Abstract
Purpose:
Glioblastomas are treated by surgical resection followed with radiotherapy (XRT) and the alkylating chemotherapeutic agent temozolomide (TMZ). Recently, inactivating mutations in the mismatch repair (MMR) gene MSH6 were identified in two glioblastomas recurrent post-TMZ. Since MMR pathway inactivation is a known mediator of alkylator resistance in vitro, these findings suggested MSH6 inactivation was causally linked to these two recurrences. However, the extent of involvement of MSH6 in glioblastoma is unknown. We sought to determine the overall frequency and clinical relevance of MSH6 alterations in glioblastomas.
Experimental Design:
The MSH6 gene was sequenced in 54 glioblastomas. MSH6 and MGMT immunohistochemistry was systematically scored in a panel of 46 clinically well-characterized glioblastomas, and the corresponding patient response to treatment evaluated.
Results:
MSH6 mutation was not observed in any pre-treatment glioblastoma (0/40), while 3/14 recurrent cases had somatic mutations (p=0.015). MSH6 protein expression was detected in all pre-treatment (17/17) cases examined but, notably, expression was lost in 7/17 recurrences from matched post-XRT+TMZ cases (41%, p=0.016). Loss of MSH6 was not associated with MGMT status. Measurements of in vivo tumor growth using 3D-reconstructed MRI demonstrated that MSH6-negative glioblastomas had a markedly increased rate of growth while under TMZ treatment (3.17 cc/month vs. 0.04 cc/month for MSH6-positive tumors, p=0.020).
Conclusions:
Loss of MSH6 occurs in a subset of post-XRT+TMZ glioblastoma recurrences, and is associated with tumor progression during TMZ treatment, mirroring the alkylator resistance conferred by MSH6 inactivation in vitro. MSH6 deficiency may therefore contribute to the emergence of recurrent glioblastomas during TMZ treatment.
Keywords: Glioblastoma, Mismatch Repair, Temozolomide, Chemotherapy Resistance
Cancers evolve during tumor progression by the clonal selection of genetic alterations in tumor cells. Successive waves of clonal outgrowth overcome a multitude of selection barriers during neoplastic development. In addition, cancers face further selection pressures once they are diagnosed and treatment is initiated, and they continue to accumulate genetic alterations that confer selective growth advantage by allowing escape from the tumoricidal effects of treatment. In this regard, glioblastomas, the most frequent primary human brain tumor, are a prototypical human neoplasia: they accumulate mutations in oncogenes (e.g., EGFR) and tumor suppressor genes (e.g., PTEN) (1), display clinical responsiveness to XRT and the alkylating chemotherapeutic agent TMZ (2), but ultimately fail these therapies and progress to fatal outcomes.
In a recent large-scale sequence analysis of malignant gliomas, somatic truncating mutations in the mismatch repair (MMR) gene MSH6 were identified in two recurrent human glioblastomas (3). Both tumors had large numbers of somatic mutations within a mutational signature consistent with alkylator treatment, and both had been treated with TMZ. Prior studies in murine embryonic stem cells (4), Chinese hamster cell lines (5), human lymphoblastoid cells (6-8), and human cancer cells (9, 10) have demonstrated that MSH6 inactivation confers tolerant cell growth under cytotoxic doses of alkylating agents in vitro. Importantly, tolerant cells can be subsequently re-sensitized to alkylating agents by reintroduction of wildtype MSH6 (9, 10), establishing the direct role of the MSH6 gene in mediating this chemoresistance effect (11). We therefore hypothesized that MSH6 inactivation in tumor subclones may have contributed to their clonal expansion during TMZ treatment and emergence as clinical recurrences in these two cases.
However, the overall frequency of MSH6 alterations in glioblastomas is unknown. Previous reports have identified homozygous germline mutations of MSH6 in isolated cases of syndromic children with brain tumors (12, 13), but heterozygous germline or somatic mutations of MSH6 have not been reported in typical adult patients with sporadic glioblastoma. Moreover, clinical phenotypic surveys of large MSH6-kindreds with familial colorectal or endometrial cancer have not identified an increased risk of glioblastoma in patients with heterozygous germline MSH6 mutations (14, 15). Nonetheless, since heterozygous germline mutations in the canonical MMR genes MSH2 and MLH1 can cause the mixed colonic-and-glial tumor spectrum of Turcot syndrome (16), it is possible that yet undiscovered germline or somatic MSH6 mutations could contribute to the development of a subset of glioblastomas.
In addition, this initial finding of MSH6 mutations in two tumors is counterbalanced by a substantial body of evidence that TMZ responsiveness in glioblastomas can be predicted prior to treatment by determining the O-6 methylguanine methyltransferase (MGMT) status (17, 18). MGMT is a well-characterized enzyme that catalyzes removal of the methyl-conjugate from O-6 methylguanine, one of the nucleotide modifications that results from TMZ treatment. High levels of MGMT activity are thought to offset alkylator modification of tumor DNA, thereby limiting TMZ anti-tumor activity. Levels of MGMT have been assessed by promoter methylation analysis (17), direct enzymatic assay (18), or immunohistochemistry (19), and elevated levels correspond to more rapid tumor progression. Further complicating the understanding of these two pathways, it appears that both can be operant in the development of alkylator resistance in vitro (20), with MMR deficiency serving as an alternative escape route when cancer cells have low levels of MGMT expression (21). It is thus possible that both pathways could contribute to clinical recurrences in glioblastoma patients receiving TMZ.
We therefore sought to determine the frequency of MSH6 mutation or loss of protein expression in glioblastomas. In addition, we examined the relationship between MSH6 loss, adjuvant treatment modality and MGMT status, and characterized the clinical consequences of MSH6 loss. To this end, we analyzed the mutational status and expression of MSH6 in both pre-treatment and post-treatment glioblastoma samples, compared this pattern with the MGMT expression status of these samples, and examined the radiologic and clinical treatment course of the cohort in detail.
Materials and Methods
Tumor and DNA stocks
Tumors and peripheral blood lymphocytes for normal controls were banked under IRB-approved informed consent at the Massachusetts General Hospital. Tumor DNA was extracted from tumor specimens or early cell-culture passage after histologic confirmation (PureGene kit, Gentra Systems, Minneapolis, MN). Tumor and normal DNA for sample xT5165 and treatment information was kindly provided by Dr. Gregory Riggins and Dr. Charles Eberhart of The Johns Hopkins Hospital. The tumors assembled for sequencing analysis were identified based on two criteria: histologically-confirmed glioblastoma (WHO grade IV) and availability of appropriate quality specimen (ie. fresh-frozen surgical resection or early cell-culture passage) for DNA-sequence-based analyses.
For the panel of tumors assembled for immunohistochemical analyses, three criteria were used to identify patients: histologically-confirmed glioblastoma (WHO grade IV), treatment with a chemotherapeutic regimen that included TMZ, and availability of appropriate formalin-fixed paraffin-embedded specimen. Five samples were analyzed by both sequencing and IHC (3 with wildtype MSH6, 2 with mutant MSH6), as can be seen in Tables 1 and 2 their results were concordant across both assessment modalities. As a control for adjuvant treatment modality, an additional 8 glioblastomas from XRT-only treated cases were also studied.
Table 1. Sequence Analysis of MSH6 in Glioblastomas.
The top-listed samples (shaded) are derived from patients that had received treatment with TMZ or unspecified chemotherapy. Details of treatment and mutation status are as noted. Sample ID list for the 40 pretreatment cases available on request. MSH6 status of samples xT155, xT924, xT2560, xT3017, xT3058, and xT3162 has been previously reported in (3).
| Sample ID | Treatment | Sample | MSH6 mutation |
|---|---|---|---|
| xT3017 | XRT+TMZ | post-treatment | Het C1453T, Q485X; Het G3907A, A1303T |
| xT3162 | XRT+TMZ, IV BCNU, thioguanine |
post-treatment | Hom delG2425, V809X |
| xT5165 | Unknown treatment | post-treatment | Het A560C, K187T; Het T794G, F265C |
| xT104 | XRT+TMZ, oncolytic virus, implantable BCNU wafer |
post-treatment | wildtype |
| xT2993 | XRT+TMZ, IV BCNU, implantable BCNU wafer |
post-treatment | wildtype |
| xT3072 | Unspecified “chemotherapy”, XRT |
post-treatment | wildtype |
| xT3296 | XRT+TMZ, radio- iodine125 EGFR MAb |
post-treatment | wildtype |
| xT3378 | XRT+TMZ, IV BCNU, radio-iodine125 EGFR MAb |
post-treatment | wildtype |
| xT3768 | XRT+TMZ | post-treatment | wildtype |
| xT155 | XRT | post-treatment | wildtype |
| xT177 | XRT+CPT11 | post-treatment | wildtype |
| xT601 | XRT | post-treatment | wildtype |
| xT1178 | XRT+PCV | post-treatment | wildtype |
| xT1201 | procarbazine | post-treatment | wildtype |
| 40 cases | - | pre-treatment | wildtype |
Table 2. MSH6 and MGMT immunohistochemical analysis.
MSH6 and MGMT status in matched samples from 17 patients' pre- and post-XRT+TMZ surgical resections. Below are 4 post-XRT+TMZ and 8 post-XRT-only samples where the pre-treatment sample was unavailable for study.
| Treatment Modality |
Pre- and post- treatment sample ID# |
Pre- treatment MGMT |
Post- XRT+TMZ MGMT |
Pre- treatment MSH6 |
Post- XRT+TMZ MSH6 |
||
|---|---|---|---|---|---|---|---|
| XRT+TMZ | xT4981/xT5089 | − | − | + | ++ | ||
| xT3546/xT4717 | + | + | ++ | ++ | |||
| xT4142/xT3692 | − | − | +++ | ++ | |||
| xT3559/xT4328 | − | − | +++ | ++ | |||
| xT2989/xT3296 | − | − | ++ | ++ | |||
| xT4107/xT3648 | + | + | ++ | +++ | |||
| xT4106/xT3307 | − | − | +++ | +++ | |||
| xT3506/xT3768 | + | + | ++ | +++ | |||
| xT4170/xT4987 | − | − | + | ++ | |||
| xT4213/xT4869 | + | + | + | + | |||
| xT4901/xT4899 | + | + | ++ | − | |||
| xT4991/xT3017 | + | + | + | − | |||
| xT3495/xT4161 | + | + | ++ | − | |||
| xT3893/xT4870 | n/s | + | + | − | |||
| xT3513/xT4718 | − | − | + | − | |||
| xT4136/xT4900 | + | + | +++ | − | |||
| xT4838/xT5065 | + | + | +++ | − | |||
| XRT+TMZ | −/xT104 | + | ++ | ||||
| −/xT5046 | n/s | − | |||||
| −/xT4803 | − | − | |||||
| −/xT3162 | − | − | |||||
| XRT-only | −/xT4986 | − | ++ | ||||
| −/xT4903 | − | ++ | |||||
| −/xT4977 | − | ++ | |||||
| −/xT4975 | + | ++ | |||||
| −/xT2422 | + | ++ | |||||
| −/xT2703 | − | ++ | |||||
| −/xT3303 | + | ++ | |||||
| −/xT4985 | + | ++ | |||||
MSH6 scored with a system of “−” = nil or scant staining, “+” = <10% of cells stained positive in the nucleus, “++” = 10-50%, and “+++” = >50% positive. MGMT scored using “+” = >20% positive staining, and “−” = <20% staining. “n/s” = assay not successfully performed.
PCR amplification and sequencing
PCR primers were derived from reference (14), covering more than 98% of the MSH6 coding sequence (sequences available on request). Amplification was performed using Taq-Platinum DNA polymerase (Invitrogen, Carlsbad, CA). One primer from each pair was tagged with M13F universal sequencing primer to facilitate subsequent fluorescent dye-terminator sequencing (Agencourt Biosciences, Beverly, MA). Of a total of 752 reactions, 705 were successfully amplified and sequenced, for a sequence coverage rate of greater than 93%.
Immunohistochemistry
Monoclonal antibodies to MSH6/GTBP (clone 44, BD Biosciences, San Jose, CA) and MGMT (MT3.1, Chemicon, Temecula, CA) were used to perform immunohistochemistry on formalin-fixed, paraffin-embedded sections. Antigen retrieval was achieved with 20-minute (MSH6) or 15-minute (MGMT) incubation in 10mM sodium citrate buffer at pH6.0. Overnight 4°C incubations with a 1:100 (MSH6) or 1:50 (MGMT) dilution were performed (Vectastatin Elite ABC kit; Vector Laboratories, Burlingame, CA). Positive control tissue consisted of nuclear staining in colonic crypt cells and the germinal centers of lymph nodes (MSH6) or cytoplasmic and nuclear stating in colon cancer and normal brain (MGMT). Scoring was performed in a blinded fashion by two reviewers (DPC and DNL).
Tumor Growth Measurements
MRI volume calculations were performed on axial imaging sections through the tumor mass using the Vitrea2 3D volumetric software (Vital Images, Inc., Minnetonka, MI). Personnel scoring tumor volume were blinded to the molecular stratification of the tumors. Tumor volume was assessed using standard criteria of T1-post-gadolinium sequence enhancing volume, correcting for acute surgical residua by exclusion of pre-gadolinium T1-sequence enhancement. Tumor growth rate was calculated on matched-pairs by determining the T1-enhancing volume from the MRI obtained at the initiation of alkylator therapy (which was typically the post-operative MRI following surgical biopsy or debulking resection), and subtracting it from the T1-enhancing tumor volume on the MRI obtained immediately prior to cessation of alkylator therapy. This change in volume was then divided by the difference in time between the two studies to calculate an overall growth rate during alkylator therapy. Similar calculations were performed for FLAIR sequences, which assess the magnitude of tumor-associated edema.
In one recurrence (corresponding to tumor specimen xT4899), a second MRI study was not obtained prior to cessation of therapy; therefore we used data calculated from an MRI scan that occurred 16 days after the last dose of TMZ, since it was still within the standard treatment window of 28 days for a single cycle of TMZ therapy. One patient (corresponding to tumor specimen xT3162) was excluded from tumor growth analyses because the follow-up MRI sequences available were obtained greater than 4 months after cessation of TMZ therapy; this patient's tumor growth data is appended to Supplementary Table 1, and can be seen as essentially comparable to the other cases, but was censored because of the length of time outside of alkylating therapy without radiologic assessment.
Statistical Analysis
Pre-treatment cases were defined as a patient's first surgical resection, and recurrences as the first surgical resection after failure of the specified treatment modality (XRT or XRT+TMZ). Two-sided, 0.05-level Fisher's exact tests were used to compare two independent proportions. An exact binomial confidence interval was calculated for the frequency of negative MSH6 expression at recurrence following TMZ treatment. An exact, two-sided, 0.05-level McNemar's test was used to compare the frequency of MSH6 negative expression at recurrence following TMZ treatment with that in the pre-treatment sample. To adjust for the length-bias induced by the sampling of recurrent cases, semi-parametric truncation-adjusted Kaplan Meier estimates (22), parametric Weibull estimates and log-rank tests were used for the analyses of age at diagnosis (right-truncated by age at recurrence), length of alkylator treatment (right-truncated by time from diagnosis to recurrence), time to recurrence (right-truncated by time to death or end of follow-up), and time from diagnosis to death (left-truncated by time to recurrence). Exact, 2-sided, 0.025-level (Bonferroni adjusted) Wilcoxon tests were used to evaluate for differences in tumor growth rates under treatment, as measured by T1 post-gadolinium enhancement and FLAIR hyperintensity. For the expression of growth rate per month, the growth rate per day was multiplied times 30 days.
Results
Sequence analysis of MSH6
Our prior sequencing study analyzed 6 glioblastomas for MSH6 alterations, finding somatic mutations in two tumors. To survey the overall spectrum of MSH6 mutation in glioblastoma, we analyzed the MSH6 coding sequence in an additional 48 glioblastomas. Thirty-seven were pre-treatment tumors, while 11 were recurrent tumors. These samples were not pre-selected based on their treatment regimen, and therefore the recurrences had previously received a range of alkylator, non-alkylator, or even no chemotherapy. The clinical details of these samples and sequence analysis results are listed in Table 1. No mutations of MSH6 were identified in the pre-treatment tumors. Therefore, germline or somatic MSH6 mutations do not appear to contribute significantly to the pre-treatment development of glioblastoma. In the recurrent samples, one tumor (xT5165) had two sequence alterations that resulted in predicted amino acid substitutions. One was an A-to-C transversion at nucleotide 560 leading to a lysine-to-threonine substitution at amino acid position 187. The other was a T-to-G transversion at nucleotide 794 resulting in a phenylalanine-to-cysteine replacement at amino acid position 265. Neither of the alterations was found in the patient's normal DNA.
Combining data from these 48 tumors with our prior findings yielded a total of 3 MSH6-mutant cases amongst 14 recurrent tumors, while none of the 40 pre-treatment tumors had mutations. Since the recurrence rate of glioblastoma is near-100%, random sampling of recurrences is unlikely to introduce significant selection bias with respect to prior pre-treatment status. With this in mind, comparison of the frequency of mutation in our pre- and post-treatment samples suggests that MSH6 mutation is associated with recurrence (p=0.015).
Immunohistochemistry for MSH6 expression
These results, in light of the known selectivity of the MMR pathway in mediating alkylator resistance in vitro, raised the possibility that selection for MSH6 inactivation could be a treatment-specific phenomenon. Moreover, the treatment regimens varied among these patients, possibly accounting for the minority of recurrent cases with detectable MSH6 mutations. We therefore sought to evaluate MSH6 in a second sample set focused on glioblastomas that had undergone adjuvant treatment with the standard-of-care alkylating agent TMZ. To maximize the assessable yield of MSH6 status, we pursued immunohistochemistry (IHC) of formalin-fixed paraffin-embedded specimens, which constitute the majority of clinically well-annotated material in glioblastoma trial settings. Most glioblastoma biopsy specimens are not sufficient to afford genomic amplification and large-scale sequence analyses; fortunately there is extensive experience validating the utility of IHC to detect MSH6 inactivation in hereditary colon and endometrial cancer (14, 15, 23, 24).
We scored both MSH6 and MGMT status in a total of 46 glioblastomas (Table 2). Within this group, 38 specimens were from 21 patients who had received treatment with XRT+TMZ. Since our study design necessitated selection of patients who had undergone two surgeries, we examined the baseline demographic characteristics of those patients treated with XRT+TMZ, to assess for selection bias. The patient cohort was 71% male, with median age at diagnosis of 53 years. These data are indicative of a patient group similar to those reported in the recent large prospective study of XRT+TMZ (2). Of the tumors studied, 34 were matched pairs derived from 17 patients' pre- and post-XRT+TMZ surgical resections, providing the additional benefit of certainty regarding the pre-treatment MSH6 status.
In the matched pre-treatment specimens, every tumor examined (17/17) stained positively for MSH6 expression. The staining displayed variability, with a range from <10% to >50% of the total tumor cell mass in any given case. However, even accounting for this variability, every pre-treatment glioblastoma evaluated was unequivocally positive for MSH6 expression (Table 2), whereas normal brain sections do not stain for MSH6 expression (Figure 1). Thus, there appears to be an induction of MSH6 protein expression in pre-treatment glioblastomas, consistent with previous IHC studies of MSH6 in other tumors types (14, 15, 23, 24), and of other MMR genes in glioblastoma (25).
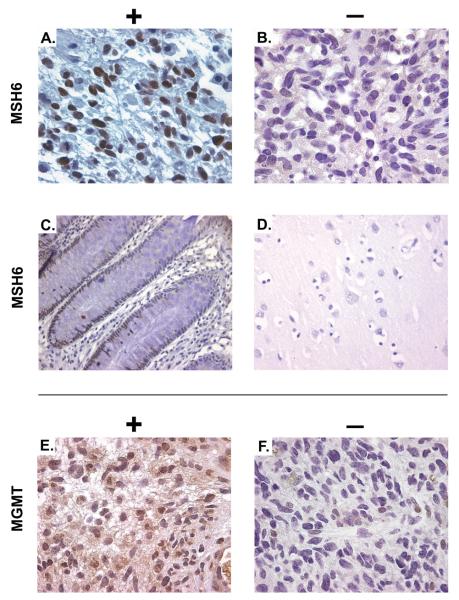
Figure 1.
MSH6 and MGMT immunohistochemistry. Immunohistochemistry was performed with monoclonal antibodies specific for MSH6 (A – sample xT3307 and B – xT3162), with controls of normal colonic epithelium (C) and normal brain (D). The larger field of view for (C) demonstrates MSH6 expression in the crypt and transit-amplifying cells, but not in differentiated colonic epithelium. MGMT immunohistochemistry (E – sample xT3506 and F – xT4142).
In matched post-XRT+TMZ tumors from the same patients, a range of MSH6 staining was also observed, with 4 cases showing an increase in staining, and 4 cases showing stable intensity. Strikingly, however, staining for MSH6 was completely absent in 7 of these cases (Figure 1). This loss of MSH6 expression at recurrence within the matched pairs was significantly different from that at diagnosis (p=0.016, based on the 7 tumor pairs with discordant pre-treatment and post-treatment expression). Furthermore, in 4 additional post-XRT+TMZ tumors, 3 had absence of MSH6 protein. Therefore, in total, 10 of 21 post-XRT+TMZ recurrent tumors displayed absence of MSH6 by IHC analysis.
To isolate the effect of TMZ chemotherapy comparatively, we analyzed an additional 8 recurrent tumors from patients who had received adjuvant treatment solely with XRT. These samples served as a treatment-modality control group; since current clinical practice does not allow withholding radiation treatment from eligible patients, it was not possible to assemble a cohort of patients who had solely received TMZ in the absence of XRT. These post-XRT-only specimens were all positive for MSH6 expression (8/8 tumors). Comparing these recurrences to post-XRT+TMZ recurrences as independent samples based on treatment modality (with the presumption of no significant selection bias amongst the recurrences with respect to their pretreatment status), there was a significant difference with respect to the frequency of MSH6 expression loss at recurrence between post-XRT+TMZ and post-XRT-only glioblastomas (p=0.012), providing evidence that MSH6 loss is specifically associated with the TMZ-alkylator component of combined adjuvant therapy.
MGMT
As noted above, decreased MGMT expression is a predictor of improved glioblastoma prognosis. It is therefore possible that acquisition of increased MGMT expression itself contributes to the emergence of resistant clones during TMZ treatment. Given the association of MGMT status with treatment response, it is also possible that MSH6 status at recurrence is linked to pre-treatment MGMT status. To determine the relationship between MGMT status, MSH6 status and post-XRT+TMZ recurrence, we assessed MGMT IHC within our panel of tumors.
MGMT staining was successfully analyzed in 44 of the 46 samples, including 16 of 17 pre-treatment tumors. We found a range of MGMT staining similar to prior estimates of the frequency of MGMT absence (17-19), with 7/16 staining negatively for MGMT and 9/16 scoring moderately or strongly positive (Figure 1 and Table 2). Similarly consistent with prior IHC analyses (19), absence of MGMT in the pre-treatment tumor displayed a trend toward prolonged overall survival (p=0.151). To evaluate changes in MGMT status during adjuvant therapy, we analyzed MGMT expression in the available matched post-XRT+TMZ specimens. For any individual patient, pre-treatment MGMT status was maintained in the post-XRT+TMZ sample, including all 7 glioblastomas that were MGMT-negative prior to TMZ treatment (Table 2). These data suggest that the induction of MGMT expression that can be seen in model systems of alkylator exposure (20) may not be frequent under the treatment dosing of TMZ in current clinical practice. Finally, we did not find a significant association between MGMT pre-treatment status and MSH6 post-treatment status (p=0.145), although our analyses suggest a possible correlative trend that might be further uncovered in larger studies.
Clinical correlation
To characterize the clinical features of patients whose tumors displayed MSH6 loss, clinical data was analyzed from patients treated with XRT+TMZ. Medical records were abstracted for the dates of initial surgery, length of treatment with TMZ and other alkylating agents, dates of recurrent surgery, and date of death. Comparing the two independent groups determined by MSH6-positive (n=11) and MSH6-negative (n=10) scoring at recurrence, we noted no significant differences in possible confounding factors such as age at diagnosis, time to initiation of alkylator treatment, length of alkylator treatment, or time to recurrent surgery (p=0.998, 0.523, 0.820, 0.698 respectively).
To determine the treatment consequences of MSH6 loss, we used MRI to perform in vivo calculations of tumor growth rates during TMZ treatment in patients stratified for MSH6 status. For all patients analyzed by IHC above, we identified matched-pairs of MRI scans, one corresponding to the initiation of alkylating chemotherapy inclusive of TMZ and one immediately prior to cessation of this treatment; appropriate matched pairs were identified for 20 of 21 patients. In aggregate, MRI data were assessed spanning 3879 days across a total alkylator treatment time of 4496 days, covering greater than 85% of the clinical treatment window. To exclude the possibility of systematic confounding biases arising during the clinical care and radiologic assessment of these patients, we examined several aspects of the timing and clinical conditions of the patients' treatment assignments and MRI schedules. We found no significant differences between MSH6-positive and MSH6-negative tumors with respect to either the time between the start of alkylator treatment and the initial MRI, or the time between cessation of alkylator treatment and the final assessment MRI. Furthermore, to exclude the possibility that tumors were being assessed at different stages of progression due to clinical factors, we examined radiologic proxy-measures of clinical performance such as the volume of tumor as assessed by T1-gadolinium-enhancing signal, or the volume of edema as assessed by FLAIR signal, and found no significant differences between the two groups when comparing either the pre-treatment or post-treatment scans (Table 3, with the full clinical-radiologic dataset available in Supplementary Table 1).
Table 3. Baseline Characteristics of Clinical and Radiologic Measures.
Radiologic assessment factors were compared between the two groups of patients with glioblastoma recurrences determined by MSH6-positive or MSH6-negative IHC staining. No significant differences were noted between potential confounding measures. Tumor growth rate, as assessed by rate of T1-post-gadolinium enhancement change, was higher in the MSH6-negative group.
| MSH6-positive (n=11) | MSH6-negative (n=9) | ||||
|---|---|---|---|---|---|
|
|
|||||
| Median | Range | Median | Range | p value |
|
| Age at recurrence |
49 | ( 23 - 70 ) | 56 | ( 31 - 67 ) | NS |
| Gender (% male) |
64% | 78% | |||
| Calculated pre-treatment tumor Volume (cc) |
11.79 | ( 1.65 - 46.59 ) | 5.12 | ( 0 - 53.73 ) | NS |
| Pre-treatment T1-post- gadolinium Enhancement Volume (cc) |
12.6 | ( 1.8 - 54.85 ) | 9.75 | ( 0 - 53.73 ) | NS |
| Pre-treatment T1 Enhancement Volume (ie. pre-gadolinium) (cc) |
1.37 | ( 0 - 41.06 ) | 0 | ( 0 - 11.33 ) | NS |
| Pre-treatment FLAIR Volume (cc) |
43.16 | ( 5 - 178.4 ) | 52.75 | ( 8.29 - 229.6 ) | NS |
| Recurrence T1-post- gadolinium Enhancement Volume (cc) |
9.15 | ( 5.2 - 44.1 ) | 16.17 | ( 4.19 - 82.83 ) | NS |
| Recurrence FLAIR Volume (cc) |
56.13 | ( 8.05 - 185.3 ) | 57.53 | ( 10.92 - 353.2 ) | NS |
| Time elapsed between initial and recurrence scans (days) |
170 | ( 71 - 364 ) | 153 | ( 57 - 432 ) | NS |
| Rate of FLAIR Change (cc/d) |
0.009 | ( −0.645 - 0.406 ) | 0.060 | ( −0.221 - 1.258 ) | NS |
| Rate of T1-post-gadolinium Enhancement Change (cc/d) |
0.001 | ( −0.087 - 0.068 ) | 0.106 | ( −0.055 - 0.288 ) | 0.020 |
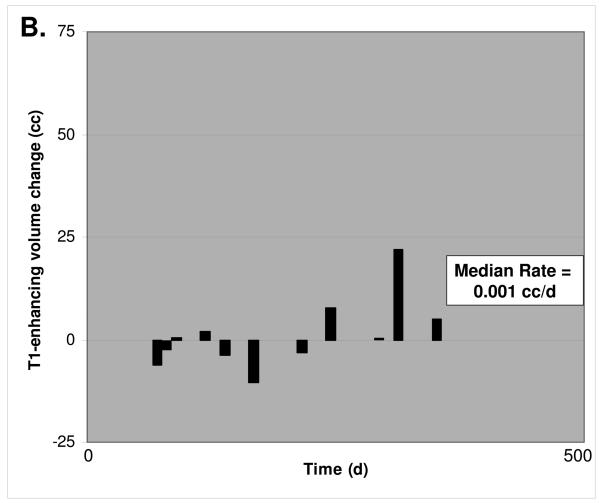
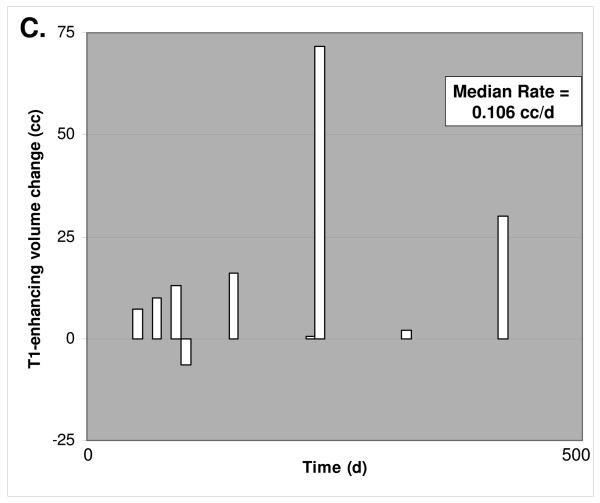
We then calculated in vivo tumor growth rates under TMZ treatment by comparing the volume of tumor on the initial scan to the volume present at cessation of TMZ treatment. Interestingly, we could not detect a correlation between pre-treatment MGMT status and the rate of tumor growth under TMZ treatment. Importantly however, MSH6-negative glioblastomas demonstrated a markedly increased rate of growth while being treated with TMZ, with a median T1-gadolinium enhancing signal change of +3.17 cc/month (0.106 cc/day), while median MSH6-positive tumor growth was only +0.04 cc/month (0.001 cc/day) under TMZ treatment (Table 3 and Figure 2). The magnitude of this difference was in accordance with in vitro studies of MSH6 function in alkylator-tolerant growth, and was significant (p=0.020). With the caveat that there may be subtle confounding factors in these comparisons that we were unable to detect due to our cohort size, this increased growth rate suggests that MSH6 loss in vivo corresponds to decreased clinical responsiveness to TMZ and subsequent recurrent tumor growth during treatment, mirroring the alkylator resistance conferred by MSH6 inactivation in vitro.
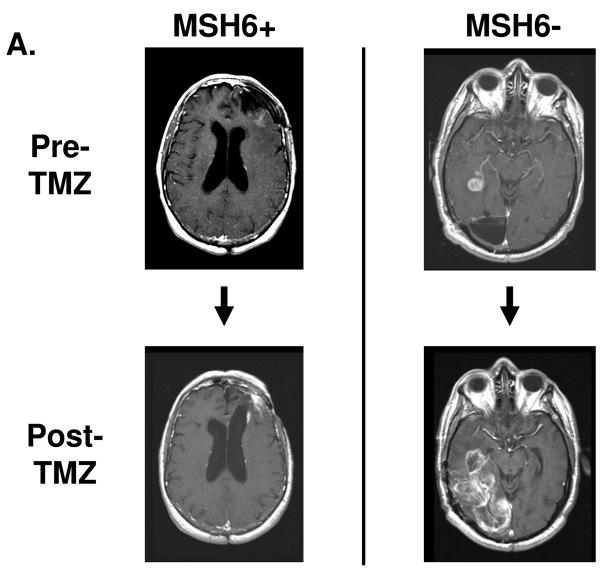
Figure 2.
3D MRI tumor growth calculations. A. Representative post-gadolinium enhancement T1 sequence axial sections through the tumor mass are shown, corresponding to the initiation of alkylating chemotherapy inclusive of TMZ and immediately prior to cessation of this treatment. The MSH6-positive case on the left shows recurrent tumor sample xT3692; 254 days elapsed between initial and final scan. The MSH6-negative case on the right shows recurrent glioblastoma samples xT4161; 249 days elapsed between initial and final scan. Multiple axial imaging sections through the tumor mass were summated to determine enhancing tumor volume. B. For 11 MSH6-positive tumors, volume change versus time elapsed between initial and followup MRI scan. The median rate of volume change was 0.001 cc/d. C. For 9 MSH6-negative tumors, volume change versus time, with a median rate of 0.106 cc/d. Full clinical/radiologic dataset available as Supplementary Table 1.
Discussion
Our findings demonstrate that loss of MSH6 expression occurs in a significant subset of post-XRT+TMZ recurrent glioblastomas, and is associated with the progressive growth of these tumors while they are under TMZ treatment. These findings parallel in vitro studies documenting the frequent emergence of MMR deficiency in cell line sub-clones after selection and outgrowth in alkylating agents (26, 27). Importantly, while many factors have been proposed to mediate therapeutic resistance based on studies of cancer cells exposed to differing doses of chemotherapeutic agents in vitro, our observations were made on patients receiving the clinical standard-of-care doses of an oral alkylating agent, indicating that MSH6 loss appears to recapitulate its known in vitro alkylator resistance role in glioblastoma patients in vivo.
This extension of in vitro mechanistic studies to an in vivo system has significant implications regarding patient:drug interactions; for example, it can be reasonably surmised that drug delivery across the blood-brain barrier and tumor vasculature is not a rate-limiting factor in TMZ treatment failure in cases with MSH6 loss, since the recurrent tumor cell mass displays the hallmarks of a selection pressure mediated by direct exposure to the drug itself. Likewise, in vitro studies indicate there is an interactive specificity between both drug and pathway in the development of resistance: MMR pathway defects can mediate up to a 100-fold resistance to alkylating agents, but only demonstrate approximately two-fold resistance to the intra-strand crosslinking agent cisplatin, and no significant resistance to agents such as topoisomerase inhibitors or inter-strand crosslinkers such as mitomycin C or CCNU (28). Our in vivo tumor growth rate measurements reflect a similar degree of difference in clinical progression between MSH6-negative and MSH6-positive glioblastomas during TMZ treatment. However, the prediction from in vitro studies would be that chemotherapeutic agents with different mechanisms (29, 30) may still prove effective in MSH6-negative glioblastomas, since the cellular resistance associated with MSH6 absence is specific for exposure to alkylators.
Previous reports have indicated that alkylator resistance can be mediated either by MGMT overexpression or MMR deficiency (20, 21, 25, 31). Importantly, our analyses of recurrent glioblastoma samples indicate that MSH6 loss occurred independently of the MGMT status of the pre-treatment tumor, indicating that MSH6 loss is not merely a proxy of MGMT status. Our data illustrate the relationship between these two mechanisms in patients who have undergone TMZ treatment, indicating that while pre-treatment tumor MGMT levels may be linked to primary resistance, acquisition of further increased expression does not appear to be a common cause of emergent TMZ resistance in the clinical setting. Interestingly, many current salvage chemotherapeutic regimens propose increasing the dosing schedule of TMZ, in variations known collectively as “dose-dense” modifications (32), in an attempt to overcome increased levels of MGMT activity. However, it appears that MSH6 loss can occur during TMZ treatment regardless of MGMT status, with subsequent clonal outgrowth leading to recurrence through an alternative route of treatment escape. Dose-dense modifications may therefore prove ineffective for patients with MSH6-negative tumors, as the window of therapeutic index available for this substantially myelotoxic therapy may have closed.
Given that there may be several operative mechanisms for functional MSH6 alteration, our IHC results may underestimate the contribution of altered MSH6 pathway function to TMZ insensitivity. Our sequencing efforts demonstrate clear evidence of somatic mutational alteration and clonal selection in some recurrent tumors. However, at the protein level, the MSH6 protein participates in a large multi-protein complex with other MMR proteins to coordinate mismatch binding, mismatch repair, and DNA damage checkpoint signaling. Recent work has demonstrated that O-6 methylguanine-thymine mismatches are specifically bound by the heterodimeric MSH2:MSH6 MutSalpha complex to drive activation of the ATR/Chk1 S/G2-phase checkpoint pathway (33). Post-translational changes in the subcellular localization of the MSH2:MSH6 complex occurs in response to genotoxic agents (34), communicating repair activity signals to checkpoint machinery (35). This checkpoint-signaling pathway is thought to mediate much of the tumoricidal effect of alkylating chemotherapy (11). Since study of MMR checkpoint pathway signaling remains an active area of investigation, it is likely that other novel candidates for analysis in treatment-tolerant tumors will emerge.
Indeed, the existence of post-XRT+TMZ glioblastomas that retain or even increase MSH6 expression highlights the probability that alternative genetic mechanisms do contribute to TMZ evasion. While it is plausible that alterations in other MMR genes such as MSH2 or MLH1 could play a similar role to MSH6 in glioblastoma, the canonical microsatellite instability typical of MSH2 or MLH1 deficiency is rarely found in adult sporadic pre-treatment or post-treatment glioblastoma specimens (36, 37). Notably, as opposed to other MMR genes, MSH6 deficiency is not associated with high levels of microsatellite instability (11). Furthermore, MSH6, but not other MMR genes, is preferentially targeted in vitro in unbiased chemotherapy resistance genetic screens (4). With this in mind, we speculate that TMZ may engender a selection pressure specifically targeting for MSH6 loss instead of other MMR genes, as tumor sub-clones negotiate a balance between the increased mutation load of MMR deficiency and the selective growth advantage of TMZ insensitivity.
Given the variability amongst salvage therapies within recurrent glioblastoma patient cohorts, further analyses in a more uniformly treated group of patients will be needed to confirm our findings and provide further insight into such gene-specific and treatment-specific hypotheses. Current clinical practice withholds surgical intervention in many cases of recurrence, due to a perceived lack of benefit relative to the risk of surgery in rapidly declining patients. With the advent of combined XRT+TMZ treatment, more glioblastoma patients are likely to undergo recurrent surgical intervention, affording important opportunities for well-controlled molecular pathologic studies of chemotherapeutic response and escape pathways. By better understanding the molecular basis for treatment evasion and subsequent clinical failure, we can envision the rational design of targeted combination therapeutic strategies that will limit the emergence of such treatment escape in future patients.
Acknowledgments
We thank the patients who participated in this research through donation of blood and tumor samples. We also thank the Massachusetts General Hospital Neuroradiology 3D Lab for use of 3D volumetric software, and Dr. Gregory Riggins and Dr. Charles Eberhart of The Johns Hopkins Hospital for providing tumor and normal DNA samples. The authors acknowledge funding support from the Institute of Cancer Research, the Wellcome Trust, a Brain Tumor Society Seth Harris Feldman Research Award, a National Brain Tumor Foundation GBM Grant, an American Brain Tumor Association Fellowship, an American Association of Neurosurgeons NREF Fellowship, and the National Institutes of Health.
References
- 1.Louis DN. Molecular Pathology of Malignant Gliomas. Annual Reviews of Pathology: Mechanisms of Disease. 2006:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New England Journal of Medicine. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–91. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–5. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 5.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci U S A. 1999;96:10764–9. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci U S A. 1993;90:6424–8. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomita-Mitchell A, Kat AG, Marcelino LA, Li-Sucholeiki XC, Goodluck-Griffith J, Thilly WG. Mismatch repair deficient human cells: spontaneous and MNNG-induced mutational spectra in the HPRT gene. Mutat Res. 2000;450:125–38. doi: 10.1016/s0027-5107(00)00020-8. [DOI] [PubMed] [Google Scholar]
- 8.Szadkowski M, Iaccarino I, Heinimann K, Marra G, Jiricny J. Characterization of the mismatch repair defect in the human lymphoblastold MT1 cells. Cancer Research. 2005;65:4525–9. doi: 10.1158/0008-5472.CAN-05-0080. [DOI] [PubMed] [Google Scholar]
- 9.Umar A, Koi M, Risinger JI, et al. Correction of hypermutability, N-methyl-N′-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Research. 1997;57:3949–55. [PubMed] [Google Scholar]
- 10.Lettieri T, Marra G, Aquilina G, et al. Effect of hMSH6 cDNA expression on the phenotype of mismatch repair-deficient colon cancer cell line HCT15. Carcinogenesis. 1999;20:373–82. doi: 10.1093/carcin/20.3.373. [DOI] [PubMed] [Google Scholar]
- 11.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 12.Hegde MR, Chong B, Blazo ME, et al. A homozygous mutation in MSH6 causes Turcot syndrome. Clinical Cancer Research. 2005;11:4689–93. doi: 10.1158/1078-0432.CCR-04-2025. [DOI] [PubMed] [Google Scholar]
- 13.Menko FH, Kaspers GL, Meijer GA, Claes K, van Hagen JM, Gille JJ. A homozygous MSH6 mutation in a child with cafe-au-lait spots, oligodendroglioma and rectal cancer. Fam Cancer. 2004;3:123–7. doi: 10.1023/B:FAME.0000039893.19289.18. [DOI] [PubMed] [Google Scholar]
- 14.Buttin BM, Powell MA, Mutch DG, et al. Penetrance and expressivity of MSH6 germline mutations in seven kindreds not ascertained by family history. Am J Hum Genet. 2004;74:1262–9. doi: 10.1086/421332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berends MJ, Wu Y, Sijmons RH, et al. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton SR, Liu B, Parsons RE, et al. The Molecular-Basis of Turcots-Syndrome. New England Journal of Medicine. 1995;332:839–47. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 17.Hegi ME, Diserens A, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New England Journal of Medicine. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 18.Silber JR, Blank A, Bobola MS, Ghatan S, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase-deficient phenotype in human gliomas: frequency and time to tumor progression after alkylating agent-based chemotherapy. Clin Cancer Res. 1999;5:807–14. [PubMed] [Google Scholar]
- 19.Nakasu S, Fukami T, Baba K, Matsuda M. Immunohistochemical study for O6-methylguanine-DNA methyltransferase in the non-neoplastic and neoplastic components of gliomas. J Neurooncol. 2004;70:333–40. doi: 10.1007/s11060-004-9170-6. [DOI] [PubMed] [Google Scholar]
- 20.Bearzatto A, Szadkowski M, Macpherson P, Jiricny J, Karran P. Epigenetic regulation of the MGMT and hMSH6 DNA repair genes in cells resistant to methylating agents. Cancer Research. 2000;60:3262. + [PubMed] [Google Scholar]
- 21.Liu LL, Markowitz S, Gerson SL. Mismatch repair mutations override alkyltransferase in conferring resistance to temozolomide but not to 1,3-bis(2-chloroethyl)nitrosourea. Cancer Research. 1996;56:5375–9. [PubMed] [Google Scholar]
- 22.Wang M-C. A semiparametric model for randomly truncated data. Journal of the American Statistical Association. 1989;84:742–8. [Google Scholar]
- 23.Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751–63. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 24.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 25.Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O-6-alkylguanine-DNA alkyltransferase analysis and response to temodal in newly diagnosed malignant glioma. Journal of Clinical Oncology. 1998;16:3851–7. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 26.Branch P, Aquilina G, Bignami M, Karran P. Defective Mismatch Binding and a Mutator Phenotype in Cells Tolerant to DNA Damage. Nature. 1993;362:652–4. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- 27.Bardelli A, Cahill DP, Lederer G, et al. Carcinogen-specific induction of genetic instability. Proc Natl Acad Sci U S A. 2001;98:5770–5. doi: 10.1073/pnas.081082898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papouli E, Cejka P, Jiricny J. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res. 2004;64:3391–4. doi: 10.1158/0008-5472.CAN-04-0513. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Taverna P, Whitacre CM, Chatterjee S, Gerson SL. Pharmacologic disruption of base excision repair sensitizes mismatch repair-deficient and -proficient colon cancer cells to methylating agents. Clinical Cancer Research. 1999;5:2908–17. [PubMed] [Google Scholar]
- 30.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Research. 2005;65:6394–400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 31.Pepponi R, Marra G, Fuggetta MP, et al. The effect of O6-alkylguanine-DNA alkyltransferase and mismatch repair activities on the sensitivity of human melanoma cells to temozolomide, 1,3-bis(2-chloroethyl)1-nitrosourea, and cisplatin. J Pharmacol Exp Ther. 2003;304:661–8. doi: 10.1124/jpet.102.043950. [DOI] [PubMed] [Google Scholar]
- 32.Vera K, Djafari L, Faivre S, et al. Dose-dense regimen of temozolomide given every other week in patients with primary central nervous system tumors. Ann Oncol. 2004;15:161–71. doi: 10.1093/annonc/mdh003. [DOI] [PubMed] [Google Scholar]
- 33.Yoshioka K, Yoshioka Y, Hsieh P. ATR Kinase Activation Mediated by MutSalpha and MutLalpha in Response to Cytotoxic O(6)-Methylguanine Adducts. Mol Cell. 2006;22:501–10. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christmann M, Tomicic MT, Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutSalpha mismatch-binding activity. Nucleic Acids Res. 2002;30:1959–66. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawn MT, Umar A, Carethers JM, et al. Evidence for a Connection between the Mismatch Repair System and the G(2) Cell-Cycle Checkpoint. Cancer Research. 1995;55:3721–5. [PubMed] [Google Scholar]
- 36.Alonso M, Hamelin R, Kim M, et al. Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res. 2001;61:2124–8. [PubMed] [Google Scholar]
- 37.Martinez R, Schackert HK, Plaschke J, Baretton G, Appelt H, Schackert G. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology. 2004;66:395–403. doi: 10.1159/000079488. [DOI] [PubMed] [Google Scholar]