Abstract
Somatically acquired epigenetic changes are present in many cancers. Epigenetic regulation is maintained via post-translational modifications of core histones. Here, we describe inactivating somatic mutations in the histone lysine demethylase, UTX, pointing to histone H3 lysine methylation deregulation in multiple tumour types. UTX reintroduction into cancer cells with inactivating UTX mutations resulted in slowing of proliferation and marked transcriptional changes. These data identify UTX as a new human cancer gene.
In the nuclei of eukaryotic cells, DNA is packed into chromatin. The basic constituent of chromatin, the nucleosome, is comprised of 147 basepairs of DNA wound around an octamer of core histone proteins. Post-translational modifications of the N-terminal tails of these core histone proteins, including methylation, acetylation, ubiquitylation and phosphorylation1, modulate chromatin configuration and mediate access to nuclear protein factors, resulting in changes in transcriptional regulation which may influence cell proliferation, differentiation and death. These epigenetic modifications are controlled by complex enzymatic machinery that includes specific histone methylases and demethylases. Deregulation of this machinery could alter chromatin configuration and disrupt normal transcriptional programs, both of which are features of cancer cells. Indeed, genomic alterations of MLL, NSD1, CREBBP(CBP) and MMSET, all encoding proteins involved in post-translational histone modification, have been implicated in acute leukaemias and myeloma (http://www.sanger.ac.uk/genetics/CGP/Census/).
As part of systematic mutational screens of the coding genome (http://www.sanger.ac.uk/genetics/CGP/Studies/), we investigated the role of mutations of the histone methylation machinery in human cancer. A combination of PCR-based resequencing of coding exons for point mutations, SNP array hybridisation for copy number changes and multiplex exonic PCR identified 39 inactivating mutations in 1390 cancer samples (Table 1, Supplementary Tables 1,2) in UTX, located on Xp11.2, encoding a histone H3 lysine 27 (H3K27) demethylase2-4.
Table 1.
UTX inactivating mutations identified
| Sample | Cancer type |
Sex | mutation - DNA | mutation - Protein |
|---|---|---|---|---|
| MONO-MAC-6 | AML | M | Hom del exons 3-10 | |
| THP-1 | AML | M | Hom del exons 1-16 | |
| RT-112 | Bl | F | Het c.3416delC | p.P1139fsX19 |
| KU-19-19 | Bl | M | Hom c.2587C>T | p.Q863X |
| HCC1806 | Br_a | F | Hom del exon 6 | |
| HCC2157 | Br_b | F | Hom del all coding exons | |
| MUTZ-1* | CML | F | Hom del exons 3-8 | |
| NALM-1 | CML | F | Hom del exons 3-8 | |
| BV-173 | CML | M | Hom del exons 3,4 | |
| LS-174T* | Colo | F | Hom c.3945_3946insA | p.E1316fsX17 |
| KM12* | Colo | F | Hom c.117delC | p.S40fs*2 |
| SNU-C2B* | Colo | F | Het c.3520_3521insT | p.W1174fsX6 |
| MFE-296* | Endo | F | Het c.1793_1794delTA | p.I598fsX6 |
| D-247MG | Gbm | F | Hom c.514C>T | p.R172X |
| D-423MG | Gbm | M | Hom Exon 4 +1 G>T | |
| HDLM-2 | HL | M | Hom del exons 3-8 | |
| L-363 | MM | F | Hom del exons 1, 2 | |
| LP-1 | MM | F | Hom c.1621C>T | p.Q541X |
| PD2916a | MM | F | Het c.1787delA | p.N596fsX3 |
| KMS-12-BM | MM | F | Hom del exons 5-12 | |
| PD2933a | MM | M | Hom c.3014delT | p.L1005fsX43 |
| SK-MM-2 | MM | M | Hom del exons 1, 2 | |
| SK-LU-1 | NSCLC | F | Het c.2029C>T | p.Q677X |
| TE-15 | Oes | F | Hom del exons 3, 4 | |
| PD3245a | Oes | M | Hom c.646G>T | p.E216X |
| KYSE-180 | Oes | M | Hom c.997C>T | p.Q333X |
| KYSE-450 | Oes | M | Hom del exons 5-8 | |
| TE-11 | Oes | M | Hom del all coding exons | |
| TE-6 | Oes | M | Hom del exons 1, 2 | |
| MIA-PaCa-2 | Panc | M | Hom del all coding exons | |
| PD2213a | RCC | F | Het c.2090delA | p.N697fsX18 |
| LB2241-RCC | RCC | M | Hom Exon 12 −1 G>T | |
| LB996-RCC | RCC | M | Hom c.2654_2663delTGTCTGTGTC |
p.M885fsX10 |
| PD3318a | RCC | M | Hom c.3445_3446delAA | p.N1149fsX1 |
| PD2147a | RCC | M | Hom c.4161_4162delTG | p.Y1387fsX1 |
| PD3577^ | RCC | F | Het c.1834C>T | p.R612X |
| NCI-H1926 | SCLC | M | Hom c.869_870insT | p.G291fsX22 |
| NCI-H128 | SCLC | M | Hom c.2122G>T | p.G708X |
| PF-382 | T-ALL | F | Het c.3835C>T | p.R1279X |
Underlined samples - verified somatic mutations
microsatellite unstable cancers
germline mutation
PDxxxx - samples are from primary cancers. Cancer type code: AML : acute myelogenous leukaemia, Bl : bladder transitional cell carcinoma, Br_a : breast acantholytic squamous carcinoma, Br_b : breast adenocarcinoma, CML : chronic myelogenous leukemia-blast crisis, Colo : colorectal adenocarcinoma, Endo : endometrial adenocarcinoma, Gbm : gliobastoma, HL : Hodgkin Lymphoma, MM : multiple myeloma, NSCLC : non-small cell lung cancer, Oes : oesophageal squamous cell carcinoma, Panc : pancreatic adenocarcinoma, RCC : renal clear cell carcinoma, SCLC : small-cell lung carcinoma,T-ALL : T-cell acute lymphoblastic leukaemia. Note all primary female cancer samples (PDxxxxx) are denoted conservatively as heterozygotes. Mutations are denoted relative to UTX sequence GenBank accession NM_021140.1 GI:10863942
Inactivating mutations in UTX included 16 homozygous (in females) or hemizygous (in males) large deletions, 9 nonsense mutations, 12 small frame-shifting insertion/deletions and 2 consensus splice site mutations which lead to aberrant splicing and premature termination codons (Table 1). All large homozygous/hemizygous deletions were confirmed by PCR (Supplementary Figure 1, Table 3). Nine were contained within the genomic footprint of UTX indicating that UTX is the target of the homozygous deletion cluster (Supplementary Figure 1).
Matching constitutional DNA was available from 12 cancers with inactivating mutations. Eleven proved to be somatic upon analysis, seven in primary cancers and four in cancer cell lines. The exception was a nonsense mutation in a renal cancer found to be a germline allele, suggesting that UTX mutations may contribute to susceptibility in rare instances. The ratio of truncating to non-truncating somatic single base substitutions (Supplemental Table 4) observed in UTX was 1:2 compared to 1:17 expected by chance, indicating selection for inactivating variants. To assess the likely provenance of the 28 inactivating variants found in samples for which no constitutional DNA was available, all coding exons of UTX were sequenced in constitutional DNA from 400 male controls. No truncating variants were identified. Furthermore, 411 normal DNAs were evaluated on Affymetrix SNP6.0 arrays for copy number changes and no instances of homozygous or hemizygous deletions were detected. It is therefore likely that most were somatically acquired.
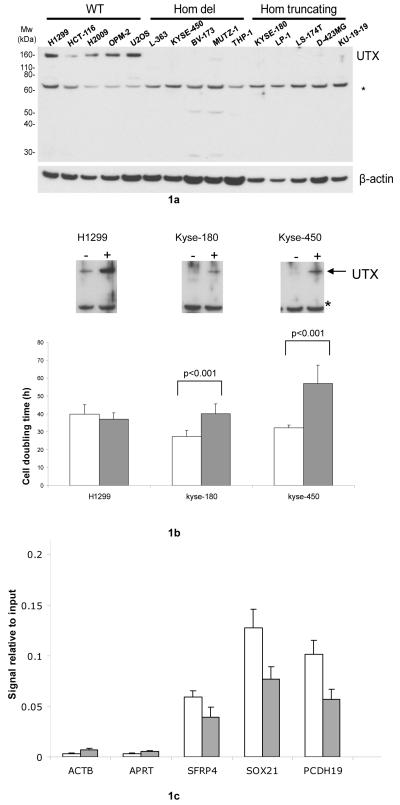
Western analysis showed no detectable protein in any of 24 cancers with homozygous inactivating mutations (Figure 1a; data not shown). In addition to the alterations described above, which are clearly predicted to inactivate the protein, two overlapping but non-identical deletions within intron 2 of UTX were detected in two cancer cell lines. One line expressed no detectable UTX transcript or protein (RPMI-8226) whilst the other (U-266) expressed UTX transcript and protein. By contrast, 14 cancers of diverse tissue origins with wildtype UTX sequence all showed readily detectable protein expression (Figure 1a; data not shown). The pattern of inactivating mutations is consistent with UTX acting as a tumour suppressor gene.
Figure 1.
1a) Western blot of samples showing lack of protein expression in UTX mutant samples.1b) Growth Effects of UTX reintroduction. Top panel -western blot confirmation of UTX expression in the transfected cell lines (UTX wildtype NCI-H1299, UTX null KYSE-180, 450). Bottom panel - significant cell doubling time increases in hours seen upon reintroduction of UTX wildtype in to UTX null lines but not in wildtype control cells. White bars- empty vector controls, grey bars – UTX reintroduction 1c) Specific reduction of H3K27me3 on genes showing differential expression after UTX introduction in KYSE180 (SOX21,PCDH19). Relative enrichment (fraction H3K27me3 bound DNA/input DNA) was quantified by real time PCR using primers against promoter regions of respective genes. Error bars represent the standard deviation from two independent transfection experiments. Primer sequences are given in supplementary Table 3. ACTB and APRT are negative controls for H3K27me3 marks and SFRP4 is a positive antibody control for H3K27me3. Controls are all taken from Schlesinger et al15. White bars - empty vector control, grey bars -UTX reintroduction
UTX mutations were found in multiple cancer types. The highest prevalence, 10% (6/58), was observed in multiple myeloma. Of note, activation of MMSET/WHSC1, which has been shown to have methylase activity towards H3K275 and other histone residues6, has also been implicated in multiple myeloma. Interestingly, the UTX null multiple myeloma samples reported here are all negative for the MMSET activating t(4;14) translocation 6-8, suggesting potential mutual exclusion of mutations. Inactivating UTX mutations were also detected in 8% (6/77) of oesophageal squamous cell carcinomas and 1.4% (6/419) of renal clear cell carcinomas. For these three cancer types, the total number of mutations reflects both an initial screening series followed by the identification of additional mutations in a larger follow-up series. Since primary tumours were not analysed for homozygous/hemizygous deletions, the prevalence of inactivating mutations is likely an underestimate. Other cancer types with inactivating mutations included myeloid leukaemias, breast and colorectal cancers and glioblastoma.
UTX is one of a limited number of genes on the X chromosome that escapes X inactivation in females9. Therefore, in contrast to the X-linked tumour suppressor gene WTX, which is subject to X inactivation10, biallelic mutations of UTX in females may be necessary to contribute to oncogenesis. There was a trend amongst the 16 cancer cell lines with inactivating mutations derived from cancers in females to lose the other UTX allele with 11/16 (65%) as compared to 111/262 (42%) UTX wildtype cancers showing LOH (P=0.039). In further support of biallelic UTX inactivation, examination of cDNA and protein from the remaining six lines with heterozygous truncating mutations revealed one, PF-382, with loss of the wildtype transcript and no UTX protein expression and one, SNU-C2B, with both wildtype and mutant transcripts present but no UTX protein. The remaining four cell lines (Table 1), expressed both mutant and wildtype transcripts, had detectable UTX protein expression and may represent passenger events.
Males have only a single copy of UTX. However, there is a paralogue on the Y chromosome, UTY, which shows 83% amino acid identity9. We investigated whether UTY may be functioning as the second tumour suppressor gene allele to UTX in males. Genomic loss of UTY in male cancer lines with inactivating UTX mutations (13/16, 81%) was significantly more frequent than UTY loss in UTX wildtype cancers (153/307, 49%) (P = 0.0142). In addition, a focal UTY deletion was found in a single myeloma line, U-266. Overall, the data support an allelic role of UTY for UTX, although in vivo studies indicate that purified UTY does not fully recapitulate the equivalent histone demethylase activity of UTX3,11. Taking the data from female and male cancers together, there is support for a two-hit model of UTX/UTY inactivation, with P= 0.0016 for loss of either UTX/UTY in samples with an inactivating UTX mutation versus those with wildtype.
UTX is one of two recently identified histone H3 lysine 27 demethylases2-4 Methylation at lysine 27 is highly correlated with genomic silencing and repression of transcription1. UTX is also a component of the Mixed-Lineage-Leukaemia (MLL)2/3 complexes that promote histone H3 lysine 4 (H3K4) methylation4,12, a mark of open and actively transcribed chromatin1. A concerted mechanism of transcriptional control involving cycles of H3K27 and H3K4 methylation linked via UTX has been proposed2,4. This transcriptional control would likely be subverted by the UTX inactivating mutations reported here.
Following from this, functional evaluation of the impact of UTX mutation was undertaken. No correlation between global H3K27me3/H3K4me3 levels measured via western blot in UTX null lines compared to UTX wildtype samples was seen (data not shown). To further investigate potential mechanisms of involvement in oncogenesis, wildtype UTX was expressed in two UTX null cell lines (KYSE-180 and KYSE-450) and a UTX wildtype line (NCI-H1299). Re-expression of UTX resulted in significant increases in cell doubling time compared to empty vector controls in the two null lines but had no discernable effect in the wildtype line, even though expression was increased over endogenous levels (Figure 1b). We then used microarray analysis to investigate if UTX reintroduction induced gene expression changes. Both null lines showed greater changes in gene expression (KYSE-180, n=327 genes, KYS3-450, n=241 genes, FDR<0.05) upon UTX re-expression compared to the NCI-H1299 UTX wildtype line (n= 46 genes FDR<0.05; Supplementary Table 5). The sets of differentially expressed genes from the transfectants were analysed for possible enrichment of genes associated with the H3K27 methylating polycomb complex and/or H3K27me3/H3K4me3 marks13,14 (Supplemental methods). There was no enrichment for any of these gene sets in the UTX wildtype control line (p >0.1 for all three datasets). However, significant enrichment (p<0.001) was found for polycomb genes in both UTX null lines transfected with wild type UTX. Of note, in both lines there was enrichment for H3K27me3 marked genes (KYSE180 p<0.0001; KYSE450 p=0.065) but not H3K4me3 (KYSE180 p=0.7; KYSE450 p=0.3). ChIP-PCR of SOX21 and PCDH19, two genes showing significant expression changes in the transfected UTX null lines, demonstrated a significant decrease in H3K27me3 levels upon UTX reintroduction (Figure 1c), suggesting that the expression changes were a direct effect of reconstituted UTX demethylase activity. These data indicate that mutational inactivation of UTX is likely to be acting, at least in part, through transcriptional control mechanisms.
Taken together, the results presented provide strong evidence for UTX as the first mutated histone demethylase that acts as a human cancer gene, indicating that genetic mechanisms may underpin a significant component of epigenetic deregulation in cancers.
Supplementary Material
Acknowledgements
Funding was provided by the Wellcome Trust. GvH is an EMBO fellow and PJC is a Kay Kendall Leukaemia Fund Fellow. RAD is supported by the Robert A. and Renee E. Belfer Foundation for Innovative Cancer Science, GT is a Special Fellow of the Leukemia and Lymphoma Society. GlaxoSmithKline provided support for the SNP 6.0 microarray analyses. UTX antibodies were a kind gift from Prof. Eli Canaani, Weizmann Institute of Science, Israel.
References
- 1.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Agger K, et al. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 3.Lan F, et al. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 4.Lee MG, et al. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 5.Kim J-Y, et al. Mol. Cell. Biol. 2008;28:2023–2034. doi: 10.1128/MCB.02130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marango J, et al. Blood. 2008;111:3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchner H, et al. Blut. 1981;43:93–97. doi: 10.1007/BF00320467. [DOI] [PubMed] [Google Scholar]
- 8.Pegoraro L, et al. Blood. 1989;73:1020–1027. [PubMed] [Google Scholar]
- 9.Greenfield A, et al. Hum. Mol. Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 10.Rivera MN, et al. Science. 2007;315:642–645. doi: 10.1126/science.1137509. [DOI] [PubMed] [Google Scholar]
- 11.Hong S, et al. Proc. Natl. Acad. Sci. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issaeva I, et al. Mol. Cell. Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genes & Development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan G, et al. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Schlesinger Y, et al. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.