Abstract
Mammalian white adipocytes have a unique structure in which nearly the entire cell volume is occupied by a single large lipid droplet, while the surrounding cytoplasm occupies minimal space. The massive cytoplasmic remodeling processes involved in the formation of this unique cellular structure are poorly defined. Autophagy is a membrane trafficking process leading to lysosomal degradation of cytoplasmic components. Here, we investigated the functional role of atg5, a gene encoding an essential protein required for autophagy, in adipocyte differentiation in a cellular model and in mice. Massive autophagy was activated when wild-type primary mouse embryonic fibroblasts (MEFs) were induced for adipocyte differentiation. Importantly, the autophagy deficient primary atg5-/- MEFs exhibited dramatically reduced efficiency in adipogenesis. Time-lapse microscopy revealed that atg5-/- MEFs initially appeared to differentiate normally; however, a majority of the differentiating atg5-/- cells ultimately failed to undergo further morphological transformation and eventually died, likely through apoptosis. Consistent with these in vitro results, histological analysis revealed that the atg5-/- late-stage embryos and neonatal pups had much less subcutaneous perilipin A-positive adipocytes. Consistently, when treated with chloroquine, a functional inhibitor of autophagy, wild-type MEFs exhibited drastically reduced efficiency of adipocyte differentiation. Taken together, these findings demonstrated that Atg5 is involved in normal adipocyte differentiation, suggesting an important role of autophagy in adipogenesis.
Keywords: Atg5, autophagy, adipogenesis, WAT, adipocyte differentiation
Introduction
Obesity has become an epidemic in the United States and other industrialized countries. The direct cause of obesity is excessive accumulation of adipose tissue. Humans and other mammals contain two types of adipose tissue: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT, the predominant type, functions as storage depots for fuel (in the form of triglyceride) and as essential regulators of whole-body energy homeostasis.1 In contrast, the principal function of BAT is to dissipate energy and to generate heat, an important defense mechanism against cold.2 Obesity is primarily associated with an expansion of subcutaneous and visceral WAT.
WAT is composed of white adipocytes, which are generally considered to arise from a mesodermal origin. Adipogenesis, or the development of adipocytes from the fibroblast-like pre-adipocytes, has been well studied. Extensive research in a white pre-adipocyte cell line, 3T3-L1, has demonstrated that the process of adipogenesis is coordinately regulated by a network of transcription factors in which PPARγ plays a central role.3-6 Upon hormone induction, primary MEFs undergo an adipogenesis process that is very similar to that of the 3T3-L1 cells, culminating in the formation of mature adipocytes with one or several large lipid droplets. The primary MEFs derived from mice of various genetic backgrounds provide an additional useful cellular model to determine the role of genes of interest in white adipogenesis.7-10
While the transcriptional network regulating adipocyte differentiation has been elucidated, the picture is less clear on the cellular processes involved in adipogenesis and how they are coordinately regulated during the complex differentiation process. Consistent with its function as a storage depot for triglycerides, a mature mammalian white adipocyte has a highly distinctive structure in which nearly the entire cell volume is occupied by one large lipid droplet, while other cellular components, including the nucleus and cytoplasm, localize peripherally and occupy minimal space. A massive cellular remodeling process is likely involved in adipogenesis, which is poorly defined at this time.
Autophagy is a major cellular degradation process.11,12 It is initiated by the emergence of a double-membrane structure in the cytoplasm which expands to engulf and sequester a portion of the cytoplasm, resulting in the formation of the hallmark double-membrane autophagosome. Fully mature autophagosomes translocate towards and fuse with lysosomes. The cargo of the autophagosome is then released to the lysosome where it is degraded. The molecular machinery of autophagy is highly conserved evolutionarily from yeast to humans.13,14 The genes encoding the essential components of the machinery, which are named atg (autophagy-related) genes, have been identified. A number of mouse models with targeted deletions of the atg genes, including beclin 1 (atg6),15,16 atg5,17,18 and atg7,19,20 have been generated. These mouse models exhibit defects in autophagy and they have been instrumental in defining the various functions of autophagy in mammals, including tumor suppression, neuronal protection and surviving the neonatal period of nutrient deprivation. Recent studies also showed that autophagy is required during differentiation of certain cell types such as erythrocytes for normal removal of organelles such as mitochondria,21-23 and it is involved in hepatocytes in lipid droplet formation24 and lipid metabolism.25
The degradation function of autophagy prompted us to postulate that autophagy might be involved in cytoplasmic remodeling during adipogenesis. More than 25 years ago an increased level of autophagosomes was observed when differentiating 3T3-L1 cells were analyzed morphologically with electron microscopy by Novikoff et al. although at that time molecular characterization of autophagy was impossible.26 The elucidation of autophagy machinery at the molecular level and the availability of various mouse genetic tools make it possible to determine the functional role of various autophagy genes in adipogenesis. The atg5 gene encodes an acceptor protein, Atg5, for the ubiquitin-like protein Atg12.27 Atg5 conjugates with Atg12, forming a multimeric structure which serves as a platform that is specifically required for the maturation of the autophagic membrane.28 In the current study we have determined the impact of targeted deletion of atg5 on adipogenesis in the MEF model and in vivo. Our study has demonstrated that atg5 deletion interferes with normal adipocyte differentiation and suggested a critical role of autophagy in adipogenesis.
Results
Autophagy was activated in wild-type MEFs during adipocyte differentiation
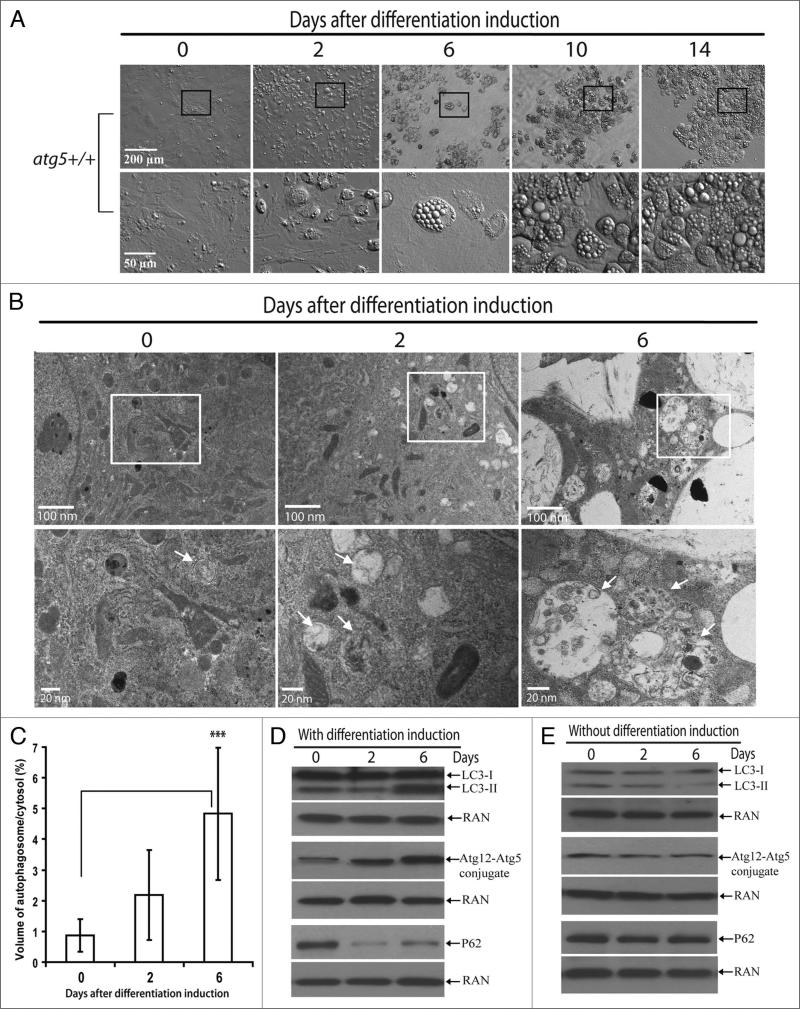
We analyzed the activation of autophagy in the primary MEFs during adipogenesis via morphology study with electron microscopy (EM) as well as by molecular characterization with autophagy-specific markers. Similar to the induction protocol of adipogenesis in 3T3-L1 cells, the primary MEFs were first grown to confluence. Two days after confluence, a cocktail of differentiation agents containing dexamethasone (DEX)/3-Isobutyl-1-methylxanthine (IBMX)/troglitazone/insulin5,29 was added to the medium to induce differentiation and the time was recorded as Day 0 of induction. Two days later (or on Day 2 of induction), the differentiation maintenance medium (containing only insulin and troglitazone) replaced the original differentiation cocktail. From then on, the fresh maintenance medium was added to the cells every two days to replace the old medium. The differentiation of cells was monitored with a microscope equipped with relief contrast lens, which was used to observe the three-dimensional structure of the cells. As shown in Figure 1A, the kinetics of adipogenesis in wild-type primary MEFs was very similar to that of 3T3-L1 cells: on Day 2 of induction, isolated cells started to “inflate” to form a spheroid morphology from the original flat morphology, and micro-size lipid droplets started to accumulate in the spheroid cells; on Day 6 of differentiation induction, small patches of the spheroid cells formed, each cell in the patch containing many small lipid droplets; as differentiation continued, more flat cells participated in differentiation and exhibited the “inflated” spheroid morphology; in the meantime small lipid droplets grew larger in size or fused with each other; on Day 14, the majority of cells formed patches of “inflated” spheroid cells, many of which contained one or several large lipid droplets.
Figure 1.
Autophagy was activated in wild-type MEFs during adipogenesis. (A) Primary atg5+/+ MEFs were induced for adipogenesis. At indicated time points, the progress of differentiation was analyzed. Cells were observed under microscope (Olympus IX70) equipped with relief contrast objectives (10X and 40X, for low and high magnification, respectively). Selected regions in pictures of low magnification (within the squares) are shown below with high magnification. (B) Electron microscopy analysis of primary atg5+/+ MEFs 0, 2 or 6 days after differentiation induction, as indicated. Upper panel shows micrographs of low magnification, and the lower panel shows high magnification of the selected regions (square) in the upper panel. Arrows indicate autophagosomes. (C) Ratio of the volume of autophagosomes to cytosol. The volume of autophagosomes and cytosol were determined by point counting of 15–20 micrographs of cells 0, 2 or 6 days after differentiation induction, as indicated. ***p < 0.001. Student t-test. (D and E). Immunoblotting assays of differentiating cells. The cells at indicated time points with (D) or without (E) differentiation induction were harvested and immunoblotting assays were performed with LC3, Atg12, p62 or RAN antibodies, as indicated. The levels of RAN served as a loading control. The data are representative results from three independent experiments.
We performed electron microscopy to analyze autophagosome formation during adipogenesis of the wild-type primary MEFs. As shown in Figure 1B and C, ultrastructure of cells on Day 0, Day 2 and Day 6 during differentiation induction was analyzed by EM and the volume of autophagosomes was quantified. Prior to induction of differentiation (Day 0), autophagosomes occupied about 1% of the cytoplasmic volume. The level of autophagosomes steadily increased as the cells underwent adipogenesis. By Day 6, around 5% of the total cytoplasmic volume (volume of lipid droplets included) was occupied by autophagosomes.
Autophagy activation during adipogenesis was further analyzed with specific autophagy molecular markers. MAP-LC3 is a mammalian homolog of the yeast Atg8 protein. During autophagy activation, it is cleaved at its C-terminus and the N-terminus portion of MAP-LC3 is conjugated with phospholipids and translocated onto the autophagosome.30 The abundance of the processed form of MAP-LC3, known as LC3-II, reflects a steady-state level of autophagosomes.30 As shown in Figure 1D (upper), the level of LC3-II dramatically increased as differentiation progressed. To confirm that the increase of autophagosomes also reflected the increase of functional autophagic degradation, autophagy flux was analyzed by measuring the levels of the p62, which is a common autophagosome cargo whose degradation reflects the levels of autophagy flux.30 As shown in Figure 1D, lower, induction of adipogenesis drastically reduced p62 levels, indicating an increase of autophagy flux.
In addition, we also measured the levels of other proteins that are specifically involved in autophagy. Interestingly, the level of the Atg5-Atg12 protein conjugate also increased significantly during adipogenesis (Fig. 1D, middle). Importantly, the increase of autophagosomes formation, autophagy flux, and Atg5-Atg12 conjugation were specifically associated with adipogenesis. As a control, when the MEFs were not induced for adipogenesis, LC3-II levels slightly reduced over time, and ATG5-ATG12 conjugate levels and p62 levels remained unchanged, as shown in Figure 1E. Taken together, these results demonstrated that autophagic activity was increased in the primary MEFs undergoing adipogenesis.
Autophagy deficient primary atg5-/- MEFs exhibited significantly reduced efficiency in adipogenesis
We then studied a functional role of autophagy in adipocyte differentiation by examining the impact of atg5 deletion on adipogenesis in the primary MEF model. Mice with homozygous atg5 deletion (atg5-/-) develop without any apparent defects and are born in normal Mendelian ratios, but die within the first day following birth in part due to failure to cope with neonatal starvation.18 Measurements of autophagy demonstrated that, as expected, the formation of autophagosomes was absent in the tissues of the atg5-/- mice.18 The wild-type and atg5-/- primary MEFs from E13.5 embryos of the same pregnant mother were induced for adipogenesis. The progression of differentiation was monitored by microscopy with relief contrast lens. In contrast to wild-type (atg5+/+) MEFs, which underwent normal adipogenesis as shown in Figure 1A, atg5-/- cells initially accumulated small lipid droplets but seemed to become inert after the initiation phase of differentiation (Fig. 2A). These cells with small lipid droplets had difficulty progressing to more advanced stages. At any given time within the 14-day differentiation, only a small portion of atg5-/- cells showed initial morphologic changes with the spheroid morphology and with the accumulation of micro-size to small-size lipid droplets. Even at the end of the 14-day differentiation, very few cells were found in the more advanced differentiation stages signified by one or a few large lipid droplets.
Figure 2.
Autophagy deficient primary atg5-/- MEFs exhibited reduced efficiency in adipogenesis. Primary atg5+/+ or atg5-/- MEFs were induced for adipogenesis. At indicated time points, the progress of differentiation was analyzed. (A) Cells were observed under microscope (Olympus IX70) equipped with relief contrast objectives (10X and 40X, for low and high magnification, respectively). Selected regions in pictures of low magnification (within the squares) are shown below with high magnification. (B) Cells were stained with the lipid dye Bodipy 493/503 and observed with microscope under phase contrast objectives (20X). (C) 14 days post-differentiation inductions, cells were stained with the lipid dye Oil Red-O and hematoxylin, and observed under phase contrast microscope. (D) Cells grown on cover slip were stained with Oil Red-O at indicated time points and Oil Red-O was extracted and measured by spectrometry. These data represent results from experiments with cells derived from four independent pairs of embryos of three independent breeding parents.
Lipid accumulation is a hallmark of adipocyte differentiation. In addition to the morphological characterization of adipocyte differentiation, we analyzed lipid accumulation in the atg5-/- cells and their wild-type counterparts following induction of differentiation. As shown in Figure 2B, cells at various time points post-differentiation induction were fixed and stained with Bodipy 493/503, a fluorescent dye that specifically stains intracellular lipid droplets. As the differentiation progressed, more and more lipid staining was observed in the wild-type cells. In contrast, very limited lipid staining in the atg5-/- cells was observed even on day 14, when the majority of wild-type cells were in advanced differentiation stages. The differences of lipid accumulation between the two types of MEFs were quantified using Oil Red-O, another dye that specifically stains the lipid droplets and can be extracted for spectrometry measurement. As shown in Figure 2C, at the end of 14 days of differentiation, a very dramatic difference in Oil Red-O staining between the wild type and the mutant cells was observed. At various time points Oil Red-O staining was performed and the dye was extracted from the cells for quantification by spectrometry. As summarized in Figure 2D, the atg5-/- MEFs exhibited a significant defect in accumulating lipid droplets upon differentiation induction, which was completely consistent with the morphological analysis (Figs. 1A and 2A).
The adipogenesis defects observed in these atg5-/- cells were not clone-specific effects. The same phenotypes were observed in four pairs of MEFs from three independent pairs of breeding parents. The primary MEFs of early passages (passages three to five) were used in the adipocyte differentiation experiments. We also tested MEFs of earlier passages (passages one to two), as well as MEFs of later passages (passages six to eight). While clear differences in adipocyte differentiation were observed between the wild-type cells and the atg5-/- cells of all passages, interestingly, it appeared that the inhibitory effect of autophagy deficiency on adipogenesis was less dramatic in the earlier passage cells than the later passage cells (data not shown). These results suggest that events secondary to autophagy deficiency may accumulate over time, which can exacerbate the direct effect of autophagy deficiency on adipocyte differentiation.
Gene expression analysis of the differentiating wild-type and atg5-/- MEFs
The mRNA levels of genes in cells prior to differentiation induction were compared to those in cells six days after differentiation induction in wild-type and atg5-/- MEFs. First, we performed gene expression profiling experiments with oligonucleotide microarray. The genes involved in adipocyte differentiation exhibited the most dramatic changes upon differentiation induction in both wild-type and atg5-/- cells (data not shown). The expression levels of a subset of those genes were confirmed by quantitative PCR analysis, as shown in Figure 3. It seemed that most genes that were upregulated in the wild-type cells were also upregulated in the atg5-/- cells; however, the extent of gene activation was less robust in the atg5-/- cells. Moreover, it appeared that the later transcriptional events of adipogenesis, such as upregulation of Fabp4 and Perilipin genes, were more severely impacted in the atg5-/- cells as compared to the early transcriptional events, such as PPARγ and CEBPα upregulation. Although these results did not pinpoint a particular event in which autophagy interacted with adipogenesis, they did confirm at the molecular level that less atg5-/- cells underwent adipogenesis.
Figure 3.
Quantitative PCR analysis of the expression of a subset of adipogenesis marker genes. mRNA were extracted from the atg5+/+ and atg5-/- cells at Day 0 or Day 6 of differentiation and analyzed by quantitative PCR. The graphic representations show relative expression levels of each adipogenesis related gene, as indicated, as compared to the normalizer gene Wbp11NORM. *denote values that were undetectable. Error bars represent one standard deviation.
Time-lapse microscopy showed that adipocyte differentiation stalls at an early stage in primary atg5-/- MEFs
To determine the stages at which adipocyte differentiation was affected and the likely cause of the defect, we generated movies of actively differentiating atg5+/+ and atg5-/- MEFs using time-lapse microscopy. Figure 4A and B show picture frames selected from two-day movies (Suppl. Material 1 and 2) of differentiating wild-type cells starting on day three. Small lipid droplets emerged and actively fused and consolidated into larger droplets during this period of time. Figure 4C and D are picture frames from movies of atg5-/- MEFs in the same time frame as the wild-type cells shown in Figure 4A and B. In contrast to the differentiating wild-type cells, most differentiating atg5-/- cells at the beginning of the movie (day three) had accumulated only numerous micro lipid droplets. Strikingly, most of these differentiating atg5-/- cells stalled at this stage and were unable to progress further morphologically. Eventually these cells slowly lost their anchorage to neighboring cells, began to rotate freely in the medium and ultimately died (Fig. 4C and D, and Suppl. Material 3 and 4). It is noteworthy in the movies that only the atg5-/- cells that underwent initial early differentiation died, while the undifferentiated cells remained normal and alive. These results suggest that the atg5 function might not be indispensable for adipogenesis initiation in accumulating micro-sized lipid droplets but it was critical for efficient progression of adipogenesis. As a result, atg5 deletion frequently led to aborted differentiation.
Figure 4.
Time-lapse microscopy analysis of adipogenesis in the atg5+/+ and atg5-/- MEFs. Primary MEFs were treated to induce adipocyte differentiation. Three days after induction, time-lapse microscopy with relief contrast lens was performed to monitor the progression of differentiation. (A–D) are picture frames taken from two-day movie clips (Suppl. Material 1–4, respectively) showing the continuous morphologic changes during differentiation. Areas in square regions of (A and C) are enlarged below to show detail in (B and D), respectively. White arrows in (B) point to a growing lipid droplet; and black arrows in (C), and (D) point to cells undergoing abortive differentiation. The data represent results from experiments performed with three independent pairs of MEFs.
The differentiating atg5-/- cells exhibited higher levels of apoptosis than the wild-type cells
To determine whether the aborted cells during differentiation died of apoptosis, we performed TUNEL assays, which detect DNA breakage/fragmention, a hallmark of apoptosis. As shown in Figure 5, the wild-type and atg5-/- primary MEFs were analyzed at various time points during adipogenesis by TUNEL assay. These cells were also co-stained with Bodipy 493/503 to monitor adipogenesis. Interestingly, essentially all TUNEL positive cells were also Bodipy 493/503 positive (Fig. 5A), indicating that only the cells that started adipogenesis and began accumulating lipid droplets were vulnerable to apoptosis. For the wild-type cells, at early stages of differentiation (Day 2), there was a small percentage of the differentiating cells underwent apoptosis as shown in Figure 5B. As differentiation progressed, the cells undergoing apoptosis decreased. In contrast, the percentage of the differentiating atg5-/- cells undergoing apoptosis at Day 6 was significantly higher than that of the wild-type cells. Moreover, the apoptotic cells as a percentage of the differentiating cells continued to increase in later time points in the atg5-/- cell. These observations were consistent with Figure 4 and suggested that in the cellular adipogenesis model system the differentiating cells that failed to mature would eventually die of apoptosis.
Figure 5.
Differentiating atg5-/- MEFs exhibited higher rates of apoptosis. (A) Primary atg5+/+ or atg5-/- MEFs were induced for adipogenesis. The progress of differentiation and apoptotic cell death was analyzed with Bodipy 493/503 staining (green), DAPI staining (blue) and TUNEL assay (red), respectively. The pictures showed cells at Day 6 post-differentiation induction. Representative low (with a scale bar of 50 μm) and higher (with a scale bar of 10 μm) magnification pictures are shown. (B) Quantification of the TUNEL positive cells as a percentage of Bodipy 493/503 positive cells at the indicated time points. Total number of Bodipy 493/503 positive cells and total number of both TUNEL positive cells and Bodipy 493/503 positive cells in randomly selected regions were counted and the percentage was calculated. The data are representative results from three independent experiments. **p < 0.01; Student t-test.
The atg5-/- late-stage embryos and neonatal pups had less subcutaneous fat cells
To determine whether atg5 deletion affects adipogenesis in vivo, we analyzed the adipocytes in the atg5-/- late-stage embryos and neonatal pups and their wild-type counterparts. As described earlier, the atg5-/- mice develop normally throughout gestation, but always die within the first day of birth partly due to the fact that these mice cannot mobilize sufficient internal nutrients to survive the neonatal starvation period.18 We analyzed the adipocytes of the E18.5 atg5+/+ and atg5-/- embryos, as well as pups within 12 hours after birth. In rodents, the white fat tissue at birth is not well developed; however, dispersed white adipocytes can be observed in subcutaneous regions. Transverse sections of the mice were cut at the level of the scapulae, where the subcutaneous white adipocytes can be analyzed. The tissues were examined by immunofluorescence microscopy with antibody against perilipin A, a protein that localizes on the membrane of lipid droplets of adipocytes. Figure 6A shows the immunostaining of tissues in subcutaneous regions of the embryos. The atg5-/- embryos had only 15% of perilipin A positive adipocytes at corresponding sub-cutaneous regions compared to their wild-type counterparts (Fig. 6A and C), suggesting adipogenesis of white adipocytes was significantly reduced. Similarly, the neonatal atg5-/- pups had drastically reduced perilipin A positive adipocytes at corresponding subcutaneous regions compared to their wild-type littermates (Fig. 6B and D). Together, these results suggest that atg5 deletion affects adipogenesis in vivo.
Figure 6.
The atg5-/- mice had fewer subcutaneous fat cells. atg5-/- embryos (E18.5) and neonatal pups (within 12 hours after birth) and their wild-type littermates were obtained and the transverse sections at the level of scapulae were analyzed by immunofluorescence microscopy with primary antibody against perilipin A and FITC-conjugated secondary antibody. (A) Subcutaneous regions of embryos showing perilipin A positive adipocytes. (B) Subcutaneous regions of neonatal pups showing perilipin A positive adipocytes. (C) quantification of (A). Total number of perilipin A positive cells in subcutaneous regions of three adjacent scapulae sections were counted and averaged. (D) quantification of (B). Total number of perilipin A positive cells in subcutaneous regions of three adjacent scapulae sections were counted and averaged. The data are representative results from three independent pairs of pups born to two independent pairs of parents and three pairs of embryos born to three pairs of parents. **p < 0.01; ***p < 0.001. Student t-test.
Autophagy inhibitor chloroquine blocked adipocyte differentiation in primary MEFs
Chloroquine, an FDA-approved drug for malaria and rheumatoid arthritis, targets the lysosome and it also blocks the fusion of the autophagosome with the lysosome.31,32 Chloroquine is an effective autophagy inhibitor which has been extensively used for autophagy inhibition both in clinical trials and in laboratory studies.31,32 We determined the effect of chloroquine on the adipocyte differentiation of primary MEFs. The wild-type primary MEFs were treated to induce adipogenesis with or without the presence of 10 μM chloroquine, a concentration that inhibits the fusion of the autophagosome with lysosome (thereby increases intracellular levels of autophagosomes but inhibits autophagy flux (reviewed in ref. 32 and Fig. 7E)). At this concentration chloroquine had little cytotoxic effect on the MEFs (Fig. 7D). As shown in Figure 7A, chloroquine significantly inhibited normal adipocyte differentiation as detected by morphological analysis. Consistently, lipid accumulation analysis using Bodipy 493/503 (Fig. 7B) or Oil Red-O (Fig. 7C) staining showed the same inhibitory effect of chloroquine on adipocyte differentiation. Together, these results indicate that chloroquine effectively inhibits adipogenesis in the primary MEF model.
Figure 7.
Chloroquine significantly reduced the efficiency of adipogenesis in primary MEFs. Wild-type primary MEFs were induced for adipogenesis with or without co-treatment of 10 μM chloroquine (CQ). Differentiation progress was then monitored by: (A) microscopy analysis; (B) lipid analysis with Bodipy 493/503 staining (14 days after differentiation induction); (C) lipid analysis by spectrometry of Oil Red-O staining (14 days after differentiation induction). (D and E) are controls that show that chloroquine was nontoxic (D) and efficacious in inhibiting autophagosome fusion with lysosome and in inhibiting autophagy flux (E) at the experimental concentration. (D) Tunel assays of wild-type MEFs treated with 10 μM chloroquine for 4 days compared with cells without chloroquine treatment. Cells treated with 10 μM staurosporine (STS) for 6 hr was used as a positive control. (E) Cells treated with/or without 10 μM chloroquine at different time points were harvested, immunoblotting assays were performed with LC3, p62 or RAN antibodies, as indicated. The levels of RAN served as a loading control. The results represent three independent experiments. *p < 0.05; Student t-test.
Discussion
White adipocytes are differentiated from the fibroblast-like pre-adipocytes and they have a unique cellular structure in which nearly the whole cell is occupied by one large lipid droplet. However, the cellular processes involved in the cytoplasmic remodeling during adipogenesis are poorly understood. In the current study, using primary MEFs, a white adipogenesis cellular model, we demonstrated that massive autophagy was induced during adipogenesis. Moreover, by using the primary MEFs derived from atg5 knockout mice, we demonstrated that atg5 deficiency significantly reduced adipogenesis efficiency. We further showed that atg5 deletion did not appear to significantly affect the early events of adipogenesis, including upregulation of PPAR-γ and CEBP genes and accumulation of micro-sized lipid droplets. However, atg5 deletion may cause adipogenesis arrest at later stages and eventually lead to apoptosis of the differentiating cells. Finally, we demonstrated that atg5 deficiency affected adipogenesis in vivo. Together, these data indicated that atg5 function is important for adipogenesis, suggesting an involvement of autophagy in adipogenesis. Consistent with this notion, pharmacologically inhibition of autophagy function by chloroquine blocks adipogenesis in a cellular model.
The exact mechanism by which the loss of Atg5 leads to abortive adipogenesis is not clear. It is likely that the stalled differentiation at later stage of adipogenesis and subsequent demise of the differentiating cells are results of failure in successful removal of cytoplasmic components, including mitochondria. However, recent reports indicate that Atg8 can localize on the surface of lipid droplets of hepatocytes24,25 and may facilitate the fusion of lipid droplets.24 We cannot rule out the possibility that a defect in Atg5-dependent Atg8 lipidation and translocation to the lipid droplets of adipocytes may result in inefficient droplet fusion, which in turn contributes to the defect in adipogenesis in the cellular model.
As reported here, adipogenesis may represent one of a few physiological processes in which autophagy activity is extremely high. At Day 6 after differentiation induction, ultrastructural study with electron microscopy indicated more than 5% of the cytoplasmic volume of the differentiating cells is autophagosome. If the “inert” volume of lipid droplet, which accounts for more than 50% of cytoplasm at the time, is subtracted from the total cytoplasmic volume, more than 10% of the cytoplasmic volume of the differentiating cells is autophagosome. Consistent with the elevated autophagosome levels, the autophagy flux increases. The mechanism by which autophagy is activated is not clear. Interestingly, we have observed a dramatic increase in Atg5-Atg12 conjugates in the differentiating cells. Usually Atg5-Atg12 conjugates are not a rate-limiting factor and an increase of autophagic activity from basal levels does not necessarily require the increase of Atg5-Atg12 conjugates.30 The dramatic increase of Atg5-Atg12 conjugates likely reflected a massive increase of autophagic activity in these differentiating cells which may have exceeded the capacity of normal cellular levels of Atg5-Atg12 conjugates in supporting autophagosome formation. It will be interesting to investigate the mechanism by which Atg5-Atg12 conjugates are upregulated and/or stabilized upon adipocyte differentiation induction.
In summary, we have for the first time demonstrated that deletion of an essential autophagy gene, atg5, impacts on normal adipogenesis in a cellular model and in vivo. This provides novel mechanistic insight into the molecular and cellular processes underlying the drastic cytoplasmic remodeling during adipogenesis. Obesity is directly caused by the excess accumulation and expansion of adipocytes. Interestingly, it is becoming clear that adipocytes are constantly renewed throughout adulthood.33 Our results that inactivation of Atg5 as well as treatment with chloroquine, a functional autophagy inhibitor, significantly reduce adipogenesis in a cellular model, may also provide a new venue for regulating the size and cellular structures of adipose tissues, thereby interfering with adipose related functions.
Material and Methods
Adipocyte differentiation of primary MEFs
The MEFs were prepared from 13.5 day embryos of atg5+/+, atg5-/- mice according to standard protocol. Briefly, whole mouse embryos were removed from the uterus, dissected and the head, tail, limbs and all internal organs were removed. The carcasses were minced, washed in PBS, and then incubated in 2 ml 0.05% Trypsin-EDTA (cat n° 25300-054, Invitrogen) at 37°C for 20 min with shaking. The digested cells were plated on a 100-mm dish in Dulbecco's modified Eagle's medium (DMEM, cat n° 11960-069, Invitrogen) with 10% fetal bovine serum (FCS, cat n° 100-106, Gemini Bio-Products) and incubated at 37°C in humidified air containing 5% CO2. Cells were grown for 24 hr until the culture was 90% confluent, and then split and passed on. The primary MEF cells of passages three through five were treated under standard protocol to induce adipocyte differentiation.29 Briefly, cells were seeded in six-well plates with cover slips and propagated to confluence. Forty-eight hours later, which was designated as Day 0, differentiation was initiated using DMEM containing 10% fetal bovine serum, 5 μg/ml insulin (cat n° I5500, Sigma), 1 μM dexamethasone (cat n° D4902, Sigma), 0.5 mM 3-Isobutyl-1-methylxanthine (IBMX, cat n° I7018, Sigma) and 10 μM troglitazone (cat n° T2573, Sigma). Two days after initiation, the medium was replaced with a maintenance medium (DMEM containing 10% fetal bovine serum, 5 μg/ml insulin, and 10 μM troglitazone). Fresh maintenance medium was replaced every two days thereafter. Cells were collected at various time points up to fourteen days from the date of induction.
For chloroquine treatment, the wild-type primary MEFs of early passages were seeded in six-well plates. The cells were treated with 10 μM chloroquine (cat n° C6628, Sigma) at 50% confluence and propagated to full confluence. Two days after confluence, adipocyte differentiation was induced with differentiation medium containing 10 μM chloroquine. The medium was replaced with maintenance medium including 10 μM CQ two days after initiation. Fresh maintenance medium with 10 μM CQ was replaced every two days thereafter.
Autophagosome quantification by EM and immunoblotting
The differentiating cells were fixed at indicated time points with 2.5% gluteraldehyde/4% paraformaldehyde in 0.1 M cacodylate buffer for two hours. The samples were processed and thin sections (90 nm) were cut on a Reichert Ultracut E microtome. Sections were viewed at 80 kV with a JEOL 1200EX transmission electron microscope. Micrographs were taken in the Philips CM12 (15–20 per sample) by random sampling with a primary magnification of X6300. The cytoplasmic volume fraction of autophagic vacuoles was quantified by point counting method.34 western blotting was carried out according to standard protocol. The sources of the antibodies are: MAP-LC3 antibody: made by Cocalico Biologicals (PA, US) using a recombinant rat MAP-LC3 protein as antigen; rabbit polyclonal Atg12 antibody: cat n° 2011, Cell Signaling Technology; rabbit polyclonal Ran antibody: cat n° sc-1156, Santa Cruz; p62: primary antibody: guinea pig anti-p62 Protein (cat n° 03-GP62-C, American Research Products, Inc.,); secondary: donkey anti-guinea pig polyclonal antibody from Jackson ImmunoResearch Laboratories, Inc., (cat n° 706-035-148).
Imaging
Live cells were observed under the Olympus IX70 microscope with relief contrast objectives (10X or 40X) and phase contrast objectives (10X). Fixed and stained cells were observed with a Universal Microscope Axioplan 2 imaging system (Carl Zeiss, NY, US) with 20X phase contrast objectives. For time-lapse microscopy, cells were plated on tissue culture plates and monitored using an Olympus IX70 microscope with a 37°C and 5% CO2 environmental chamber under 10X relief contrast objectives. Images were collected with a CCD video camera (Model MicroMax; Princeton Instruments, Trenton, NJ, US) at 5 min intervals and saved as image stacks using IPLab software (BD Biosciences Bioimaging, Rockville, MD, US). Images were processed using Image J software (NIH, Bethesda, MD, US).
Lipid droplet staining
Bodipy
The differentiating cells were stained with BODIPY 493/503 (cat n° D-3922, Invitrogen) as described.35 Briefly, the cell culture slides were washed once with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde in PBS for 30 min at room temperature, washed four times with PBS, stained with 10 μg/ml Bodipy 493/503 at room temperature for 15 min in darkness and then mounted with Vectashield Mounting Medium (cat n° H-1000, Vector Laboratories).
Oil red-O
The cells on cover slides were stained with Oil Red-O (cat n° O0625, Sigma) according to Kim et al. with some modification.36 Oil Red-O was dissolved in isopropanol to 3.5 mg/ml. Before using, dilute six parts Oil Red-O stock with four parts H2O, sit at room temp for 20 min, and filter through 0.2 μm filter. The slides were washed once with PBS, fixed with PBS buffered formalin (cat n° 00740, Surgipath Medical Industries, Inc.,) for 1 hr, and then washed twice with 60% isopropanol for five minutes each. Cells were then air dried and stained with Oil Red-O working solution for 30 min at 25°C. For Figure 2C, slides were counterstained with hematoxylin for 1 min. To quantify staining, Oil Red-O was extracted from cells on the slides with isopropanol containing 4% NP-40, and optical density (OD) was then measured at a wavelength of 520 nm.
cDNA amplification and quantitative real-time PCR
RNA was extracted from cell culture lysates using TRIzol reagent (cat n° 15596-026, Invitrogen) according to standard protocol. Subsequent RNA quality assessment, cDNA amplification, and quantitative RT-PCR reactions were carried out by the Bionomics Research and Technology Center (BRTC) of the Environmental and Occupational Health Science Institute (EOSHI) at Rutgers University, Piscataway, NJ (detailed protocols available at http://eohsi-brtc.com). RNA quality was assessed by electrophoresis using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and by spectrophotometric analysis prior to cDNA synthesis. Between 5 and 20 ng of total RNA from each sample was used to generate high fidelity cDNA for quantitative PCR. The Ribo-SPIA (NuGen, San Carlos, CA) linear amplification process was used to generate “antisense” cDNA. The SPIA process (NuGen, San Carlos, CA) was used to amplify cDNA and a 1:200 dilution of amplified cDNA product was used for subsequent QPCR analysis.
Six genes relevant to adipocyte differentiation were selected for quantitative real-time PCR analysis to validate the findings of the microarray analysis. Gpam, Cebpa, Pparg, Fabp4, Agpat2 and Plin were identified as candidate genes for analysis and two further genes, Ube23DNORM and Wbp11NORM, were selected as normalizers for the PCR reactions. Gene expression was examined using Taqman chemistry with probes and primers designed using the Roche Universal Probe Library design algorithm (www.universalprobelibrary.com). Results of the probe design are as follows: Gpam, left primer GAG GCA AGG ACA TTT ATG TGG and right primer GGT GCT TTC ACA ATC ACT CG; Cebpa, left primer CTG GCT CTG GGT CTG GAA and right primer AGC CAC AGG GGT GTG TGT A; Pparg, left primer CTC TCA GCT GTT CGC CAA G and right primer CAC GTG CTC TGT GAC GAT CT; Fabp4, left primer GCA CGG TCT CTC TGC AAT C and right primer ACA ATC AAT CAG CGC AGG A; Ucp1, left primer CCA GTG GAT GTG GTA AAA ACA A and right primer CAC AGC TTG GTA CGC TTG G; Agpat2, left primer TTC CCA CCT CAA GCC TGT and right primer TGC CTT GTG GTC TTG TGG; Plin, left primer CTC CGG CCT TTC CTC TCT A and right primer GGG GGA GTG ATG ACA TGG; Ube23NORM, left primer TTA GTG ATT TGG CCC GTG A and right primer TGG CTT GCC AAT GAA ACA T; and Wbp11NORM, left primer GAG CAA TGT CCA CTG TCA GG and right primer ATC CCA GCA GGC AAA CAT. The following dye combinations for probe generation were used for detection and data normalization: FAM (for the genes of interest), HEX (for normalizer genes) and BHQ1 (non-fluorescent quencher) and ROX (reference). All probes were 8mer MGB probes selected from the Roche Universal Probe Library as appointed per each assay design. Prior to comparative analysis, a validation experiment was performed in order to determine the relative efficiency of the assays designed for the genes of interest and Wbp11NORM and Ube23DNORM which were subsequently used as reference genes for comparative analysis. All reactions were performed in triplicate and the experiments were replicated three times. All reactions were run in an ABI 7900 (Applied Biosystems, Foster City, CA) with the following cycle parameters: 1 cycle of 50°C (2 min) followed by 95°C (10 min), 40 cycles of 95°C (15 sec) followed by 60°C (1 min). Data was collected at every temperature phase during every cycle and analyzed using the Sequence Detection Software (Applied Biosystems, Foster City CA) while relative quantitation using the comparative threshold cycle (CT) method was performed. The expression levels of the genes of interest were presented as the relative levels to the mRNA level of the control gene Wbp11NORM.
TUNEL assay
The differentiating cells growing on cover slides were fixed with PBS buffered formalin for 15 min. The TUNEL assay was conducted with In Situ Cell Death Detection Kit, TMR red (cat n° 12 156 792 910, Roche Applied Science) according to the instruction of the manufacturer. The slides were counter-stained with BODIPY 493/503 (10 μg/ml) and DAPI (1 μg/ml, cat n° D9564, Sigma) for 10 min before mounting and taking picture under the microscope. In randomly selected areas, the differentiating cell with Bodipy 493/503 signal (green) were counted and cells with TUNEL positive signal (red) were counted among these cells. The percentage of apoptotic cells among differentiating cells were calculated. For chloroquine treatment experiments, the apoptotic cell populations were quantified by Coulter Cytomics FC500 Flow Cytometer (Beckman Coulter, CA, US).
Immunofluorescence analyses
E18.5 mouse embryos and neonatal mice (within 12 hr after birth) were sacrificed, fixed with PBS buffered formalin, and embedded in paraffin. After genotyping via PCR,18 transverse sections were cut at the level of scapulae.37 Immunofluorescence was performed according to standard protocol. Briefly, sections were deparaffinized with Xylene and rehydrated through graded ethanol. Antigen unmasking was carried out in 10 mM sodium citrate buffer (pH 6.0) at 95–99°C for 10 min. Slides were allowed to cool down at room temperature for 30 min and rinsed with H2O and PBS. Specimens were blocked with 5% goat serum in PBS/Triton for 1 hr, followed by incubating with Perilipin A antibody (cat n° P 1998, Sigma, 1:50 dilution) at 4°C overnight. For immunostaining detection, slides were incubated with secondary antibody FITC-Goat Anti-Rabbit IgG (cat n° 62-6111, Invitrogen, 1:100 dilution) for 1 hr, rinsed with PBS, and mounted with Vectashield Mounting Medium (cat n° H-1000, Vector Laboratories). Pictures were taken with a Universal Microscope Axioplan 2 imaging system (Carl Zeiss, NY, US) with 100X phase contrast objectives. The diameters of lipid droplet in the pictures were measured with Adobe Photoshop software (Adobe Systems, Inc., San Jose, CA).
Acknowledgements
We thank Dr. Noboru Mizushima for providing atg5 knockout mice, Drs. Loren Runnels and Li-Ting Su for assistance with the microscopy, Dr. Dawn L. Brasaemle for help with adipocyte differentiation, Raj Patel for technical assistance with the EM, Adam Oberstein for making recombinant MAP-LC3 protein, Dr. Andrew Brooks for Real-time PCR analyses, and all Jin lab members for productive discussions. R.B. is supported by NIH 5 F31 GM078857-02, Y.Z., P.C. and S.J. are supported by NIH 1R01 CA116088-01A1 and 1R01AG030081-01A1.
Footnotes
References
- 1.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–53. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 3.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–73. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen JB, Kristiansen K. Regulatory circuits controlling white versus brown adipocyte differentiation. Biochem J. 2006;398:153–68. doi: 10.1042/BJ20060402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–96. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 6.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–307. [PubMed] [Google Scholar]
- 7.Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 8.Kubota N, Terauchi Y, Miki H, et al. PPARgamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 9.Sue N, Jack BH, Eaton SA, et al. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol Cell Biol. 2008;28:3967–78. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, Rosen ED, Brun R, et al. Cross-regulation of C/EBP alpha and PPARgamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–8. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 11.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 14.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–6. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 18.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008 doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibata M, Yoshimura K, Furuya N, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–23. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol. 1980;87:180–96. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998 Sep 24;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–9. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 29.Banks AS, Li J, McKeag L, et al. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J Clin Invest. 2005;115:2462–71. doi: 10.1172/JCI23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13:7271–9. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 32.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spalding KL, Arner E, Westermark PO, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–7. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 34.Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–94. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiDonato D, Brasaemle DL. Fixation methods for the study of lipid droplets by immunofluorescence microscopy. J Histochem Cytochem. 2003;51:773–80. doi: 10.1177/002215540305100608. [DOI] [PubMed] [Google Scholar]
- 36.Kim YK, Choi HY, Kim NH, et al. Reversine stimulates adipocyte differentiation and downregulates Akt and p70(s6k) signaling pathways in 3T3-L1 cells. Biochem Biophys Res Commun. 2007;358:553–8. doi: 10.1016/j.bbrc.2007.04.165. [DOI] [PubMed] [Google Scholar]
- 37.Oishi Y, Manabe I, Tobe K, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]