Abstract
Objective:
The cellular innate immune response to HIV-1 is poorly characterised. In view of HIV-1 tropism for macrophages, which can be activated via pattern recognition receptors (PRR) to trigger antimicrobial defenses, we investigated innate immune responses to HIV-1 by monocyte derived macrophages.
Design:
In a model of productive HIV-1 infection, cellular innate immune responses to HIV-1 were investigated, at the level of transcription factor activation, specific gene expression and genome-wide transcriptional profiling. In addition, the viral determinants of macrophage responses and the physiological effect of innate immune cellular activation on HIV-1 replication were assessed.
Results:
Productive HIV-1 infection did not activate NF-κB and IRF3 transcription factors or interferon gene expression (IFN), and caused remarkably small changes to the host cell transcriptome, with no evidence of inflammatory or IFN signatures. Evasion of IFN induction was not dependent on HIV-1 envelope mediated cellular entry, inhibition by accessory proteins or reverse transcription of ssRNA that may reduce innate immune cellular activation by viral RNA. Furthermore, IFNβ priming did not sensitize responses to HIV-1. Importantly, exogenous IFNβ or stimulation with the RNA analogue poly I:C to simulate innate immune activation invoked HIV-1 restriction.
Conclusions:
We conclude that macrophages lack functional PRRs for this virus and that HIV-1 tropism for macrophages helps to establish a foothold in the host without triggering innate immune cellular activation which would otherwise block viral infection effectively.
Keywords: HIV-1, Innate immunity, Macrophages, Interferons, Pattern recognition receptors
Introduction
Many lines of evidence indicate that macrophages contribute significantly to the ability of HIV-1 to establish infection in a new host [1]. The HIV-1 population during primary infection is predominantly macrophage tropic. Evidence for HIV-1 infection of macrophages in vivo has been reported, the ability of HIV-1 to infect monocyte-derived macrophages in vitro is well established and HIV-1 infected macrophages have been shown to augment T cell infection through a virological synapse [2]. Importantly, macrophages are sentinel cells of innate immunity, that can be activated by a wide range of pathogen associated molecular patterns (PAMP) and orchestrate local and systemic immune responses [3]. However, the innate immune response to HIV-1 infection in these cells and its physiological significance has not been investigated systematically. Viruses induce cellular innate immune responses via pattern recognition receptors (PRR) that invoke antiviral defenses through activation of NF-κB, interferon regulatory factor (IRF)3 and IRF7 transcription factors, induction of type 1 interferons (IFN) and of IFN-stimulated genes (ISG) [4]. It is evident that a number of ISGs, such as APOBEC3G, tetherin and TRIM5α can restrict HIV-1 infection, and that the virus has evolved mechanisms to overcome these [5-9].
PRRs that may mediate innate immune cellular activation in response to RNA viruses are mainly members of the Toll-like receptor (TLR) family, TLR3, TLR7 or TLR8, and members of the RIG-Iike RNA helicase (RLH) family, RIG-I and MDA-5. Uridine-rich oligonucleotides derived from HIV-1 RNA have been shown to induce innate immune activation of murine macrophages, human peripheral mononuclear cells and the human myeloid leukaemia cell line THP-1 primed with IFNγ [10, 11]. In addition, the principle chemokine receptor for HIV-1 infection of macrophages, CCR5, may also mediate activation of NF-κB and mitogen-activated protein kinase (MAPK) pathways when stimulated by recombinant HIV-1 envelope gp-120 [12, 13], and transcriptional responses following gp120 stimulation of macrophages have been described [14]. The physiological significance of these observations in HIV-1 infection of macrophages is not known.
In view of the role of macrophages as key cellular targets in early HIV-1 infection, it is critically important to understand their ability to mount an antiviral innate immune response and to establish whether this is an important determinant of HIV-1 tropism for macrophages. We show that productive HIV-1 infection of macrophages is critically dependent on evasion of innate immune cellular activation and not evasion of inducible antiviral effector mechanisms. Our data show that macrophages lack constitutive and IFN-inducible expression of a functional PRR for HIV-1 and suggest that HIV-1 exploits a uniquely permissive niche in a host cell that is otherwise adept at innate immune detection of pathogens.
Methods
Peripheral blood mononuclear cells and monocyte derived macrophages
Human blood samples were obtained from healthy volunteers for isolation of peripheral blood mononuclear cells (PBMC) and production of MDM cultures as previously described [15]. The study was approved by the joint University College London/University College London Hospitals National Health Service Trust Human Research Ethics Committee and written informed consent was obtained from all participants. Adherent monocytes were differentiated into macrophages using one of three conditions: RPMI 1640 (GIBCO Invitrogen) with 20% autologous heat-inactivated human serum (HS), RPMI 1640 with 10% HS and either 20 ng/mL macrophage colony stimulating factor (M-CSF) (R&D systems) or 100 ng/mL granulocyte macrophage colony stimulating factor (GM-CSF) (gift from Schering-Plough Research Institute, Kenilworth, NJ) for 3 days (d). The medium was then refreshed (RPMI 1640 with 10% HS), removing any remaining non-adherent cells. After 6 days culture 10% autologous HS was replaced with 5% type AB HS (Sigma-Aldrich-Aldrich). Replicate experiments were performed with cells derived from different donors.
HIV-1 strains and cell culture infections
The CCR5 tropic HIV-1 strains, Ba-L, ADA-M, JR-CSF, JR-FL from National Institute for Biological Standards and Controls (NIBSC) and a primary HIV-1 isolate (designated MM25-2) derived from a clinical blood sample from a patient with primary HIV-1 infection [16] were propagated in peripheral blood lymphocytes (PBL) [15]. The CCR5 tropic HIV-1 strain Yu2 and CXCR4 tropic strains NL4-3 were derived from infectious clones by transient transfection of HEK293t cells [15]. HIV-1 vif deficient Yu2 packaged with APOBEC3G was produced by co-transfection (ratio 1:1) of HEK293t cells with the molecular clone for each (gifts from Kate Bishop, Kings College London). Vesicular stomatitis virus (VSV)-G envelope pseudotyped replication deficient HIV-1 was produced by co-transfection of HEK293t cells with equal amounts of VSV-G, p8.91 HIV-1 gag-pol and HIV-1 derived pHRSIN-CSGW expression vectors encoding eGFP [9]. All these viruses were ultracentrifuged through a 20% sucrose buffer and resuspended in RPMI 1640 with 5% NHS, for subsequent infection of MDM. Replication competent HIV-1 virus preparations were titrated on the NP2 astrocytoma cell line stably transfected with CD4 and CCR5 or CXCR4 [15]. The titre of VSV-G pseudotyped HIV-1 vector was measured by flow cytometric detection of GFP in HEK293t cells [9]. The concentrations of non-replicative HIV-1 vif deficient Yu2 packaged with APOBEC3G were assessed by an ELISA assay for p24 (below) standardised with titrated replication competent HIV-1 virus strains. Influenza A /Duck /England /62 (H4N6) was propagated and titrated in MDCK cells [17]. High titre (>8 ×109 plaque forming units /mL) Influenza A virus were used to allow >1000-fold dilutions, hence minimising risk of significant contamination with immunologically active moieties from MDCK cell cultures. Viral infection of macrophages was performed by 3 hour (h) inoculation of 6 day old MDM cultures using doses equivalent to multiplicity of infection (MOI) of 3.
Detection of extracellular and intracellular p24
Cell-free HIV-1 gag (p24) concentrations were quantified by ELISA with recombinant standards (kit version 5, from AIDS Vaccine Programme, National Cancer Institute- Fredrick). Intracellular detection of p24 was performed as previously described [15].
Reverse transcriptase (RT) PCR detection of HIV-1 transcripts
MDM culture lysates collected in RLT buffer (Qiagen) were used to purify total RNA with RNAeasy spin columns (Qiagen). cDNA was generated using oligo dT primers (First Strand cDNA synthesis kit, New England Biolabs). PCR was performed on the heat inactivated (95°C, 5 min) product using primers for spliced HIV-1 (413MOD and P659) [18] and β actin (Supplementary Table S1). No PCR product was evident without reverse transcription and relative amounts of HIV-1 transcript were assessed semi-quantitatively by densitometry and limiting dilution PCR, normalised to β actin levels.
Innate immune and cytokine stimulation of MDM
Poly I:C, ultrapure lipopolysaccharide from E. coli O111:B4 and CL075 (3M002) (Invivogen) were used as minimal innate immune stimuli. Recombinant human interferon (IFN)β was obtained from Merck Serono. Recombinant human IFNγ was obtained from Peprotech.
Quantitative confocal immunofluorescence analysis of NF-κB and IRF3 nuclear translocation
Nuclear:cytoplasmic ratios of NF-κB RelA and IRF3 transcription factors were derived from image analysis (Metamorph v7.17, Molecular devices) of confocal immunofluorescence stains obtained with the Leica SP2 confocal microscope using rabbit polyclonal anti NF-κB RelA (C-20) (Santa Cruz Biotechnology) or rabbit polyclonal anti IRF3 (FL 425) (Santa Cruz Biotechnology) (Supplementary Figure S2) [15, 19].
Quantitative PCR detection of gene transcription
Total RNA was subjected to on-column DNase treatment (Qiagen) or the Turbo DNA free kit (Ambion), and cDNA generated as described above. Taqman qPCR using the Mastercycler Realplex4 (Eppendorf) or ABI Prism 7000 (Applied Biosystems) was performed for selected genes using primer /probe sequences (Table S1). The reaction mixture for each qPCR reaction was performed with 200 ng cDNA, 7.5 μM primers and probes and Platinum Quantitative PCR SuperMix-UDG w/ROX (Invitrogen). qPCR products were quantified using standards (cDNA plasmid clones for GAPDH, IFNβ, IP10 and TRIM5α) or by 2−ΔΔCt method (STAT1 and IRF7) after normalization for GAPDH.
Transcriptional profiling by DNA microarray
Total RNA integrity and concentrations were analysed electrophoretically [20]. Cy5 labelled cRNA was generated (Agilent Low RNA Input Linear Amplification Kit), mixed in equally with Cy3 labelled reference cRNA derived similarly from a universal human reference RNA mix (Stratagene), and used hybridised with Agilent 4×44K whole human genome cDNA microarrays (www.agilent.com). Array images were acquired with Aglient's dual-laser microarray scanner G2565BA and signal data were collected with Agilent Feature Extraction software (v9.5.1). Data were normalised as previously described [15] and gene expression values were compared by unpaired T-test (p<0.05) using MultiExperiment Viewer v4.0 [21]. Gene lists of interest were annotated using DAVID functional annotation clustering (http://david.abcc.ncifcrf.gov) [22, 23]. This analysis was restricted to genes with refseq accession numbers for which contemporary functional annotation is available. Multidimensional scaling was used to compare relative similarity /distance between normalized expression data for selected gene lists, by generating a 3 dimensional representation of the Euclidean distance matrix, to give stress values of ≤0.1 (XLSTAT v2008.7.01). MIAME compliant microarray data have been submitted to the ArrayExpress database (www.ebi.ac.uk/arrayexpress), accession number: E-MEXP-2032.
Results
Productive HIV-1 infection in MDM
An established model of HIV-1 infection in MDM was used in this study [15]. Viral replication of the CCR5-tropic HIV-1 strain Ba-L is evident within 24 h by detection of spliced HIV-1 transcript (Supplementary Figure S1A-B) and HIV-1 release into the culture supernatant is seen after 24 h (Supplementary Figure S1C). Virus replication increases to reach a plateau at about 7 d after infection, at which time MDM inoculated with HIV-1 Ba-L show uniform positive intracellular p24 staining (Supplementary Figure S1D). MDM do not support productive infection with T cell tropic CXCR4 using HIV-1 strains such as NL4-3 (Supplementary Figure S1C-D).
Measurement of NF-κB activation in HIV-1 infection of MDM
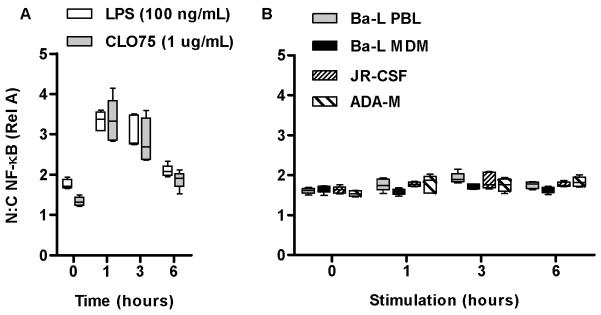
Activation of the classical NF-κB pathway leading to nuclear translocation of the NF-κB p65 (RelA) transcription factor is a key event in innate immune cellular activation [24] and has been reported to occur in response to macrophage stimulation with recombinant gp120 from HIV-1 Ba-L or 2,2′-dithiodipyridine denatured HIV-1 Ba-L virions [12]. We therefore tested activation of this pathway in M-CSF differentiated MDM following inoculation with live HIV-1 Ba-L. In a time course study (0-6 h), stimulation of MDM via specific ligands for TLR4, or innate immune receptors for ssRNA TLR7/8, induced NF-κB RelA translocation that was maximal at 1 h (Figure 1A). In contrast stimulation of MDM with HIV-1 Ba-L or other CCR5-tropic strains propagated in peripheral blood lymphocytes (PBL) using inocula that establish productive infection, did not activate NF-κB RelA translocation (Figure 1B). Therefore, lack of classical NF-κB activation was not specific to virus strain or the host-cell derivation of the virus. Stimulation of MDM with HIV-1 similarly failed to induce nuclear translocation of IRF3 (Supplementary Figure S3A).
Figure 1. HIV-1 does not induce NF-κB nuclear translocation or type 1 IFN responses in MDM.
MDM stimulation with LPS (100 ng/mL) or CL075 (1 μg/mL) induces significant (p<0.0001, ANOVA) nuclear translocation of NF-κB RelA that is maximal at 1 h (A). In contrast, HIV-1 Ba-L derived from MDM or PBL and additional macrophage tropic strains ADA-M and JR-CSF propgated in PBL do not induce any detectable NF-κB activation (B) Median, interquartile and full ranges derived from approximately 500 cells are shown. Data are representative of 3 replicate experiments.
Measurement of innate immune IFN responses by MDM
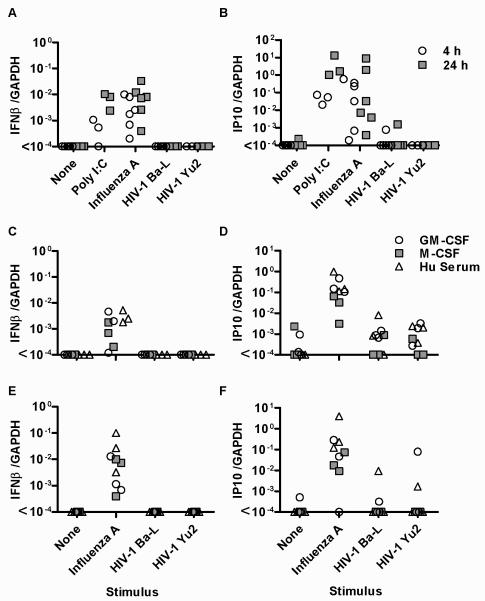
NF-kB and IRF3 independent activation of IFN responses has been reported in macrophages [25]. Therefore, we measured mRNA levels of IFNβ by quantitative (q)PCR, as well as the IFN-sensitive gene, the chemokine CXCL10 also known as interferon-inducible peptide (IP)10. In response to stimulation of MDM with the synthetic dsRNA ligand poly I:C, or with a prototypic ssRNA virus Influenza A, IFNβ and IP10 transcription were upregulated (Figure 2A-B). However, in keeping with our data on transcription factors, we did not detect any consistent response to HIV-1 strains. Importantly, all IFNs increase IP10 expression, therefore the absence of stochastic IP10 induction secondary to an IFN response indicated a lack of induction of any functional IFN activity. In contrast, virus preparations that were not subjected to sucrose purification did induce upregulation of IP10 in macrophages (Supplementary Figure S3B), suggesting that unpurified virus preparations contain contaminating immunoreactive soluble factors from the producer cells that may explain some of the inconsistency in reports of macrophage responses to HIV-1 infection. Only sucrose purified virus was used in all other experiments in this study.
Figure 2. HIV-1 does not induce type 1 IFN responses in MDM.
Stimulation of M-CSF differentiated MDM with poly I:C (10 μg/mL) or Infuenza A (MOI 3) induces siginificantly increased gene expression of IFNβ (A) and IP10 (B) quantified by qPCR (p<0.01, Wicoxon sign-ranked test of paired samples). Neither macrophage tropic HIV-1 strains induced detectable expression of either of these genes. Comparable observations are evident in alternatively differentiated MDM after 4 h stimulation (C-D) and after 24 h stimulation (E-F). Individual data paints are shown for each experiment.
Alternatively differentiated MDM exhibit differences in both innate immune responses and their permissiveness to HIV-1. M-CSF differentiated MDM are reported to show less innate immune pro-inflammatory responses [26] and to be more permissive to HIV-1 infection than GM-CSF differentiated MDM [27-29], but we found IFNβ and IP10 responses to influenza A and to purified HIV-1 to be comparable between MDM differentiated using M-CSF, GM-CSF or autologous HS only (Figure 2C-F). Hence, deficiency of the IFN responses to HIV-1 measured here was a consistent feature of the different MDM models as well as different virus strains.
Genome-wide innate immune responses during HIV-1 infection of MDM
Innate immune cellular activation of MDM induces wide-ranging transcriptional responses that can be comprehensively assessed by genome-wide transcriptional profiling, rather than by measurement of specific candidates. Using this strategy, we previously showed that HIV-1 infection of macrophages did not significantly affect the host cell response to LPS at a systems level [15]. The focus of the present study was to investigate macrophage responses to HIV-1 itself.
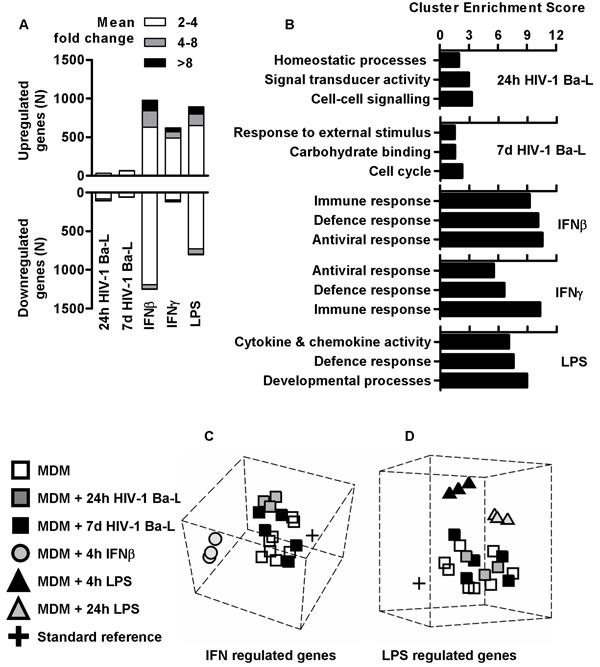
Whole genome cDNA microarrays were used to compare the transcriptomes of control and HIV-1 infected MDM. In contrast to changes in microarray transcriptional profiles induced by stimulation of uninfected MDM with lipopolysaccharide (LPS) or IFNs, both acute (within 24 h) and sustained (after 7 d) gene expression changes in HIV-1 infected MDM were small (Figure 3A and Supplementary Figure S4). Furthermore, in functional clustering analysis of gene expression changes using gene ontology associations (Figure 3B and Supplementary Table S2), LPS and IFN stimulated gene expression changes in control MDM showed highly significant enrichment of functionally related groups of genes with cytokine and chemokine activity, and genes involved in immune, defence and antiviral responses reflecting co-ordinated inflammatory and anti-viral host defence responses. In marked contrast, MDM infected with HIV-1 for 24 h or 7 d showed no such enrichment for host defence gene expression (Figure 3B and Supplementary Figure S5), suggesting that in this model, HIV-1 does not induce co-ordinated changes to host cell gene expression at the systems level, either during acute or established infection.
Figure 3. HIV-1 induces modest transcriptional changes in MDM.
(A) Significant gene expression changes (p<0.05, t-test) detected by whole genome microarray transcriptional profiling in HIV-1 Ba-L infected M-CSF differentiated MDM (at 24 h and 7 d after infection) appear quantitatively modest compared to 4 h stimulation with IFNβ (2 ng/mL), IFNγ (10 ng/mL) or LPS (10 ng/mL). Data are derived from 3 separate microarray experiments for each stimulus compared to 8 unstimulated MDM microarrays. (B) Functional annotation clustering analysis for significant gene expression differences identified by microarray transcriptional profiling in MDM infected with HIV-1 or stimulated for 4 hours with LPS (10 ng/mL), IFNβ (2 ng/ml) or IFNγ (10 ng/ml). Multidimensional scaling of the top 1000 (C) IFN- or (D) LPS-regulated genes shows that transcriptional profiles of 24 h and 7 d HIV-1 Ba-L infected M-CSF differentiated MDM cluster together with uninfected /unstimulated control MDM. Each node is derived from separate experiments indexed in the left-hand panel.
In addition, multidimensional scaling (MDS) was used to plot the relative similarity /dissimilarity of expression profiles for selected gene sets. In both IFNβ-regulated and LPS-regulated gene sets (Figure 3C-D), we found that HIV-1 infected MDM (both at 24 h and 7 d) cluster together with uninfected /unstimulated MDM. Collectively, these bioinformatic approaches unequivocally illustrate the lack of inflammatory or IFN-associated transcriptional responses upon MDM infection with HIV-1. To validate and extend these results, we used qPCR to quantify expression changes for selected IFN-regulated genes, 3 h to 7 d after HIV-1 infection and found no clear evidence for induction of any of these after HIV-1 infection of MDM (Supplementary Figure S6).
Biological significance of deficient IFN responses to HIV-1 infection in MDM
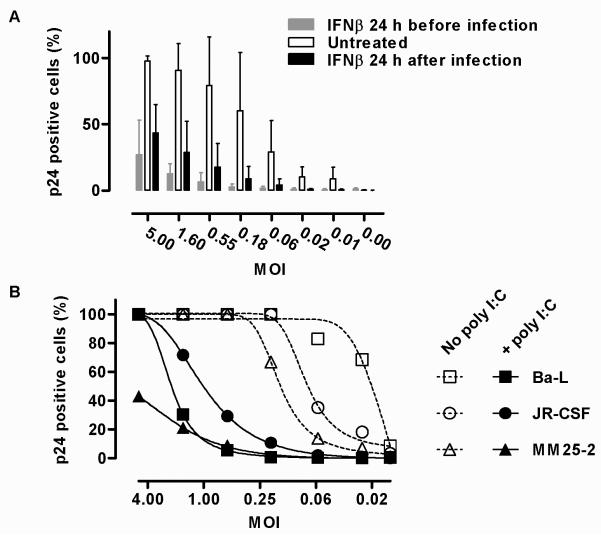
IFNs induce antiviral effects with very broad range specificity [4] and are known to restrict HIV-1 infection in macrophages [15, 30-32]. Accordingly, we found that stimulation of MDM with IFNβ 24 h before HIV-1 infection or 24 h afterwards to simulate an endogenous type 1 IFN response to viral infection reduced the HIV-1 titre by 50-100 fold (Figure 4A). This effect was evident against commonly used laboratory macrophage-tropic HIV-1 strains as well as a clinical HIV-1 isolate (Supplementary Figure S7). Concurrent stimulation of MDM with poly I:C, to invoke an endogenous innate immune IFN-response at the time of virus inoculation, also induced potent HIV-1 restriction (Figure 4B). Poly I:C mediated restriction of HIV-1 in dendritic cells has recently been shown to be dependent on type 1 IFN responses [30], but whether this effect in macrophages is wholly mediated by IFNs is not assessed further in this study. These data show that the full complement of HIV-1 mechanisms for evasion of IFN-inducible antiviral factors is insufficient to escape host cell defences, and that evasion of IFN-induction significantly enhances productive infection of MDM.
Figure 4. IFNβ and poly I:C restrict productive HIV-1 infection in MDM.
(A) Titration of HIV-1 Ba-L infection in M-CSF differentiated MDM assessed by the proportion of p24 positive cells at 7 d, shows a significant (p<0.001, ANOVA) reduction in MDM stimulated with IFNβ (2 ng/mL) 24 h before or 24 h after viral infection. Mean±SD of 3 separate experiments are shown. (B) Similar significant (p<0.001, ANOVA) reduction of HIV-1 infection (for each of the strains indicated) is induced by stimulation of MDM with poly I:C (10 μg/mL) at the time of inoculation with HIV-1 (mean of 2 experimental replicates).
Macrophages lack a functional innate immune receptor for HIV-1
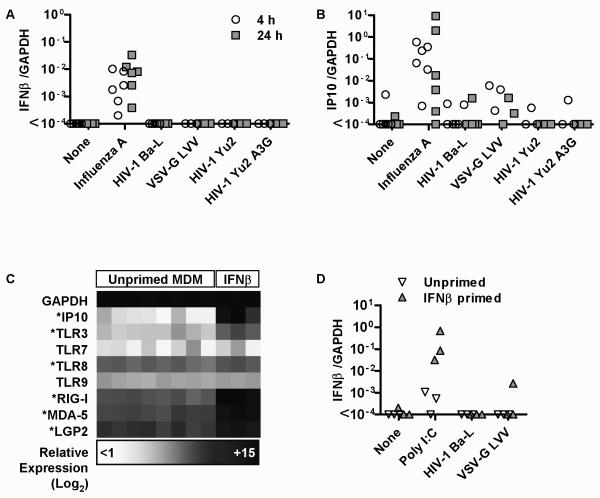
In order to investigate the mechanism for our observations, we next tested MDM responses to a VSV-G pseudotyped HIV-1 lentiviral vector deficient in envelope and accessory proteins but packaged with gag-pol products that allow transcription and integration. The ability of these vectors to enter macrophages is established [33], but like wild type HIV-1 strains did not invoke IFNβ and IP10 responses (Figure 5A-B). We also tested the possibility that rapid reverse transcriptase activity on viral entry may limit host cell exposure to ssRNA. In the absence of the HIV-1 Vif accessory protein, APOBEC3G, a host cytidine deaminase is packaged within the virion and inhibits subsequent reverse transcription [34]. We found no induction of IFNβ or IP10 expression in MDM inoculated with a vif-defective macrophage tropic HIV-1 strain containing human APOBEC3G (Yu2 A3G, Figure 5A-B).
Figure 5. MDM lack constitutive or IFN-inducible expression of a functional PRR for HIV-1.
IFNβ (A) and IP10 (B) gene expression levels, measured by qPCR and normalized to GAPDH, are shown for M-CSF differentiated MDM at 4 h and 24 h and at 4 h for IFNβ-primed (2ng/mL 24 h before infection) and unprimed control MDM (D) after infection or stimulation with the panel of reagents indicated. In contrast to the significant response to Infuenza (p<0.01, Wicoxon sign-ranked test of paired samples), no significant IFNβ or IP10 response was evident to any of the lentiviruses. IFNβ priming consistently enhanced the response to poly I:C stimulation but not HIV-1 or VSV-G pseudotyped lentiviral vector. Values for at least 3 separate experiments shown. (C) IFNβ-induced upregulation of IP10 and selected viral PRRs is shown in an expression matrix, derived from microarray transcriptional profiling of 4 h IFNβ-stimulated and control M-CSF differentiated MDM. * denotes genes that are significantly upregulated by IFNβ (p<0.05, t-test).
IFNβ stimulation upregulated expression of a number of host cell PRRs for viruses (Figure 5C), but did not significantly induce its own expression. Therefore we tested the hypothesis that type 1 IFN priming of MDM enhances their sensitivity to recognise and respond to HIV-1 infection. In keeping with upregulation of TLR3 and RIG-I expression in IFN-primed cells, the subsequent IFNβ response to poly I:C stimulation did show increased gene expression, but had no effect on deficiency of MDM responses to HIV-1 or the VSV-G pseudotyped LVV (Figure 5D). Collectively these data suggest that HIV-1 does not actively inhibit innate immune cellular activation, but that macrophages lack a constitutively expressed or IFN-inducible PRR for HIV-1 that is functionally active.
Discussion
Our findings clearly show the ability of HIV-1 to infect and replicate in macrophages without invoking innate immune cellular activation, and extend our understanding of the importance of HIV-1 tropism for these cells. The functional significance of deficient macrophage innate immune cellular activation and IFN responses to HIV-1 is highlighted by type 1 IFN mediated restriction of a range of wild type HIV-1 strains, that can be replicated by stimulation of macrophages with poly I:C at the time of viral inoculation. This suggests that if macrophages did mount an innate immune IFN response to HIV-1, the infection would be aborted despite the full complement of viral evasion mechanisms for IFN-inducible defences.
Both induction and deficiency of type 1 interferon responses to HIV-1 infection in macrophages have been reported in the early literature [35-39]. In addition, others have reported host-cell transcriptional responses using both microarray and qPCR technology during HIV-1 infection of MDM, that include a wide selection of inflammatory and IFN-related genes [13, 14, 40-43]. There are a number of possible experimental reasons for these inconsistencies. Importantly, we used sucrose purified virus preparations to minimise the risk of contamination with soluble factors from host cells used to produce virus, such as cytokines that may elicit confounding immunological responses. In addition, we used high enough MOI to better model synchronous infection and avoid bystander cell effects. Our findings were also consistent in different MDM models, and using a range of macrophage-tropic laboratory adapted HIV-1 strains as well as a primary clinical isolate.
The present study focuses on innate immune signaling and transcriptional responses but it is equally important that despite active viral replication, established HIV-1 infection of macrophages by 7 days was associated with quantitatively modest genome-wide changes to steady state transcriptional profiles and statistically weak enrichment for functionally related gene clusters. Importantly, uniform infection of cell cultures at 7 days avoids confounding by bystander or averaging effects in transcriptional profiling. These data suggest that established HIV-1 infection in macrophages does not have class effects on gene expression and casts doubt on the functional significance of the changes detected. This interpretation adds further support for an adapted host-pathogen relationship between HIV-1 and macrophages.
Identification of the mechanism of deficient macrophage responses to HIV-1 may provide therapeutic opportunities to correct the deficit and restrict HIV-1 transmission /propagation at critical points in the natural history of infection. We considered the possibilities that HIV-1 trafficking into the cell avoids interaction with PRRs or that a packaged component within the virion actively inhibits activation of PRR pathways. Comparable findings for the VSV-G pseudotyped LVV and full length HIV-1 strains indicate that neither HIV-1 envelope dependent cellular entry nor the HIV-1 components for which the LVV is defective are responsible. Together with the observation that IFNβ priming upregulates a number of known viral PRRs, but does not sensitise cells to incoming HIV-1, our findings show that macrophages do not express a constitutively active or inducible, functional PRR for this virus. Innate immune IFN responses to HIV-1 by plasmacytoid dendritic cells (pDC) that express TLR7 [44, 45], together with conspicuously low levels of constitutive or inducible TLR7 expression by macrophages in our transcriptional data point to TLR7 as a key innate immune receptor for HIV-1. However, unlike macrophages, pDC are not resident cells at mucosal sites of infection. If innate immune cellular activation in the mucosa does occur in HIV-1 infection, as suggested by data from primate models of simian immunodeficiency virus infection [46], the host cell and molecular mechanism that mediate initial innate immune recognition of the virus and may serve to simultaneously recruit antiviral host defences and permissive target cells remain to be verified.
This study provides a foundation to the body of literature suggesting that HIV-1 inhabits a uniquely adapted niche in macrophages, which is likely to be a key step to eventual pathogenesis of HIV-1 associated disease. Further study of this host-pathogen interaction, promises new insights into the pathophysiology of HIV-1 infection and into the determinants of lentiviral tropism for macrophages in general.
Supplementary Material
Acknowledgements
JT, and MN designed the experiments with intellectual input from BMC, RFM, WB, GJT and DRK. JT, BW and MN performed the experiments. BMC and MN performed microarray data analysis. WB provided key reagents. JT and MN wrote the manuscript with input from all other authors. All authors have agreed to all the content in the manuscript.
Funding: This work was supported by Wellcome Trust fellowships to MN (WT077161) and to GJT (WT076608).
Footnotes
Parts of this work have been presented in the following meetings: Keystone Symposia: HIV Immunobiology: From Infection to Immune Control (X4) Keystone Colorado March 2009 Abstract 349, and Keystone Symposia: Pattern Recognition Molecules and Immune Sensors of Pathogens (Z1) Banff Alberta Canada April 2009, Abstract 438
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Noursadeghi M, Katz DR, Miller RF. HIV-1 infection of mononuclear phagocytic cells: the case for bacterial innate immune deficiency in AIDS. Lancet Infect Dis. 2006;6(12):794–804. doi: 10.1016/S1473-3099(06)70656-9. [DOI] [PubMed] [Google Scholar]
- 2.Groot F, Welsch S, Sattentau QJ. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood. 2008;111(9):4660–4663. doi: 10.1182/blood-2007-12-130070. [DOI] [PubMed] [Google Scholar]
- 3.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A. 2002;99(3):1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakuma R, Mael AA, Ikeda Y. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J Virol. 2007;81(18):10201–10206. doi: 10.1128/JVI.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 7.Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203(1):41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besnier C, Takeuchi Y, Towers G. Restriction of lentivirus in monkeys. Proc Natl Acad Sci U S A. 2002;99(18):11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, et al. TLR7 Is Involved in Sequence-Specific Sensing of Single-Stranded RNAs in Human Macrophages. J Immunol. 2008;180(4):2117–2124. doi: 10.4049/jimmunol.180.4.2117. [DOI] [PubMed] [Google Scholar]
- 11.Meier A, Alter G, Frahm N, Sidhu H, Li B, Bagchi A, et al. MyD88-Dependent Immune Activation Mediated by Human Immunodeficiency Virus Type 1-Encoded Toll-Like Receptor Ligands. J Virol. 2007;81(15):8180–8191. doi: 10.1128/JVI.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fantuzzi L, Spadaro F, Purificato C, Cecchetti S, Podo F, Belardelli F, et al. Phosphatidylcholine-specific phospholipase C activation is required for CCR5-dependent, NF-kB-driven CCL2 secretion elicited in response to HIV-1 gp120 in human primary macrophages. Blood. 2008 doi: 10.1182/blood-2007-08-104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C, Liu QH, Tomkowicz B, Yi Y, Freedman BD, Collman RG. Macrophage activation through CCR5- and CXCR4-mediated gp120-elicited signaling pathways. J Leukoc Biol. 2003;74(5):676–682. doi: 10.1189/jlb.0503206. [DOI] [PubMed] [Google Scholar]
- 14.Cicala C, Arthos J, Selig SM, Dennis G, Jr., Hosack DA, Van RD, et al. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc Natl Acad Sci U S A. 2002;99(14):9380–9385. doi: 10.1073/pnas.142287999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noursadeghi M, Tsang J, Miller RF, Straschewski S, Kellam P, Chain BM, et al. Genome-wide innate immune responses in HIV-1-infected macrophages are preserved despite attenuation of the NF-kappaB activation pathway. J Immunol. 2009;182(1):319–328. doi: 10.4049/jimmunol.182.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.aasa-Chapman MM, Aubin K, Williams I, McKnight A. Primary CCR5 only using HIV-1 isolates does not accurately represent the in vivo replicating quasi-species. Virology. 2006;351(2):489–496. doi: 10.1016/j.virol.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol. 2006;80(16):8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brussel A, Sonigo P. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J Virol. 2004;78(20):11263–11271. doi: 10.1128/JVI.78.20.11263-11271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noursadeghi M, Tsang J, Haustein T, Miller RF, Chain BM, Katz DR. Quantitative imaging assay for NF-kappaB nuclear translocation in primary human macrophages. J Immunol Methods. 2008;329(1-2):194–200. doi: 10.1016/j.jim.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noursadeghi M, Tsang J, Miller RF, Katz DR. Comment on “Transcription factor FOXO3a mediates apoptosis in HIV-1-infected macrophages”. J Immunol. 2008;180(12):7783–7784. doi: 10.4049/jimmunol.180.12.7783. [DOI] [PubMed] [Google Scholar]
- 21.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 22.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 23.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci U S A. 2006;103(15):5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reimer T, Brcic M, Schweizer M, Jungi TW. poly(I:C) and LPS induce distinct IRF3 and NF-kappaB signaling during type-I IFN and TNF responses in human macrophages. J Leukoc Biol. 2008;83(5):1249–1257. doi: 10.1189/jlb.0607412. [DOI] [PubMed] [Google Scholar]
- 26.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177(10):7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 27.Gruber MF, Weih KA, Boone EJ, Smith PD, Clouse KA. Endogenous macrophage CSF production is associated with viral replication in HIV-1-infected human monocyte-derived macrophages. J Immunol. 1995;154(10):5528–5535. [PubMed] [Google Scholar]
- 28.Kedzierska K, Maerz A, Warby T, Jaworowski A, Chan H, Mak J, et al. Granulocyte-macrophage colony-stimulating factor inhibits HIV-1 replication in monocyte-derived macrophages. AIDS. 2000;14(12):1739–1748. doi: 10.1097/00002030-200008180-00008. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda S, Akagawa K, Honda M, Yokota Y, Takebe Y, Takemori T. Suppression of HIV replication in human monocyte-derived macrophages induced by granulocyte/macrophage colony-stimulating factor. AIDS Res Hum Retroviruses. 1995;11(9):1031–1038. doi: 10.1089/aid.1995.11.1031. [DOI] [PubMed] [Google Scholar]
- 30.Trapp S, Derby NR, Singer R, Shaw A, Williams VG, Turville SG, et al. Double-Stranded RNA Analog Poly(I:C) Inhibits Human Immunodeficiency Virus Amplification in Dendritic Cells via Type I Interferon-Mediated Activation of APOBEC3G. J Virol. 2009;83(2):884–895. doi: 10.1128/JVI.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol. 2009;83(8):3834–3842. doi: 10.1128/JVI.01773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meylan PR, Guatelli JC, Munis JR, Richman DD, Kornbluth RS. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology. 1993;193(1):138–148. doi: 10.1006/viro.1993.1110. [DOI] [PubMed] [Google Scholar]
- 33.Neil S, Martin F, Ikeda Y, Collins M. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J Virol. 2001;75(12):5448–5456. doi: 10.1128/JVI.75.12.5448-5456.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francis ML, Fan XS, Meltzer MS. Loss ability to produce IFN-alpha in response to HIV-1 as monocytes differentiate into macrophages. Induction through a mechanism independent of double-stranded RNA. J Immunol. 1996;156(7):2481–2487. [PubMed] [Google Scholar]
- 36.Szebeni J, Dieffenbach C, Wahl SM, Venkateshan CN, Yeh A, Popovic M, et al. Induction of alpha interferon by human immunodeficiency virus type 1 in human monocyte-macrophage cultures. J Virol. 1991;65(11):6362–6364. doi: 10.1128/jvi.65.11.6362-6364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gendelman HE, Friedman RM, Joe S, Baca LM, Turpin JA, Dveksler G, et al. A selective defect of interferon alpha production in human immunodeficiency virus-infected monocytes. J Exp Med. 1991;173(1):277. doi: 10.1084/jem.173.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gessani S, Borghi P, Fantuzzi L, Varano B, Conti L, Puddu P, et al. Induction of cytokines by HIV-1 and its gp120 protein in human peripheral blood monocyte/macrophages and modulation of cytokine response during differentiation. J Leukoc Biol. 1997;62(1):49–53. doi: 10.1002/jlb.62.1.49. [DOI] [PubMed] [Google Scholar]
- 39.Gessani S, Puddu P, Varano B, Borghi P, Conti L, Fantuzzi L, et al. Induction of beta interferon by human immunodeficiency virus type 1 and its gp120 protein in human monocytes-macrophages: role of beta interferon in restriction of virus replication. J Virol. 1994;68(3):1983–1986. doi: 10.1128/jvi.68.3.1983-1986.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coberley CR, Kohler JJ, Brown JN, Oshier JT, Baker HV, Popp MP, et al. Impact on genetic networks in human macrophages by a CCR5 strain of human immunodeficiency virus type 1. J Virol. 2004;78(21):11477–11486. doi: 10.1128/JVI.78.21.11477-11486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woelk CH, Ottones F, Plotkin CR, Du P, Royer CD, Rought SE, et al. Interferon gene expression following HIV type 1 infection of monocyte-derived macrophages. AIDS Res Hum Retroviruses. 2004;20(11):1210–1222. doi: 10.1089/aid.2004.20.1210. [DOI] [PubMed] [Google Scholar]
- 42.Giri MS, Nebozhyn M, Showe L, Montaner LJ. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J Leukoc Biol. 2006;80(5):1031–1043. doi: 10.1189/jlb.0306157. [DOI] [PubMed] [Google Scholar]
- 43.Brown JN, Kohler JJ, Coberley CR, Sleasman JW, Goodenow MM. HIV-1 activates macrophages independent of Toll-like receptors. PLoS ONE. 2008;3(12):e3664. doi: 10.1371/journal.pone.0003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fonteneau JF, Larsson M, Beignon AS, McKenna K, Dasilva I, Amara A, et al. Human immunodeficiency virus type 1 activates plasmacytoid dendritic cells and concomitantly induces the bystander maturation of myeloid dendritic cells. J Virol. 2004;78(10):5223–5232. doi: 10.1128/JVI.78.10.5223-5232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manches O, Munn D, Fallahi A, Lifson J, Chaperot L, Plumas J, et al. HIV-activated human plasmacytoid DCs induce Tregs through an indoleamine 2,3-dioxygenase-dependent mechanism. J Clin Invest. 2008;118(10):3431–3439. doi: 10.1172/JCI34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009 doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.