Abstract
In an effort to develop octahedral metal complexes as chemotherapeutic and diagnostic agents targeted to DNA, it is critical to optimize the properties of their cellular uptake. Appending d-octaarginine has been found to improve both the uptake and nuclear localization efficiency of these complexes, but the increased positive charge interferes with selective DNA binding and hence activity. Herein, we evaluate the nuclear entry of a series of luminescent ruthenium peptide conjugates of shorter sequence and lower charge. As is the case for the d-octaarginine conjugate (Ru-D-R8), the tetrapeptide RrRK (where r = d-arginine) facilitates nuclear localization of the ruthenium complex above a threshold concentration, though the threshold is higher for this conjugate (Ru-RrRK) than for Ru-D-R8. Furthermore, appended fluorescein, which lowers the threshold concentration for Ru-D-R8, does not improve nuclear entry of Ru-RrRK, indicating that fluorescein conjugation is not a general strategy for modulating the distribution of cell-penetrating peptides. Similarly, the concentration required for nuclear entry of Ru-RrRK is much higher than has been reported for a thiazole orange RrRK conjugate, demonstrating the influence of payload on the efficiency of uptake and localization of cell-penetrating peptides.
Keywords: ruthenium complex, metal-DNA interaction, peptide conjugate, cellular uptake, nuclear targeting
1. Introduction
Peptide conjugation is a widely used and effective method for improving both cellular and nuclear entry of a variety of cargo molecules.1–3 We have successfully delivered our rhodium(III) 5,6-chrysenequinone diimine (chrysi) and ruthenium(II) dipyridophenazine (dppz) complexes to the nucleus through covalent attachment of d-octaarginine.4,5 Without the peptide, these compounds localize in the cytoplasm, as seen by microscopy studies on the luminescent Ru(II) dppz complexes.6 Similarly, others have linked octaarginine to a ruthenium bis(dipyridophenazine) complex, resulting in greatly increased cellular and nuclear accumulation versus the parent complex.7 In general, ruthenium polypyridyl complexes exhibit poor nuclear permeability without assistance of a targeting moiety, though there are exceptions.8
The chrysi complexes of rhodium(III) bind single-base mismatches in DNA,9,10 but the added +8 charge imparted by octaarginine increases the nonspecific binding of the metal-peptide conjugate, due to electrostatic association with the negatively charged DNA backbone.4 In order to enhance the nuclear accumulation of our chrysi complexes of rhodium(III) without significant impairment of their specificity for mismatches, we have now investigated appending shorter peptides possessing less overall charge than octaarginine, but using luminescent ruthenium analogues.
Studies by Kelley and coworkers have demonstrated the feasibility of using very short peptides to target small molecules to the nucleus or mitochondria.11,12 Thiazole orange (TO) conjugated to the tetrapeptide RrRK (r = d-arginine) accumulates primarily in the nucleus of HeLa cells, whereas TO-FrFK localizes mainly in the mitochondria. Both conjugates are reported to cross the plasma membrane with efficiencies approaching that of the longer Tat peptide (RKKRRQRRR).11 Using RrRK as the nuclear targeting signal for metal complexes reduces the positive charge added by the peptide in half compared to octaarginine, and thus should reduce the amount of nonspecific DNA binding. Here, we use dppz complexes of Ru(II) functionalized with short peptides, luminescent analogues of our rhodium complexes, to evaluate the cellular uptake.
2. Results and discussion
2.1. Synthesis of the conjugates
A series of Ru(II) dipyridophenazine (dppz) conjugates were prepared by solid-phase coupling of Ru(phen)(bpy′)(dppz)2+ to the N-terminal amine of the peptide. The dppz complexes of ruthenium(II) serve as luminescent analogues of our chrysi complexes of rhodium(III).13 The complexes bind duplex DNA by intercalation.14-16 In aqueous solution the luminescence is quenched through hydrogen bonding, but when intercalated and protected from water, the complex luminesces brightly.14,17 More generally, the ruthenium complexes function as light switches for non-aqueous environments, luminescing only when bound to the hydrophobic regions of membranes, nucleic acids, and other macromolecules. Furthermore, the ruthenium complexes are stable in aqueous solution, making them useful cellular probes.
2.2 Cellular uptake of Ru-RrRK
We conjugated Ru(phen)(bpy′)(dppz)2+ to the nuclear targeting signal RrRK (r = d-arginine)(Figure 1). HeLa cells incubated for 2 h with 5–20 μM Ru-RrRK show punctate cytoplasmic luminescence, but no staining of the nucleus (Figure 2A, Table 1). Interestingly, increasing the incubation time to 24 h does not change the subcellular localization of 20 μM Ru-RrRK. This staining pattern is similar to that previously observed for the d-octaarginine conjugate of this ruthenium complex (Ru-D-R8) at 5–10 μM.5 This distribution also implicates endocytosis, a proposed mechanism of uptake for oligoarginine cell penetrating peptides,18 as the mode of entry, though this remains to be confirmed by mechanistic studies. As expected, cellular uptake of the peptide conjugate is enhanced compared to the unconjugated complex Ru(phen)2dppz2+, as observed by direct comparison of the two complexes by confocal microscopy following identical incubation conditions (10 μM, 2 h).
Figure 1.
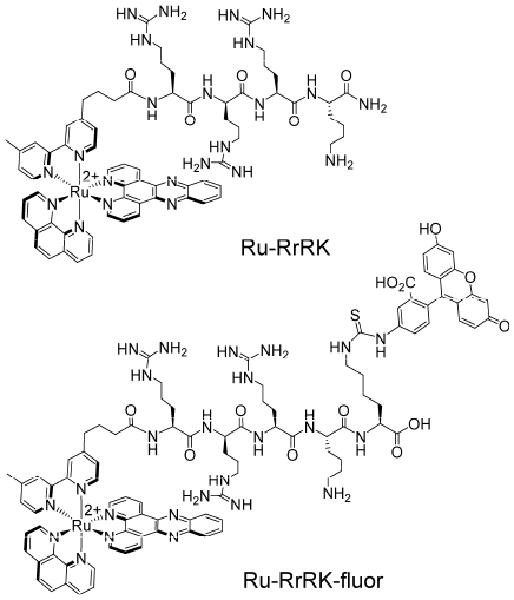
Structures of Ru-RrRK conjugates.
Figure 2.
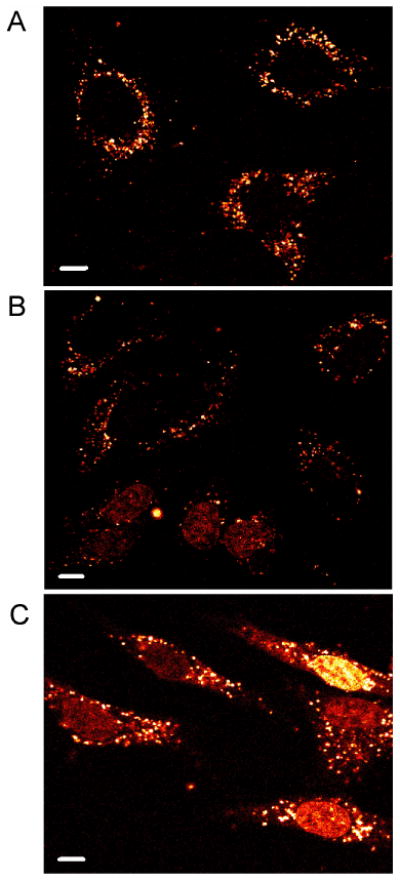
Subcellular distribution of Ru-RrRK. HeLa were incubated with (A) 20, (B) 40, or (C) 100 μM Ru-RrRK in complete medium for 2 h. At 20 μM, only punctate staining of the cytoplasm is present. At higher concentrations, some cells show additional nuclear staining. Scale bars are 10 μm.
Table 1.
Percentage of cells with nuclear staining by Ru-RrRKa
| concentration (μM) | + serum | - serum |
|---|---|---|
| 10 | 0% | 0% |
| 20 | 0% | 2% |
| 30 | 7% | 53% |
| 40 | 14% | 65% |
| 100 | 74% | n.d. |
HeLa cells were incubated with Ru conjugate for 2 h at 37 °C in medium +/- serum, then rinsed with HBSS and analyzed by confocal microscopy. Dead cells were excluded by their morphology. Data not determined are indicated by n.d.
The same enhancement in cellular uptake for Ru-RrRK versus Ru(phen)2dppz2+ is seen by flow cytometry analysis. Cells treated with Ru-RrRK have a 1.7-fold increase in mean luminescence compared to those exposed to Ru(phen)2dppz2+ (Table 2). Given that the luminescence of Ru-RrRK bound to DNA is inherently 61% of Ru(phen)2dppz2+ bound to DNA, based upon titrations with calf thymus DNA, the uptake is enhanced 3-fold with the appended short peptide. Significantly, however, the cellular uptake of Ru-RrRK is only ∼25% that for Ru-D-R8 (Table 2). This observation is consistent with previous results showing that short oligoarginines are less effective at promoting the cellular entry of fluorescein than longer ones.19 In fact, based upon cell luminescence, Ru-D-R8 shows a significantly higher level of luminescent uptake in comparison to several ruthenium conjugates functionalized with slightly shorter peptides (Table 2).
Table 2.
Cellular uptake of ruthenium conjugates assayed by flow cytometry
| complex | mean luminescence (a.u.)a |
|---|---|
| Ru(phen)2dppz2+ | 79 ± 17 |
| Ru-D-R8 | 328 ± 11 |
| Ru-RrRK | 133 ± 7 |
| Ru-PKKKRKV | 86 ± 4 |
| Ru-KSKKQK | 73 ± 8 |
HeLa cells were incubated with 10 μM ruthenium conjugate for 2 h at 37 °C. Ruthenium complexes were excited at 488 nm, with emission observed at 600–620 nm. The mean luminescence intensity of cells not treated with complex is 20. Each data point is the mean ± the standard deviation of three samples.
There is evidence in the literature that fluorescein-conjugated cell-penetrating peptides can adhere to the cellular exterior,20 artificially increasing the apparent uptake when measured by flow cytometry. Although trypsinization is recommended to reduce the membrane-bound material, we did not use trypsin in these experiments, as it would preferentially cleave at the l-amino acids, thus increasing the apparent amount of Ru-D-R8 cellular uptake versus our other conjugates. Furthermore, the lack of defined staining of the cellular periphery in our confocal microscopy experiments indicates that either our conjugates do not accumulate at the membrane, or that the luminescence of such bound species is quenched.
At higher concentrations, the distribution of Ru-RrRK within cells changes, and the cell population becomes heterogeneous. In addition to the punctate cytoplasmic structures, the complex localizes in the nucleus in a small percentage of cells when incubated at 30–40 μM (Figure 2B, Table 1). The fraction of cells with nuclear staining increases with concentration of the complex. At 100 μM, the complex is located in the nucleus in 74% of cells (Figure 2C, Table 1).
This population heterogeneity has been observed previously for fluorescein-nonaarginine21,22 and for our ruthenium-octaarginine conjugate lacking fluorescein.5 A notable difference is that Ru-RrRK requires a higher concentration (30 μM vs. 15 μM) to accumulate inside the nucleus than Ru-D-R8, and a higher threshold (>40 μM vs. 20 μM) is necessary for the majority of cells to exhibit nuclear staining. Thus RrRK is less effective at promoting nuclear uptake of our ruthenium complex than d-octaarginine.
A shortened oligoarginine, Ru-tetraarginine (Ru-D-R4) was also examined, and it was found to have similar cellular uptake characteristics to Ru-RrRK. When incubated at 20 μM for 2 h, Ru-D-R4 is limited to punctate structures in the cytoplasm (Figure 3). At 30 μM, no cells had nuclear staining, in contrast to Ru-RrRK which reached the nucleus in 7% of cells. These two conjugates were synthesized with different C-termini, an amide for Ru-RrRK and a carboxylic acid for Ru-D-R4. This could play a small role in their differences in cellular internalization, however the amide- and carboxy-terminated versions of Ru-RrRK at 20 μM and 24 h show similar uptake.
Figure 3.
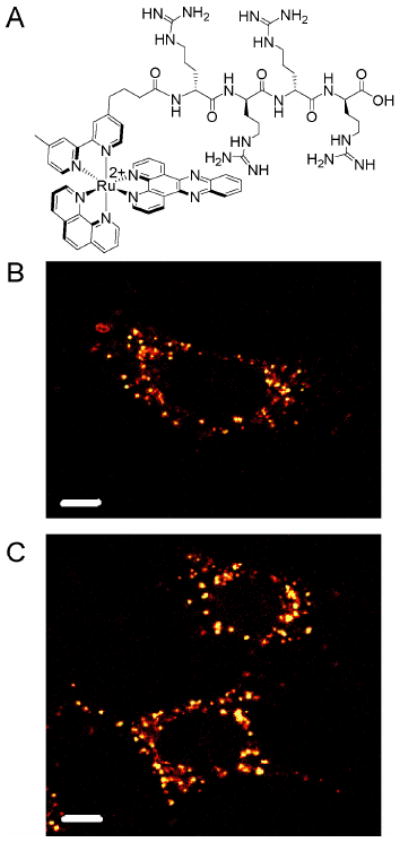
Subcellular distribution of Ru-D-R4. The structure of the conjugate is shown in (A). HeLa cells were incubated with (B) 20 or (C) 30 μM Ru-D-R4 in complete medium for 2 h. Punctate staining of the cytoplasm is observed. Scale bars are 10 μm.
We also evaluated the cellular accumulation of Ru-RrRK in serum-free medium (Figure 4), to allow comparison to the previously described experiments on the thiazole-orange conjugate (TO-RrRK), which were performed in the absence of serum.11 Not surprisingly, Ru-RrRK enters cells more readily under these conditions, and the concentration required for nuclear staining is reduced. At only 30 μM complex for 2 h, half of the cells show nuclear staining (Table 1). However, Ru-RrRK exhibits less efficient nuclear entry than TO-RrRK, which localizes in the nucleus at a lower incubation concentration (5 μM for 1.5 h) in the same cell line.11 Hence, the ability of RrRK to impart nuclear localization is affected by the nature of the cargo, with the larger and more positively charged ruthenium complex being more difficult to direct than thiazole orange.
Figure 4.
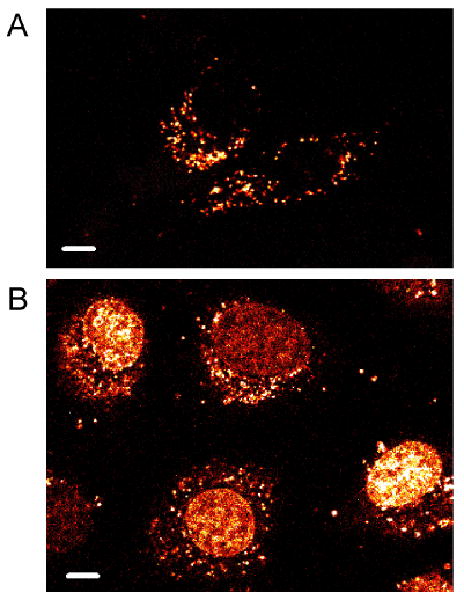
Subcellular distribution of Ru-RrRK in serum-free medium. HeLa were incubated with (A) 20 or (B) 40 μM Ru-RrRK in for 2 h. At 20 μM in medium without serum, the most cells show only punctate cytoplasmic staining, while at 40 μM, the majority of cells exhibit additional nuclear labeling. Scale bars are 10 μm.
In earlier experiments, we observed that attachment of fluorescein to Ru-D-R8 influences its subcellular distribution, allowing the conjugate with fluorescein to enter the nucleus under conditions for which the complex without fluorescein is excluded.5 Interestingly, an appended fluorescein does not have the same effect on Ru-RrRK; instead, cellular uptake is impaired by the dye. At concentrations up to 30 μM and 2 h incubation, only punctate cytoplasmic luminescence is seen (Figure 5), whereas the conjugate lacking fluorescein localizes in the nucleus in 7% of cells under these conditions (Table 1). The lack of benefit from fluorescein could be due the stronger relative effect of its negative charge on this shorter peptide. The fluorescein-tagged peptide also contains a carboxylic acid terminus, rather than the amide, which adds another negative charge. With reduced positive charge, the conjugate is less able to use the membrane potential as a driving force for entry; membrane potential has been shown to be an important factor in the cellular uptake of guanidinium-rich peptides.23
Figure 5.
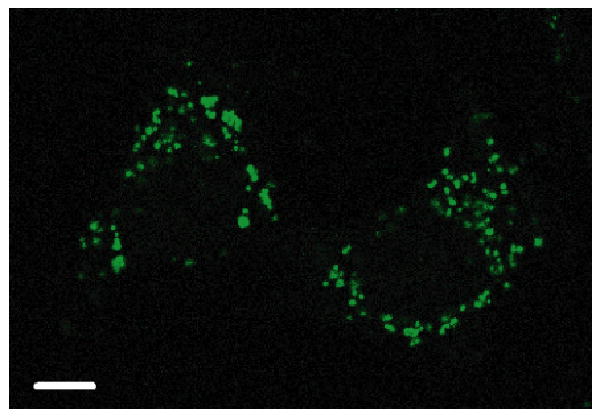
Subcellular distribution of Ru-RrRK-fluor. HeLa were incubated with 30 μM Ru-RrRK-fluor for 2 h in complete medium. Punctate staining of the cytoplasm is observed. Scale bar is 10 μm.
2.3 Effect of sequence variations on short peptides
It is known in the literature that charge is not the sole determinant in the uptake of cell-penetrating peptides; oligoarginines enter cells much more effectively than oligolysines.21 To confirm this for our system, we examined the cellular entry of Ru-KKKK. We also synthesized Ru-SrSr, which has even less total charge, but still contains two arginines. For Ru-KKKK and Ru-SrSr, only faint luminescence in the cytoplasm was observed after incubation at 40 μM for 2 h. Increasing the incubation concentration and time (100 μM, 4 h) leads to brighter, punctate cytoplasmic staining, and a small percentage of cells (17% for Ru-KKKK and 5% for Ru-SrSr) exhibit additional nuclear staining (Figure 6). Ru-KKKK luminescence in the cells is a little more intense than Ru-SrSr, indicating that its increased positive charge gives a small advantage over the two arginines of Ru-SrSr. As expected, both Ru-KKKK and Ru-SrSr are poorly internalized compared to Ru-RrRK.
Figure 6.
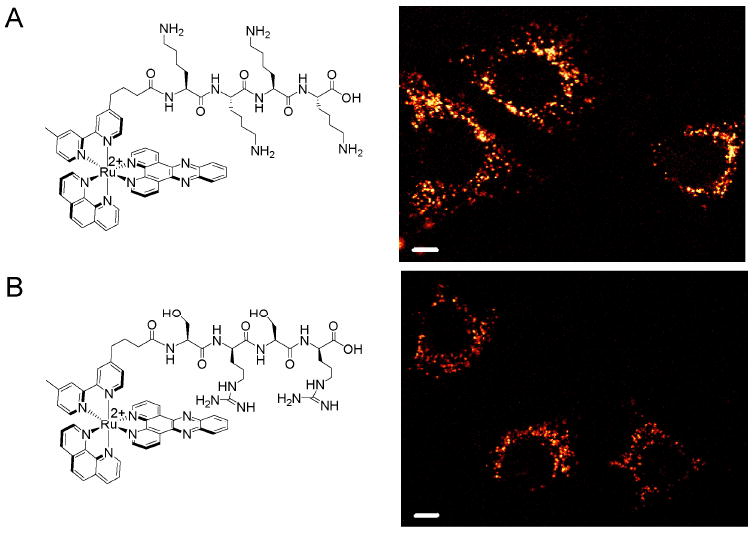
Subcellular distribution of Ru-KKKK and Ru-SrSr. HeLa cells were incubated for 4 h with 100 μM (A) Ru-KKKK or (B) Ru-SrSr in complete medium. Structures of the conjugates are shown at left. The cells shown display punctate staining of the cytoplasm. Scale bars are 10 μm.
Two longer peptides that correspond to known nuclear localization signals (NLSs) were also studied, PKKKRKV and KSKKQK.24 NLSs promote active transport through the nuclear pore complex, but the use of an NLS does not guarantee nuclear uptake. They must reach the cytosol in order to access the nuclear import machinery. If the NLS conjugates enter by endocytosis, they could become trapped in endosomes. However, a cobaltocenium cation has previously been successfully targeted to the nucleus using PKKKRKV.25 Furthermore, the chosen peptides possess less overall charge than octaarginine, and thus are suitable candidates in our efforts to reduce the nonspecific binding of our metal-peptide conjugates to DNA.
Treatment of HeLa cells with 10 μM Ru-PKKKRKV or Ru-KSKKQK for 2 h reveals only faint punctate luminescence in the cytoplasm and no nuclear staining. Increasing the incubation time to 19 h provides the same result (Figure 7). Hence, neither NLS is more effective at promoting nuclear localization of our ruthenium complex than d-octaarginine, which is also excluded from the nucleus at 10 μM. In fact, analysis by flow cytometry reveals that cellular accumulation is even less than that for Ru-RrRK, despite having similar charge (Table 2). Presumably, at higher concentrations, these Ru-NLS conjugates will accumulate in the nucleus, similar to Ru-RrRK.
Figure 7.
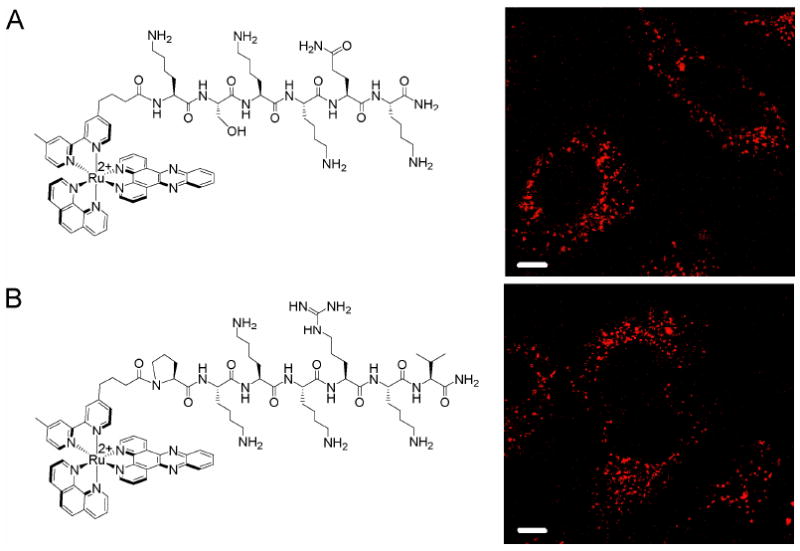
Subcellular distribution of Ru-NLS conjugates. HeLa cells were incubated for 19 h with 10 μM (A) Ru-KSKKQK or (B) Ru-PKKKRKV in complete medium. Structures of the conjugates are shown at left. Punctate staining of the cytoplasm is observed. Scale bars are 10 μm.
3. Conclusion
The large positive charge of octaarginine-metal complex conjugates both improves uptake and interferes with selective DNA binding. To resolve this issue, we studied our luminescent ruthenium complex tethered to the shorter and less charged peptide RrRK, which efficiently addresses the organic fluorophore thiazole orange to the nucleus.11 We found that this peptide was far less capable for delivery of the ruthenium complex than it was for thiazole orange, illustrating again the importance of payload to the accumulation and distribution of cell-penetrating peptides. Furthermore, the low positive charge of short peptide conjugates abrogates the benefits from fluorescein attachment that we previously observed for Ru-octaarginine. Nevertheless, RrRK conjugation increases cellular uptake as compared to analogous unconjugated complexes, and, above a threshold concentration of 30 μM, this peptide targets the ruthenium complex to the nucleus.
4. Experimental
4.1. Materials and instrumentation
Media, cell culture supplements, Hanks' balanced salt solution, and TO-PRO®-3 iodide were purchased from Invitrogen (Carlsbad, CA).
MALDI measurements were performed on an Applied Biosystems Voyager 6215. Absorption spectra were recorded on a Beckman DU 7400 spectrophotometer. HPLC was performed on an HP1100 system equipped with a diode array detector using a Vydac C18 reversed-phase semipreparative column.
4.2. Synthesis of Ru-peptide conjugates
Peptides, protected and resin-bound, were purchased from Anaspec (Fremont, CA). Rink resin was used to produce amide-terminated peptides. Ru(phen)(bpy′)(dppz)2+ was coupled to the peptide in an analogous manner to that previously described (where phen = 1,10-phenanthroline, bpy′ = 4-(3-carboxypropyl)-4′-methyl-2,2′-bipyridine, and dppz = dipyrido[3,2-a:2′,3′-c]phenazine).4,26 Ru-D-R8, which we earlier examined,5 was made with a C-terminal carboxylic acid. The Ru-RrRK conjugate was synthesized in both the amide- and carboxy-terminated versions for comparison; unless otherwise noted, Ru-RrRK refers to the amide-terminated form. A fluorescein-labeled version, Ru-RrRK-fluor, was synthesized by methods analogous to those reported for Ru-D-R8 tagged with fluorescein (Ru-D-R8-fluor).5 As with Ru-D-R8-fluor, fluorescein was appended off a C-terminal lysine, and the peptide contains a carboxylic acid at its C-terminus. Ru-KSKKQK and Ru-PKKKRKV were synthesized with C-terminal amides, and Ru-D-R4, Ru-KKKK and Ru-SrSr have the C-terminal carboxylic acid. Ruthenium-peptide conjugates were purified by reversed-phase HPLC using a water (0.1% trifluoroacetic acid)/acetonitrile gradient and characterized by MALDI-TOF mass spectrometry; Ru-RrRK: 1416.3 m/z (M+) obsd, 1415.6 m/z (M+) calcd, Ru-RrRK-COOH: 1417.6 m/z (M+) obsd, 1416.6 m/z (M+) calcd, Ru-RrRK-fluor: 1935.6 m/z (M+) obsd, 1933.7 m/z (M+) calcd, Ru-D-R4: 1443.6 m/z (M+) obsd, 1444.6 m/z (M+) calcd, Ru-KKKK: 1333.0 m/z (M+) obsd, 1332.6 m/z (M+) calcd, Ru-SrSr: 1307.1 m/z (M+) obsd, 1306.4 m/z (M+) calcd, Ru-PKKKRKV: 1683.8 m/z (M+) obsd, 1683.8 m/z (M+) calcd, Ru-KSKKQK: 1546.6 m/z (M+) obsd, 1546.7 m/z (M+) calcd. All conjugates employed in this study were used as their trifluoroacetate salts. Concentrations were determined by the absorption of Ru(phen)(bpy′)(dppz)2+; for Ru-RrRK-fluor, 361 nm, which is not obscured by fluorescein, was used (ε440= 19,000 M-1 cm-1; ε361= 19,469 M-1 cm-1). Ru(phen)2dppz2+, used for comparison to the conjugates in uptake studies, was synthesized as described previously;6 ε440 = 21,100 M-1 cm-1.27
4.3. Cell culture
HeLa cells (ATCC, CCL-2) were maintained in minimal essential medium alpha with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were grown in tissue culture flasks at 37 °C under 5% CO2 atmosphere.
4.4. Confocal Microscopy
HeLa were seeded using 4000 cells in wells of a glass-bottom 96-well plate (Whatman Inc.) and allowed to adhere overnight. The complexes were incubated with HeLa cells at 37 °C in complete medium (minimal essential medium alpha with 10% fetal bovine serum) or medium without serum, as indicated. Imaging was performed using a 63x/1.4 oil immersion objective on a Zeiss LSM 510 or a Zeiss LSM 5 Exciter inverted microscope. The optical slice was set to 1.1 μm. Ru-peptide conjugates were excited at 488 nm, with emission observed at 560+ nm. For Ru-RrRK-fluor, the emission was collected as the combined emission of Ru and fluorescein (505+ nm), both of which are excited at 488 nm.
4.5. Flow cytometry
Cells were detached from culture with EDTA (0.48 mM in phosphate-buffered saline) and incubated at 1×106 cells/mL with 10 μM ruthenium complex in Hanks' balanced salt solution (HBSS) supplemented with 2.5 mg/mL bovine serum albumin fraction V (BSAV) at 37 °C for 2 h, then rinsed with buffer and placed on ice. TO-PRO-3 was added at 1 μM immediately prior to flow cytometry analysis to stain dead cells. The fluorescence of ∼20,000 cells was measured using a BD FACS Aria at the Caltech Flow Cytometry Facility. Ruthenium complexes were excited at 488 nm, with emission observed at 600–620 nm. TO-PRO-3 was excited at 633 nm, with emission observed at 650–670nm. Cells exhibiting TO-PRO-3 fluorescence were excluded from the data analysis.
Acknowledgments
We are grateful to the National Institutes of Health (GM33309) for their financial support. We also acknowledge the Caltech Biological Imaging Center and the Caltech Flow Cytometry Facility for their expert assistance and facilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stewart KM, Horton KL, Kelley SO. Org Biomol Chem. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- 2.Fischer R, Fotin-Mleczek M, Hufnagel H, Brock R. ChemBioChem. 2005;6:2126–2142. doi: 10.1002/cbic.200500044. [DOI] [PubMed] [Google Scholar]
- 3.Goun EA, Pillow TH, Jones LR, Rothbard JB, Wender PA. ChemBioChem. 2006;7:1497–1515. doi: 10.1002/cbic.200600171. [DOI] [PubMed] [Google Scholar]
- 4.Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 5.Puckett CA, Barton JK. J Am Chem Soc. 2009;131:8738–8739. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puckett CA, Barton JK. J Am Chem Soc. 2007;129:46–47. doi: 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrave L, Devocelle M, Forster RJ, Keyes TE. Chem Commun. 2010;46:103–105. doi: 10.1039/b918611e. [DOI] [PubMed] [Google Scholar]
- 8.Gill MR, Garcia-Lara J, Foster SJ, Smythe C, Battaglia G, Thomas JA. Nat Chem. 2009;1:662–667. doi: 10.1038/nchem.406. [DOI] [PubMed] [Google Scholar]
- 9.Zeglis BM, Pierre VC, Barton JK. Chem Commun. 2007:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst RJ, Song H, Barton JK. J Am Chem Soc. 2009;131:2359–2366. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahon K, Potocky T, Blair D, Roy M, Stewart K, Chiles T, Kelley S. Chem Biol. 2007;14:923–930. doi: 10.1016/j.chembiol.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Horton KL, Steward KM, Fonseca SB, Guo Q, Kelley SO. Chem Biol. 2008;15:375–382. doi: 10.1016/j.chembiol.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Murphy CJ, Barton JK. Methods Enzymol. 1993;226:576–594. doi: 10.1016/0076-6879(93)26027-7. [DOI] [PubMed] [Google Scholar]
- 14.Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. J Am Chem Soc. 1990;112:4960–4962. [Google Scholar]
- 15.Dupureur CM, Barton JK. Inorg Chem. 1997;36:33–43. [Google Scholar]
- 16.Jenkins Y, Friedman AE, Turro NJ, Barton JK. Biochemistry. 1992;31:10809–10816. doi: 10.1021/bi00159a023. [DOI] [PubMed] [Google Scholar]
- 17.Olson EJC, Hu D, Hormann A, Jonkman AM, Arkin MR, Stemp EDA, Barton JK, Barbara PF. J Am Chem Soc. 1997;119:11458–11467. [Google Scholar]
- 18.Nakase I, Takeuchi T, Tanaka G, Futaki S. Adv Drug Delivery Rev. 2008;60:598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci U S A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. J Peptide Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 22.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 23.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. J Am Chem Soc. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 24.Pouton C. Adv Drug Del Rev. 1998;34:51–64. doi: 10.1016/s0169-409x(98)00050-7. [DOI] [PubMed] [Google Scholar]
- 25.Noor F, Wustholz A, Kinscherf R, Nolte N. Angew Chem Int Ed. 2005;44:2429–2432. doi: 10.1002/anie.200462519. [DOI] [PubMed] [Google Scholar]
- 26.Copeland KD, Lueras AMK, Stemp EDA, Barton JK. Biochemistry. 2002;41:12785–12797. doi: 10.1021/bi020407b. [DOI] [PubMed] [Google Scholar]
- 27.Holmlin RE, Stemp EDA, Barton JK. Inorg Chem. 1998;37:29–34. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]


