earlier findings (32). Importantly, these mutations would not have been distinguished from WT p53 in typical functional assays where p53 is expressed at high levels from a constitutive promoter. Thus, by reducing the level of transcription with the rheostatable promoter, it was possible to unmask subtle transcriptional discrepancies. These mutants might have unique properties in terms of biological consequences. Possibly, the mutants function similarly to WT p53 for gene expression from target genes under high stress and chemotherapeutic conditions, but differently under conditions of low p53 expression. Also, other transcription factors could further modify the response (3).
Although mutations in p53 are often associated with nuclear accumulation of the protein, recent evidence indicates this may not always apply in vivo. For example, Tsuda and Hirohashi (55) showed with immunohistochemical analysis of over 50 human breast-cancer tissue specimens that nuclear accumulation of the p53 protein was dependent on the type and position of the mutation, where missense mutations did not always result in stabilization of the protein. Similarly, in a study comparing immunohistochemical analysis with cDNA-based sequencing using ~300 primary breast tumor samples (56), in over 30% of the cases where a mutation was observed through sequencing there was not a corresponding accumulation of p53 protein.
Recent results with knock-in mouse models indicate levels of mutant p53 can be regulated in both normal and some tumor cells (57, 58). The accumulation of mutant protein does not occur until additional mutations are acquired in genes which may disrupt the p53 degradation pathway such as MDM2 or p16INK4a (59). Although the subtle altered-function mutations identified in the present study retain the ability to function from the MDM2 RE, Lukashchuck and Vousden (60) have shown that the ability of p53 to transactivate MDM2 is not essential for degradation of the mutant p53 protein. Rather, additional E3 ubiquitin ligases, such as CHIP (C terminus of Hsc70-interacting protein), can target mutant p53 for degradation independent of ubiquitination by Mdm2, where Mdm2 plays a role in delivering the ubiquitinated proteins to proteasomes.
The functional relevance of p53 at low expression levels is beginning to be elucidated and highlights the need to understand mutants under such conditions. Espinosa et al. (61) observed that p53 occupies some target REs, including p21, prior to overall p53 stabilization. There is transcriptional initiation from these REs, but the transcriptional machinery stalls prior to elongation. Such regulation may be required for a rapid response to cellular stress. In addition, there is a transcriptional-dependent role for p53 in promoting cell survival, as well as modulating glucose metabolism and reactive oxygen species at basal levels of p53 expression (62, 63). For example, p53 has been found to target the sestrins (i.e., SESN1 and SENS2) and TIGAR (TP53-induced glycolysis and apoptosis regulator) to stimulate antioxidant and pro-survival signals (62, 63). The loss of transactivation function at low levels, as observed for some of the subtle, altered-function mutants, might lead to alternative modes of promoter selectivity, and/or provide the opportunity for competing transcription factors to bind promiscuously to p53 target elements. Furthermore, p53 has also been found to promote, presumably in a transcriptional-independent fashion, global chromatin relaxation (64). This appears to influence genomic repair in response to UV stress at exposures that are lower than those required for its activation as a transcription factor.
It is possible that mutations that subtly affect transactivation are acquired early in tumor development and when combined with mutations in other genes contribute in an additive fashion to the complex cancer disease. For example, the altered-function P190L and H214R mutants that were indistinguishable from WT at higher levels of galactose-induced expression are associated with germline BRCA1 and BRCA2 mutations, respectively. Such mutations in p53 may be an underlying contributor to the genomic instability observed in BRCA1-associated breast cancer cases and the functional status of individual p53 missense mutations may impact the degree of genetic imbalance. Interestingly, a recent hierarchical clustering analysis using immunohistochemistry profiling to determine the relatedness of tumors has established that the extent of genomic instability correlates with specific breast cancer subtypes, where the basal subtype (the subtype in which p53 mutations are frequently observed) had the highest number of genomic aberrations in both sporadic and familial BRCA-associated cases (65-67).
In addition, inherited p53 mutations may influence tumor type and penetrance of the disease (57, 58, 68, 69). LFS and LFL germline disorders--which often harbor a p53 mutation--display an array of early onset, tissue specific tumors of which breast tumors are among the most frequent observed (7, 68). Recent studies which have assessed the functional status of p53 germline mutations using yeast-based assays have related severity of inherited p53 missense mutations in terms of transcription functionality with clinical manifestations (7, 70). Partial deficiency alleles, defined by the ability to transactivate from at least one RE to 25% of the levels obtained by WT p53, are associated with a less severe family history, lower number of tumors, later onset of disease in comparison to severe deficiency (loss-of-function) alleles and a higher risk of breast tumors.
In another recent study (71), the frequency and average size (bp deletion or duplication) of DNA copy number variation is enriched in carriers of germline TP53 mutations within LFS families in comparison to those with WT p53 or in a healthy population. The clinical phenotypes which arise in later generations may correlate with greater genomic instability, as well as specific germline p53 mutation. Thus, the wide spectrum of transcription potentials of the 12 p53 missense mutations associated with germline disorders in the current study (ranging from nonfunctional to altered and subtle, or fully functional) can be expected to result in varying phenotypes where the severity of the disease is likely influenced by the extent of p53 functionality.
Functional status and clinical response
We examined transcriptional functional status of p53 missense mutations found in breast cancers in relation to clinical manifestations (Table 2 and Supplemental Table 3). While the number of breast cancer-associated functional p53 mutants was small, there were trends described in the Results that suggest differences in presentation and outcome between functional versus nonfunctional missense mutations. The nonfunctional missense mutations are associated with clinical responses similar to those in our studies with null mutations (data not shown) which are known to have poorer prognosis and reduced survival as compared to patients with tumors that are WT for p53 (72). We have found that functional p53 mutations appear to have better disease-free survival compared with loss-of-function mutations (Table 2).
In terms of clinical response to chemotherapy, we observed in the present study a higher response rate for tumors expressing loss-of-function p53 mutations which may seem counterintuitive. However, tumors with p53 mutations in general are more chemo-sensitive, possibly because the breast cancer subtypes which usually have a higher proportion of p53 mutations (e.g. the basal-like) are more highly proliferative or have aberrant DNA repair functions (23, 25). Also, the cumulative response to anthracycline/cyclophosphamide followed by taxane therapy was determined in this study. While there is some indication from the literature that p53 mutant status could confer different responses to these single agents (73-75), it is difficult to predict responses to the sequential treatment. While previous studies have investigated correlations between p53 mutations and pathologic variables in breast cancers, few studies have attempted to correlate the transcriptional activity of specific mutations with clinical phenotypes. In a large scale study of a cohort of approximately 1,800 woman (72), the presence of a p53 mutation was associated with high grade, positive node status, loss of hormone receptors and greater risk of death due to breast cancer within a 10 year follow-up. However, when the functional status of the missense mutations was taken into consideration, no correlation was found between p53 transactivation activity and patient survival. Importantly, the functional status of the missense mutations was determined with another yeast-based functional assay developed by Kato et al. (12) that uses a high p53 expression plasmid along with a high copy RE reporter plasmid system.
Functional analysis based on the Kato et al. system is available at the IARC p53 mutation database for 49 of the 50 mutations examined in the present study (7). Interestingly, a comparison between the two data sets shows less than 65% agreement in terms of overall transactivation functionality (Supplemental Table 2). Only 8/49 or 16% of the missense mutations were previously identified as functional as compared to 21 mutations in the present study system. The large discrepancy between the two systems may be in part due to the method of classifying a mutation as retaining function (Supplemental Table 2) (7, 12). For example, we concluded that the R337C was an altered-function missense mutation whereas within the IARC database it is considered to be a nonfunctional mutation although it did display transactivation for 3 of the 8 REs analyzed (WAF1, MDM2 and P53R2) (7). Furthermore, in a separate analysis, the Kato et al. group (37) found that greater than 140 missense mutations were temperature sensitive for transactivation capacity towards at least one RE; this indicated there may be cellular conditions under which these mutations may become functional. Interestingly, 5 of these temperature sensitive mutations which were reported as loss-of-function in the IARC database were classified as altered-function in the present study (P151A, H214R, V272L, R283P and E285K). Another mutation Y220C had also been categorized as loss-of-function; however, we have diagnosed this as a weak altered-function protein and previously it was reported as temperature sensitive in mammalian cell assays (35).
Conclusions
Given the heterogeneity of breast cancers, understanding the consequences of altered-function mutations on the p53 transcriptional network will help elucidate how specific mutations predispose and/or contribute to the development, penetrance and phenotype of breast cancers. Among fifty p53 mutations identified in breast cancers, 21 had altered function—not simply complete loss—towards at least one RE. Although the transcriptional effects associated with these mutations are often subtle, we found that by reducing the level of transcription with the rheostatable promoter, it is possible to address the retained functions of mutant p53 and novel features in transcriptional networks. It is important to emphasize that the yeast-based results indicate the potential for transactivation and that many factors can come into play in p53 mediated transactivation in human cells. Because of the wide range of p53 expression, the present study also provides greater opportunity to identify change-of-spectrum mutants, as well as subtle changes in transactivation.
While beyond the scope of the present study, this system may also be used to address the biological activity of the multiple p53 isoforms from specific response elements. Given that p53 isoforms are known to be differentially expressed in breast tumors in comparison to normal breast tissue (76), the ratios at which these isoforms are expressed, which can change with the presence of a mutation, may alter the transactivation profiles of wild type and/or mutant p53. For example, similar to the p53 family members p63 and p73, specific p53 isoforms can influence promoter selection or functionality from specific response elements through either synergistic or antagonistic mechanisms (76, 77).
Results obtained with the yeast functional assay appear predictive of whether a mutation will also have a biological impact in mammalian cells (16). While the yeast-based assay can predict the potential for specific p53 mutations to display altered function, assays in mammalian cells can ascertain the full impact of the functional mutations in the presence of p53 transcriptional cofactors and in the endogenous chromatin context of the RE. However, we propose that assessments of functional fingerprints for p53 missense mutations associated with breast cancer in yeast provide diagnostic value and with further study may also be used as a prognostic tool for implementing chemotherapeutic treatment. This would be particularly relevant to the tailoring of individual therapies, especially when the treatment agents impact p53-dependent biological responses. Furthermore, the technique of functional fingerprinting will be useful in determining if p53 dysfunction as a transcription factor can also be corrected by agents that reverse its structural stability, such as the carbazole derivative PhiKan083 acting on Y220C (78).
Supplementary Material
Supplemental Figure 1. Functional fingerprint of 21 altered-function p53 missense mutants. Transactivation capacities of WT and mutant p53 were assessed from 11 REs with the ADE2 plate color assay at variable levels of p53 induction (shown are 0.008, 0.016, 0.032, and 0.128% galactose). Transactivation was assessed by change in colony pigmentation and scored according to the extent of transactivation from weak to strong (see color chart and also Figure 2). Relative transactivation capacities revealed unique functional fingerprints for the altered-function mutants that differentiate the specific mutant from WT p53 as well as other mutants. Consistent with previous results (2), the change-in-spectrum mutant T125R suppressed growth at low expression levels in the diploid yeast (0.032-0.064% galactose).
Supplemental Figure 2. Differential transactivation capacities of WT and mutant p53 towards REs at variable expression using the plate color assay. The ADE2 reporter provides a qualitative assay for determining transactivation potential through the accumulation over several days of pigmentation in stationary cells. Variable levels of WT or mutant p53 cDNA is expressed from the GAL1 promoter dependent on the amount of galactose in the media. A. The color assay can be used to differentiate WT p53 transactivation potential from different REs at comparable levels of galactose induction (32). Presented is the ability of WT p53 to transactivate from 5 different response elements at 0.004% galactose. B. Colony pigmentation from red (i.e., no detectable transactivation) to white of the mutant p53 relative to WT p53 was scored visually using 2-3 biological replicates per plate. To ensure consistency in evaluation of transactivation potential, the ADE2 assay was repeated at least twice on galactose plates prepared on different days. Presented is the ability of WT, R337C, and R337H to transactivate from the 14-3-3σ RE on plates containing raffinose plus 0.016% (left panel) or 0.032% galactose (right panel). Colony pigmentation was determined using a scale of 1 to 5, where 1 is no apparent transactivation (deep red colonies; R337C at 0.016% galactose) and 5 is strong transactivation (white colonies; WT at 0.016% and 0.032% galactose). Intermediate levels of transactivation were assessed qualitatively as follows: “2” were less red than “1”; a value of “3” corresponded to a distinct pink; and “4” was a tinted white with pink undertones.
Supplemental Figure 3. WT and mutant p53 expression. A. To assess if the variation in transactivation capacity towards the various REs by mutant p53 was due to differences in p53 protein levels, the relative expression of p53 mutants was compared to WT p53 at 0.024% galactose (p21-5′ RE-luciferase reporter strains were used). The p53 protein was detected with a mix of DO7 (BD BioSciences Pharmenigen) and pAb1801 (Santa Cruz) antibodies. Membranes were stained with Ponceau S to determine efficiency of protein loading. B. Longer exposures show there are detectable levels of p53 protein with the Y220C, M237I, E385K and R337C mutants. N239D is a loss-of-function mutation.
Supplemental Figure 4. Transactivation from the p21-5′ RE by p53 missense mutants with reduced protein expression. A. Western analysis was performed on the p53 mutants expressing reduced levels of p53 in comparison to WT and R337H using the combined D0-7 (BD BioSciences Pharmenigen) and pAb240 (Santa Cruz) antibodies to detect p53. The pAb240 antibody recognizes different epitope from those used in Supplemental Figure 3. Similar to results in Supplemental Figure 3, the protein expression was detectable, but reduced in comparison to WT p53. B. Transactivation from the p21-5′ RE was assessed for WT and p53 mutants with the luciferase assay. Transactivation from the p21-5′ (assessed with the same samples used in the Western blots) was significantly reduced for the L194P, Y220C, M237I and P278A mutants in comparison to WT p53, which reflected the decreased expression of protein. However, C141W and E285K induced activity from the p21-5′ RE to ~25 and 40% of the levels of WT p53, respectively which remained disproportionate to the levels of protein expressed. Based on the transactivation and protein expression results, there is no obvious explanation for the observation that these mutants can transactivate from the majority of the REs analyzed with such a compromised protein level and would suggest the mutants were more active than WT p53 based on unit of protein. However, Schärer and Iggo (81) published similar results concerning E285K where the levels of protein in yeast were decreased in comparison to WT p53, yet when assessed for transactivation from a 33 base pair sequence at 29°C, E285K activity was ~75% of WT p53; this finding was attributed to protein instability.
Acknowledgements
JJJ was supported by a Department of Defense Breast Cancer Research Program Predoctoral Traineeship Award (BC051212). This work was partially supported by the Italian Association for Cancer Research, AIRC (to AI), the Breast Cancer Specialized Program of Research Excellence award from the NCI (CA58223) to the University of North Carolina and NIH M01RR00046 (to LAC), the Department of Defense award DAMD 17-02-1-0521 (to LAC), an Avon/Partners for Progress grant (to LAC) and intramural research funds from NIEHS to MAR, project Z01-ES065079.
Abbreviations
- ACC
adrenocortical carcinoma
- BRCA1/2
breast cancer susceptibility gene
- CORE
COunterselectable REporter cassette
- DBD
DNA binding domain
- ER
estrogen receptor
- FH
familial history
- HER2
human epidermal growth factor-2
- IARC
International Agency for Research on Cancer
- LFL
Li-Fraumeni-like Syndrome
- LFS
Li-Fraumeni Syndrome
- pCR
pathologic complete response
- PR
progesterone receptor
- REs
response elements
- WT
wild-type
Footnotes
Trial registration: CALGB 150007, CDR0000069280
Competing Interests: No competing interests exist.
References
- 1.Ko LJ, Prives C. p53: puzzle and paradigm. Genes & development. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci U S A. 2003;100:9934–9. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menendez D, Inga A, Jordan JJ, Resnick MA. Changing the p53 master regulatory network: ELEMENTary, my dear Mr Watson. Oncogene. 2007;26:2191–201. doi: 10.1038/sj.onc.1210277. [DOI] [PubMed] [Google Scholar]
- 4.Jordan JJ, Menendez D, Inga A, Nourredine M, Bell D, Resnick MA. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet. 2008;4:e1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–23. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer research. 1994;54:4855–78. [PubMed] [Google Scholar]
- 7.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 8.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–93. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann RK, Yu K, Eby Y, Pavletich NP, Boeke JD. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. Embo J. 1998;17:1847–59. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisz L, Oren M, Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–11. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- 11.Kim E, Deppert W. Transcriptional activities of mutant p53: when mutations are more than a loss. J Cell Biochem. 2004;93:878–86. doi: 10.1002/jcb.20271. [DOI] [PubMed] [Google Scholar]
- 12.Kato S, Han SY, Liu W, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–9. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowan S, Ludwig RL, Haupt Y, et al. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. Embo J. 1996;15:827–38. [PMC free article] [PubMed] [Google Scholar]
- 14.Campomenosi P, Monti P, Aprile A, et al. p53 mutants can often transactivate promoters containing a p21 but not Bax or PIG3 responsive elements. Oncogene. 2001;20:3573–9. doi: 10.1038/sj.onc.1204468. [DOI] [PubMed] [Google Scholar]
- 15.Flaman JM, Robert V, Lenglet S, Moreau V, Iggo R, Frebourg T. Identification of human p53 mutations with differential effects on the bax and p21 promoters using functional assays in yeast. Oncogene. 1998;16:1369–72. doi: 10.1038/sj.onc.1201889. [DOI] [PubMed] [Google Scholar]
- 16.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Molecular and cellular biology. 2006;26:2297–308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer research. 2001;61:4092–7. [PubMed] [Google Scholar]
- 18.Crook T, Crossland S, Crompton MR, Osin P, Gusterson BA. p53 mutations in BRCA1-associated familial breast cancer. Lancet. 1997;350:638–9. doi: 10.1016/S0140-6736(05)63327-2. [DOI] [PubMed] [Google Scholar]
- 19.Scata KA, El-Deiry WS. p53, BRCA1 and breast Cancer chemoresistance. Adv Exp Med Biol. 2007;608:70–86. doi: 10.1007/978-0-387-74039-3_5. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Qiao W, Linke SP, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–71. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 21.Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006;8:R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chappuis PO, Estreicher A, Dieterich B, et al. Prognostic significance of p53 mutation in breast cancer: frequent detection of non-missense mutations by yeast functional assay. International journal of cancer. 1999;84:587–93. doi: 10.1002/(sici)1097-0215(19991222)84:6<587::aid-ijc8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Langerod A, Zhao H, Borgan O, et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9:R30. doi: 10.1186/bcr1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 26.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troester MA, Herschkowitz JI, Oh DS, et al. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer. 2006;6:276. doi: 10.1186/1471-2407-6-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carey LA, Metzger R, Dees EC, et al. American Joint Committee on Cancer tumor-node-metastasis stage after neoadjuvant chemotherapy and breast cancer outcome. Journal of the National Cancer Institute. 2005;97:1137–42. doi: 10.1093/jnci/dji206. [DOI] [PubMed] [Google Scholar]
- 29.Esserman LJ, Perou C, Cheang M, et al. Breast cancer molecular profiles and tumor response of neoadjuvant doxorubicin and paclitaxel: The ISPY Trial (CALGB 150007/150012, ACRIN 6657) J Clin Oncol. 2009;27(18s) [Google Scholar]
- 30.Geisler S, Borresen-Dale AL, Johnsen H, et al. TP53 gene mutations predict the response to neoadjuvant treatment with 5-fluorouracil and mitomycin in locally advanced breast cancer. Clin Cancer Res. 2003;9:5582–8. [PubMed] [Google Scholar]
- 31.Geisler S, Lonning PE, Aas T, et al. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer research. 2001;61:2505–12. [PubMed] [Google Scholar]
- 32.Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Molecular and cellular biology. 2002;22:8612–25. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monti P, Campomenosi P, Ciribilli Y, et al. Tumour p53 mutations exhibit promoter selective dominance over wild type p53. Oncogene. 2002;21:1641–8. doi: 10.1038/sj.onc.1205250. [DOI] [PubMed] [Google Scholar]
- 34.Mathe E, Olivier M, Kato S, Ishioka C, Vaisman I, Hainaut P. Predicting the transactivation activity of p53 missense mutants using a four-body potential score derived from Delaunay tessellations. Hum Mutat. 2006;27:163–72. doi: 10.1002/humu.20284. [DOI] [PubMed] [Google Scholar]
- 35.Di Como CJ, Prives C. Human tumor-derived p53 proteins exhibit binding site selectivity and temperature sensitivity for transactivation in a yeast-based assay. Oncogene. 1998;16:2527–39. doi: 10.1038/sj.onc.1202041. [DOI] [PubMed] [Google Scholar]
- 36.Bullock AN, Henckel J, Fersht AR. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19:1245–56. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 37.Shiraishi K, Kato S, Han SY, et al. Isolation of temperature-sensitive p53 mutations from a comprehensive missense mutation library. J Biol Chem. 2004;279:348–55. doi: 10.1074/jbc.M310815200. [DOI] [PubMed] [Google Scholar]
- 38.Wasielewski M, Elstrodt F, Klijn JG, Berns EM, Schutte M. Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines. Breast cancer research and treatment. 2006;99:97–101. doi: 10.1007/s10549-006-9186-z. [DOI] [PubMed] [Google Scholar]
- 39.Concin N, Zeillinger C, Tong D, et al. Comparison of p53 mutational status with mRNA and protein expression in a panel of 24 human breast carcinoma cell lines. Breast cancer research and treatment. 2003;79:37–46. doi: 10.1023/a:1023351717408. [DOI] [PubMed] [Google Scholar]
- 40.Fan LZ, Cherian MG. Potential role of p53 on metallothionein induction in human epithelial breast cancer cells. British journal of cancer. 2002;87:1019–26. doi: 10.1038/sj.bjc.6600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. International journal of cancer. 1998;78:766–74. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 42.Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem. 2005;280:16030–7. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 43.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–55. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 44.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 45.Joerger AC, Fersht AR. Structure-function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–42. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 46.Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci U S A. 2006;103:15056–61. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 48.Davison TS, Yin P, Nie E, Kay C, Arrowsmith CH. Characterization of the oligomerization defects of two p53 mutants found in families with Li-Fraumeni and Li-Fraumeni-like syndrome. Oncogene. 1998;17:651–6. doi: 10.1038/sj.onc.1202062. [DOI] [PubMed] [Google Scholar]
- 49.Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat. 2003;21:313–20. doi: 10.1002/humu.10185. [DOI] [PubMed] [Google Scholar]
- 50.Achatz MI, Olivier M, Le Calvez F, et al. The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett. 2007;245:96–102. doi: 10.1016/j.canlet.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 51.Ribeiro RC, Rodriguez-Galindo C, Figueiredo BC, et al. Germline TP53 R337H mutation is not sufficient to establish Li-Fraumeni or Li-Fraumeni-like syndrome. Cancer Lett. 2007;247:353–5. doi: 10.1016/j.canlet.2006.04.008. author reply 6-8. [DOI] [PubMed] [Google Scholar]
- 52.Palmero EI, Schuler-Faccini L, Caleffi M, et al. Detection of R337H, a germline TP53 mutation predisposing to multiple cancers, in asymptomatic women participating in a breast cancer screening program in Southern Brazil. Cancer Lett. 2008;261:21–5. doi: 10.1016/j.canlet.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 53.DiGiammarino EL, Lee AS, Cadwell C, et al. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat Struct Biol. 2002;9:12–6. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 54.Lee AS, Galea C, DiGiammarino EL, et al. Reversible amyloid formation by the p53 tetramerization domain and a cancer-associated mutant. J Mol Biol. 2003;327:699–709. doi: 10.1016/s0022-2836(03)00175-x. [DOI] [PubMed] [Google Scholar]
- 55.Tsuda H, Hirohashi S. Association among p53 gene mutation, nuclear accumulation of the p53 protein and aggressive phenotypes in breast cancer. International journal of cancer. 1994;57:498–503. doi: 10.1002/ijc.2910570410. [DOI] [PubMed] [Google Scholar]
- 56.Sjogren S, Inganas M, Norberg T, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. Journal of the National Cancer Institute. 1996;88:173–82. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- 57.Lang GA, Iwakuma T, Suh YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Olive KP, Tuveson DA, Ruhe ZC, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Terzian T, Suh YA, Iwakuma T, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes & development. 2008;22:1337–44. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Molecular and cellular biology. 2007;27:8284–95. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–27. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 62.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–91. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–13. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allison SJ, Milner J. Remodelling chromatin on a global scale: a novel protective function of p53. Carcinogenesis. 2004;25:1551–7. doi: 10.1093/carcin/bgh212. [DOI] [PubMed] [Google Scholar]
- 65.Melchor L, Honrado E, Garcia MJ, et al. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2008;27:3165–75. doi: 10.1038/sj.onc.1210975. [DOI] [PubMed] [Google Scholar]
- 66.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–40. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 67.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Olivier M, Goldgar DE, Sodha N, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer research. 2003;63:6643–50. [PubMed] [Google Scholar]
- 69.Birch JM, Hartley AL, Tricker KJ, et al. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer research. 1994;54:1298–304. [PubMed] [Google Scholar]
- 70.Monti P, Ciribilli Y, Jordan J, et al. Transcriptional functionality of germ line p53 mutants influences cancer phenotype. Clin Cancer Res. 2007;13:3789–95. doi: 10.1158/1078-0432.CCR-06-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shlien A, Tabori U, Marshall CR, et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci U S A. 2008;105:11264–9. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olivier M, Langerod A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–67. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 73.Kandioler-Eckersberger D, Ludwig C, Rudas M, et al. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin Cancer Res. 2000;6:50–6. [PubMed] [Google Scholar]
- 74.Di Leo A, Tanner M, Desmedt C, et al. p-53 gene mutations as a predictive marker in a population of advanced breast cancer patients randomly treated with doxorubicin or docetaxel in the context of a phase III clinical trial. Ann Oncol. 2007;18:997–1003. doi: 10.1093/annonc/mdm075. [DOI] [PubMed] [Google Scholar]
- 75.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 76.Bourdon JC, Fernandes K, Murray-Zmijewski F, et al. p53 isoforms can regulate p53 transcriptional activity. Genes & development. 2005;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Janicke RU, Graupner V, Budach W, Essmann F. The do’s and don’ts of p53 isoforms. Biological chemistry. 2009;390:951–63. doi: 10.1515/BC.2009.093. [DOI] [PubMed] [Google Scholar]
- 78.Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci U S A. 2008;105:10360–5. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods in enzymology. 2006;409:329–45. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 80.Conway K, Edmiston SN, Cui L, et al. Prevalence and spectrum of p53 mutations associated with smoking in breast cancer. Cancer research. 2002;62:1987–95. [PubMed] [Google Scholar]
- 81.Scharer E, Iggo R. Mammalian p53 can function as a transcription factor in yeast. Nucleic Acids Res. 1992;20:1539–45. doi: 10.1093/nar/20.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Functional fingerprint of 21 altered-function p53 missense mutants. Transactivation capacities of WT and mutant p53 were assessed from 11 REs with the ADE2 plate color assay at variable levels of p53 induction (shown are 0.008, 0.016, 0.032, and 0.128% galactose). Transactivation was assessed by change in colony pigmentation and scored according to the extent of transactivation from weak to strong (see color chart and also Figure 2). Relative transactivation capacities revealed unique functional fingerprints for the altered-function mutants that differentiate the specific mutant from WT p53 as well as other mutants. Consistent with previous results (2), the change-in-spectrum mutant T125R suppressed growth at low expression levels in the diploid yeast (0.032-0.064% galactose).
Supplemental Figure 2. Differential transactivation capacities of WT and mutant p53 towards REs at variable expression using the plate color assay. The ADE2 reporter provides a qualitative assay for determining transactivation potential through the accumulation over several days of pigmentation in stationary cells. Variable levels of WT or mutant p53 cDNA is expressed from the GAL1 promoter dependent on the amount of galactose in the media. A. The color assay can be used to differentiate WT p53 transactivation potential from different REs at comparable levels of galactose induction (32). Presented is the ability of WT p53 to transactivate from 5 different response elements at 0.004% galactose. B. Colony pigmentation from red (i.e., no detectable transactivation) to white of the mutant p53 relative to WT p53 was scored visually using 2-3 biological replicates per plate. To ensure consistency in evaluation of transactivation potential, the ADE2 assay was repeated at least twice on galactose plates prepared on different days. Presented is the ability of WT, R337C, and R337H to transactivate from the 14-3-3σ RE on plates containing raffinose plus 0.016% (left panel) or 0.032% galactose (right panel). Colony pigmentation was determined using a scale of 1 to 5, where 1 is no apparent transactivation (deep red colonies; R337C at 0.016% galactose) and 5 is strong transactivation (white colonies; WT at 0.016% and 0.032% galactose). Intermediate levels of transactivation were assessed qualitatively as follows: “2” were less red than “1”; a value of “3” corresponded to a distinct pink; and “4” was a tinted white with pink undertones.
Supplemental Figure 3. WT and mutant p53 expression. A. To assess if the variation in transactivation capacity towards the various REs by mutant p53 was due to differences in p53 protein levels, the relative expression of p53 mutants was compared to WT p53 at 0.024% galactose (p21-5′ RE-luciferase reporter strains were used). The p53 protein was detected with a mix of DO7 (BD BioSciences Pharmenigen) and pAb1801 (Santa Cruz) antibodies. Membranes were stained with Ponceau S to determine efficiency of protein loading. B. Longer exposures show there are detectable levels of p53 protein with the Y220C, M237I, E385K and R337C mutants. N239D is a loss-of-function mutation.
Supplemental Figure 4. Transactivation from the p21-5′ RE by p53 missense mutants with reduced protein expression. A. Western analysis was performed on the p53 mutants expressing reduced levels of p53 in comparison to WT and R337H using the combined D0-7 (BD BioSciences Pharmenigen) and pAb240 (Santa Cruz) antibodies to detect p53. The pAb240 antibody recognizes different epitope from those used in Supplemental Figure 3. Similar to results in Supplemental Figure 3, the protein expression was detectable, but reduced in comparison to WT p53. B. Transactivation from the p21-5′ RE was assessed for WT and p53 mutants with the luciferase assay. Transactivation from the p21-5′ (assessed with the same samples used in the Western blots) was significantly reduced for the L194P, Y220C, M237I and P278A mutants in comparison to WT p53, which reflected the decreased expression of protein. However, C141W and E285K induced activity from the p21-5′ RE to ~25 and 40% of the levels of WT p53, respectively which remained disproportionate to the levels of protein expressed. Based on the transactivation and protein expression results, there is no obvious explanation for the observation that these mutants can transactivate from the majority of the REs analyzed with such a compromised protein level and would suggest the mutants were more active than WT p53 based on unit of protein. However, Schärer and Iggo (81) published similar results concerning E285K where the levels of protein in yeast were decreased in comparison to WT p53, yet when assessed for transactivation from a 33 base pair sequence at 29°C, E285K activity was ~75% of WT p53; this finding was attributed to protein instability.