Abstract
The p53 homologue p63 encodes multiple protein isoforms either with (TA) or without (ΔN) the N-terminal transactivation domain. Accumulating evidence indicates that TAp63 plays an important role in various biological processes, including cell proliferation, differentiation, and apoptosis. However, how TAp63 is regulated remains largely unclear. In this study, we demonstrate that NF-κB induces TAp63 gene expression. The responsible elements for NF-κB-mediated TAp63 induction are located within the region from –784 to –296 bp in the TAp63 promoter, which contains two NF-κB binding sites. Ectopic expression of RelA stimulates TAp63 promoter-driven reporter activity and increases endogenous TAp63 mRNA levels. Inhibition of NF-κB by IκBα super-repressor or with a chemical inhibitor leads to down regulation of TAp63 mRNA expression and activity. In addition, mutations in the critical NF-κB-binding sites significantly abolish the effects of NF-κB on TAp63. Activation of NF-κB by TNFα enhances p50/RelA binding to the NF-κB binding sites. Furthermore, we show that an Sp1 site adjacent to the NF-κB sites plays a role in NF-κB-mediated upregulation of TAp63. Taken together, these data reveal that TAp63 is a transcriptional target of NF-κB, which may play a role in cell proliferation, differentiation and survival upon NF-κB activation by various stimuli.
Keywords: p63, p53, NF-κB, Sp1, CORTICAL NEURON, TRANSCRIPTION
p63, a p53 homologue, encodes six isoforms derived from differential promoter usage and alternative splicing [Mills et al., 1999; Yang et al., 1999]. The transactivation (TA) isoforms, which resemble p53, are generated by the use of an upstream promoter and consist of several domains, including an acidic N-terminal transactivation domain, a central DNA binding domain and a C-terminal oligomerization domain. The ΔN isoforms, produced from an intronic promoter, lack the N-terminal transactivation domain and are capable of forming protein complexes with other p53 family members to inhibit their function [Yang et al., 1998; Rocco et al., 2006]. Furthermore, both TAp63 and ΔNp63 isoforms undergo alternative splicing, yielding different C-terminal tails (TAp63α, β, γ and ΔNp63α, β, γ isoforms).
Unlike p53, which is dispensable for normal development, p63 is critical for the development of stratified epithelial tissues, such as epidermis, breast and prostate [Yang et al., 1998; Yang et al., 1999; Mills et al., 1999]. p63–/– mice lack stratified epithelia and their ectodermal derivatives, including epidermal appendages, mammary, lacrimal, and salivary glands. Due to the absence of an epidermal barrier, these mice dehydrate and die shortly after birth. In addition, p63–/– mice have major defects in limb and craniofacial development, including limb truncation, and cleft lip and palate [Yang et al., 1999; Mills et al., 1999]. Several autosomal dominant inherited human syndromes have been mapped to the p63 gene. These syndromes are characterized by various combinations of limb malformations fitting the split hand-split foot spectrum, orofacial clefting, and ectodermal dysplasia [Brunner et al., 2002].
TAp63 has been implicated in regulation of cell proliferation, apoptosis and differentiation. Overexpression of TAp63 inhibits anchorage-independent cell growth and induces apoptosis in human lung, gastric and pancreatic cancer cells [Kunisaki et al., 2006]. TAp63α can stimulate expression of proapoptotic genes, including Bax, BCL2L11, RAD9, DAP3, and APAF1, and induce apoptosis through death receptor CD95 and the mitochondrial apoptosis pathways [Gressner et al., 2005]. It was also shown that TAp63α is induced by several chemotherapeutic drugs and that inhibition of TAp63 function leads to chemoresistance [Gressner et al., 2005]. Despite the observation that ectopic expression of TAp63 induces apoptosis and cell growth arrest, its physiological functions remain elusive. Recently, it was reported that TAp63 is expressed in sympathetic neurons during neuronal development, and its expression is induced by NGF withdrawal. Upregulation of TAp63 leads to neuronal cell death in a p53-independent fashion [Jacobs et al., 2005]. Furthermore, TAp63α is constitutively expressed in female germ cells during meiotic arrest and is dispensable for ovary development. However, TAp63α is essential for DNA damage-induced oocyte death [Suh et al., 2006].
The transcription factor NF-κB is a key mediator of a wide variety of cellular processes involving growth, differentiation and apoptosis [Wu and Kral, 2005]. NF-κB transcription factors consist of five subunits (RelA/p65, c-Rel, RelB, p50, and p52) that bind to DNA as hetero- or homodimers [Sonenshein, 1997]. In resting cells, NF-κBis mainly retained in the cytoplasm by the IkappaB (IκB) family of proteins, which mask the nuclear translocation signal. Upon stimulation, IκB proteins are phosphorylated, triggering their ubiquitination and subsequent proteosomal degradation. The NF-κB proteins are then released for translocation to the nucleus, where they induce expression of NF-κB-dependent genes. Many different extracellular stimuli can induce NF-κB, which in turn regulates gene expression. Activation of NF-κB leads to cell survival in response to a variety of apoptotic signals [Wu et al., 1996; Bours et al., 2000]. Under certain circumstances, NF-κB functions to promote apoptosis [Chan et al., 1999; Ryan et al., 2000; Zheng et al., 2001; Aleyasin et al., 2004]. Induction of p53 causes activation of NF-κB and apoptosis in Saos-2 cells expressing inducible p53, while inhibition of NF-κB abrogates p53-mediated apoptosis [Ryan et al., 2000]. It was also showed that acute inhibition of NF-κB protects cortical neuron from DNA damage-induced apoptosis [Aleyasin et al., 2004].
Although the function and downstream targets of TAp63 have been studied extensively, the regulation of TAp63 at the transcriptional level is largely unknown. Here we show that TAp63 is a transcriptional target of NF-κB in vitro and in vivo. Ectopic expression of RelA stimulates TAp63 reporter activity and increases endogenous TAp63 mRNA levels. Inhibition of NF-κB by IκBα super-repressor or a chemical inhibitor leads to down-regulation of TAp63 mRNA expression and activity. In addition, mutations in the critical NF-κB-binding sites located on the upstream p63 promoter significantly abolish the effects of NF-κB on TAp63, whereas activation of NF-κB by TNFα enhances p50/RelA binding to these NF-κB binding sites.
MATERIALS AND METHODS
CELL CULTURE AND DRUG TREATMENT
Human mammary epithelial MCF10A cells were maintained in 1:1 mixture of Dulbecco's modified Eagle's medium (DMEM) and Ham's F12 medium with reduced Ca2+ (0.04 mM) (Invitrogen Inc., Carlsbad, CA), 20 ng/ml epidermal growth factor (Invitrogen), 100 ng/ml cholera toxin (Sigma, St. Louis, MO), 10 μg/ml insulin (Sigma), 500 ng/ml (95%) hydrocortisone (Sigma) and 5% of Chelex-treated horse serum (Invitrogen). Human embryonic kidney 293 cells and NIH3T3 cells were maintained in DMEM containing 4.5 g/L glucose supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen). Human breast cancer Hs578T cells were cultured in DMEM supplement with 10% FBS and 10 μg/ml of insulin. All media contain 100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulfate (Invitrogen). Primary cultures of cortical neurons were prepared from E15.5 mouse embryos as described [Wartiovaara et al., 2002]. Briefly, the meninges were removed, and cortical tissue was transferred into cold Dulbecco's phosphate buffered saline (DPBS; Invitrogen). The tissue was triturated into a single-cell suspension and the cells were resuspended in neurobasal medium containing 500 μM glutamine, 2% B27 supplement (Gibco, Life Technologies, Rockville, MD), and 1% penicillin-streptomycin, then plated at 2.4 × 105/cm2 in poly-d-lysine-coated 12-well plates (Becton Dickinson Labware, Bedford, MA). After 3 days in vitro, half of the media was replaced with fresh media supplemented with 7 μM cytosine arabinoside (CA). Three days later, all media was replaced with fresh media without CA. The cells were subjected to experimentation at day 6.
GENERATION OF MUTATIONS IN THE HUMAN TAp63 PROMOTER
pGL3-TAp63 luciferase (TAp63-FL) was kindly provided by Dr. Matthias Dobbelstein from Institut für Virologie in Germany [Waltermann et al., 2003]. To generate mutants including the 5′-deletions, internal deletion (Δ –655 to –611), and point mutations, we used the QuickChange Site-Directed Mutagenesis Kit (Stratagene Inc., La Lolla, CA) according to manufacturer's protocol. Primers used for mutagenesis are listed in Supplementary Table I. All of the constructs were confirmed by DNA sequencing.
ELECTROPHORETIC MOBILITY SHIFT ASSAY (EMSA)
Nuclear extracts were prepared as described [Dignam et al., 1983]. Briefly, cells were washed with cold PBS and lysed in RSB buffer (10 mM Tris pH 7.6, 10 mM NaCl, 3 mM MgCl2 and 0.5% NP40). The nuclear pellets were collected and lysed in DR buffer (20 mM HEPES pH 7.9, 420 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA and 20% glycerol) supplemented with a cocktail of protease inhibitors (0.5 mM PMSF, 1 μg/ml leupeptin, and 1 mM DTT). The cell debris was removed by centrifugation and the supernatant was saved as nuclear extract and stored at –80°C. For EMSA, 20,000 cpm of labeled probe were incubated with 5–10 μg of nuclear extract in 5× binding buffer (10 mM HEPES pH 7.5, 4 mM DTT, 0.5% Triton X-100, 1 μg/ml poly(dI-dC), and 2.5% glycerol) for 20 min at room temperature. For supershift experiments, 1 μg of primary antibody (anti-p50: sc-114 or anti-RelA: sc-109, Santa Cruz Biotechnology, Santa Cruz, CA) was added to the nuclear extract and incubated overnight at 4°C prior to the addition of labeled probe. DNA/protein complexes were separated in a 4% polyacrylamide gel. After electrophoresis, the gels were dried and autoradiographed to X-ray films. Oligonucleotides used for EMSA are listed in Supplementary Table I.
TRANSFECTION AND REPORTER ACTIVITY ASSAYS
Cells in 12-well plates were co-transfected with 0.2 μg of TAp63 firefly luciferase reporter construct (TAp63-luc), 10 ng of Renilla-luc (Promega, CA), 0.2 μg each of p50 and RelA [Sovak et al., 1997], or 0.4 μg of empty vector (pcDNA) using FuGENE 6 reagent (Roche, Switzerland). 24–48 h post-transfection, cells were harvested and subjected to firefly and Renilla luciferase activity assays using a Dual-Luciferase Reporter Assay System (Promega).
QUANTITATIVE POLYMERASE CHAIN REACTION
Total RNA was extracted from cultured cells using TRIzol (Gibco) according to manufacturer's protocol. Aliquots of RNA (5 μg) were reverse-transcribed using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative-PCR (Q-PCR) was performed in 7300 Real-Time PCR System (Applied Biosystems, CA) using a QuantiTect SYBR Green PCR Kit (Qiagen, Netherlands). The oligonucleotide primers used for Q-PCR are listed in Supplementary Table I.
RESULTS
NF-κB ACTIVATES TAp63 REPORTER ACTIVITY IN A DOSE-DEPENDENT MANNER
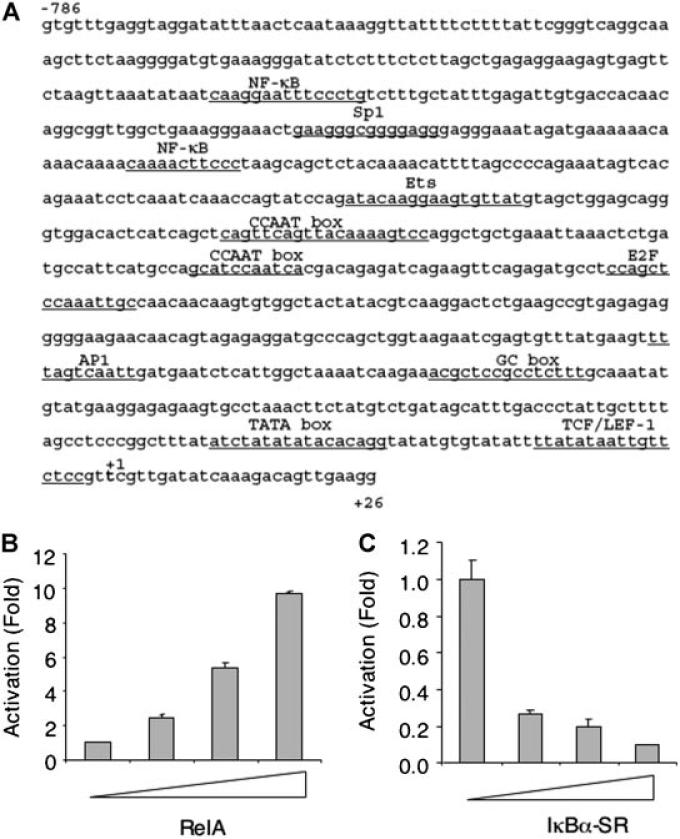
p63 plays a critical role in various cellular processes, including cell proliferation, apoptosis, senescence and differentiation. We were interested in elucidating the upstream signaling that regulates p63 expression. We analyzed the human TAp63 promoter sequence (–2335 to +26) by TRANSFAC software (www.biobase-international.com/pages/index.php?id¼transfac) to identify putative regulatory elements. Notably, there are several putative sites for the NF-κB, Sp1, TCF/LEF-1, and E2F transcription factors. The putative NF-κB binding sites are located at –537 to –527 and –648 to –634. Interestingly, an Sp1 site (–581 to –567) is located between the two NF-κB sites (Fig. 1A).
Fig. 1.
NF-κB activates TAp63 reporter activity. A: The proximal region (–786 to +26) of the human TAp63 promoter contains a number of putative transcription factor-binding sites, as analyzed by TRANSFAC software. B: NIH3T3 cells were co-transfected with TAp63 luciferase reporter (TAp63-luc) containing the TAp63 promoter (–2335 to +26), Renilla-luc, 0.2 μg of human p50 and increasing amounts (0.1. 0.2, 0.5 μg) of human RelA expression plasmids. Forty-eight hours later, cells were lysed and luciferase activities were measured using the Dual-luciferase Reporter Assay Kit (Promega). TAp63-luciferase activities were normalized to Renilla-luciferase activities and presented as fold activation (mean ± SE). At least three independent experiments in triplicate were performed. C: Hs578T cells were co-transfected with TAp63-luc, Renilla-luc and increasing amounts (0.1. 0.2, 0.5 μg) of IκB-SR expression plasmids. Luciferase activities were measured as in (B).
Since NF-κB is a central player in integrating various extracellular signals, we focused on whether NF-κB plays a role in regulating TAp63 expression. We performed co-transfection experiments to examine the effects of NF-κB on TAp63-luciferase reporter activity in NIH3T3 cells. Ectopic expression of RelA led to an increase in the reporter activity of up to 10-fold in a dose-dependent manner (Fig. 1B). By contrast, inhibition of endogenous NF-κB by expression of an IκBα super-repressor [IκBα-SR; Brown et al., 1995; Shin et al., 2006] in Hs578T breast cancer cells, which show constitutively activated NF-κB [Sovak et al., 1997], resulted in a dose-dependent reduction of TAp63 reporter activity (Fig. 1C). These data indicate that NF-κB can activate TAp63 reporter activity.
TWO NF-κB SITES ARE RESPONSIBLE FOR NF-κB-MEDIATED ACTIVATION OF THE TAp63 REPORTER
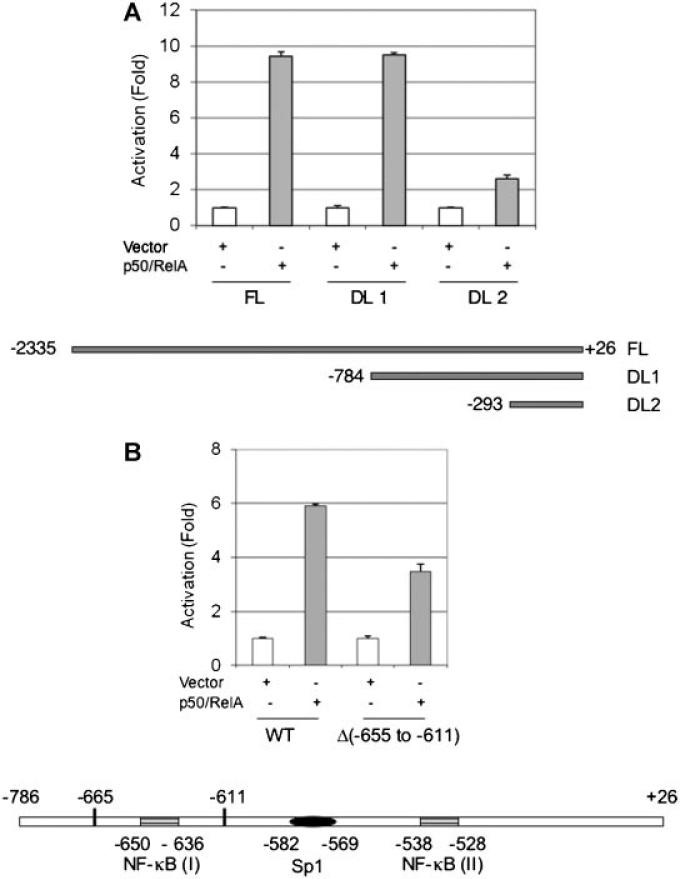
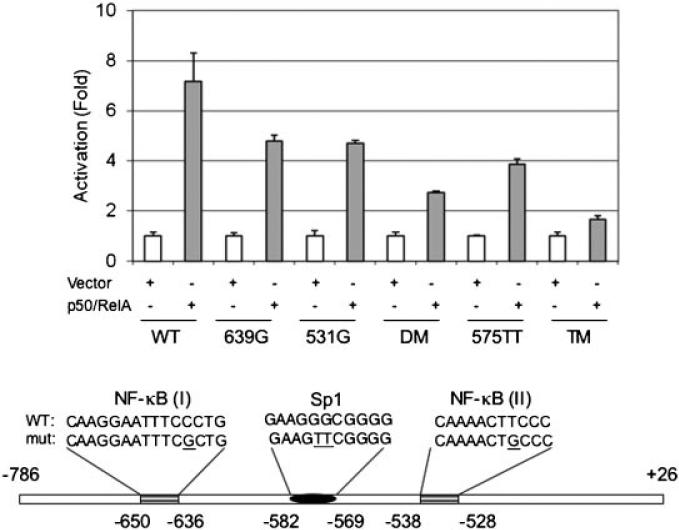
We next investigated whether the putative NF-κB sites in the proximal TAp63 promoter region are responsible for this NF-κB-mediated effect. We generated deletion mutants in the promoter region and examined their responsiveness to NF-κB. As expected, co-expression of p50 and RelA stimulated a 10-fold increase in reporter activity driven by the full-length TAp63 promoter (FL, –2335 to +26). Deletion of the –2335 to –785 region (DL1, –784 to +26) had little effect on NF-κB induction. However, further deletion of –784 to –294 (DL2, –293 to +26) led to a dramatic loss in response to NF-κB, indicating that this fragment (–784 to –294) is pivotal for NF-κB activation (Fig. 2A). Of note, this fragment contains two putative NF-κB sites (Fig. 2B). We therefore generated a deletion mutant lacking the distal NF-κB site [NF-κB (I), Δ(–665 to –611)]. As shown in Figure 2B, deletion of NF-κB (I) partially impaired the promoter activity in response to NF-κB, suggesting that other elements are also responsive to NF-κB. To avoid possible detrimental structural changes due to deletion, we generated point mutations in the critical NF-κB binding sites [Akama et al., 1998] in NF-κB (I) (639G) and NF-κB (II) (531G) as well as a double mutation (DM, 639G/531G) in the TA-p63 reporter (–784 to +26) (Fig. 3, bottom panel). Under similar experimental settings, a single point mutation in either NF-κB (I) (639G) or NF-κB (II) (531G) partially reduced reporter activity, while the double point mutation (639G/531G) further reduced the reporter activity (Fig. 3). Notably, the double mutation in both NF-κB binding sites (639G/531G) does not completely eliminate the responsiveness to NF-κB, suggesting that other elements may play a role in the NF-κB-mediated effects. Since Sp1 sites have been shown to contribute to NF-κB-mediated effects [Pazin et al., 1996; Boekhoudt et al., 2003], and there is a putative Sp1 element adjacent to the NF-κB sites (Fig. 3), we therefore examined whether this Sp1 element contributes to NF-κB-mediated TAp63 expression. We generated a double mutation (575TT), which is reported to inactivate Sp1-binding [Tamaki et al., 1995]. Interestingly, even in the presence of intact NF-κB binding elements, the reporter activity in response to NF-κB from the 575TT mutant is impaired, indicating that the Sp1 element is important for NF-κB-mediated transcription of TAp63. To confirm the role of Sp1 in the NF-κB action, we generated a triple mutant (TM, 639G/531G/575TT). As shown in Figure 3, this triple mutant exhibited almost complete loss of activity when compared to the double NF-κB mutant. Together, these data indicate that Sp1 is important in NF-κB-mediated activation of TAp63.
Fig. 2.
Deletion of a putative NF-κB binding site reduces TAp63 promoter response to NF-κB. A: 5′-deletion mutants of the TAp63 promoter were constructed by using SiteDirect Mutagenesis Kit. NIH3T3 cells were co-transfected with the indicated full-length or deletion mutants of TAp63 promoter and Renilla-luc in the presence or absence of 0.2 μg each of p50/RelA. In the absence of p50/RelA, an equal amount of vector plasmid DNA (pcDNA) was used. B: NIH3T3 cells were co-transfected with either WT TAp63-luc or TAp63-luc (Δ –665 to –611) with Renilla luciferase reporter in the presence or absence of p50/RelA. Twenty-four hours after transfection, cells were lysed and subjected to luciferase activity assay. The relative activity was normalized to the vector control and presented as fold activation (mean ± SE). At least three independent experiments in triplicate were performed. Schematic presentation is not to the scale.
Fig. 3.
Mutations in the NF-κB and Sp1 sites reduce NF-κB effects on TAp63 promoter activity. A: NIH3T3 cells were co-transfected with wild type or a TAp63-luc mutant bearing either a single site mutation, double site mutations (DM, 639G531G) or triple site mutations (TM, 639G 531G 575TT), as indicated. Twenty-four hours after transfection, cells were lysed and subjected to luciferase activity assay. The relative activity was normalized to the vector control and presented as fold activation (mean ± SE). At least three independent experiments in triplicate were performed. Schematic presentation is not to the scale.
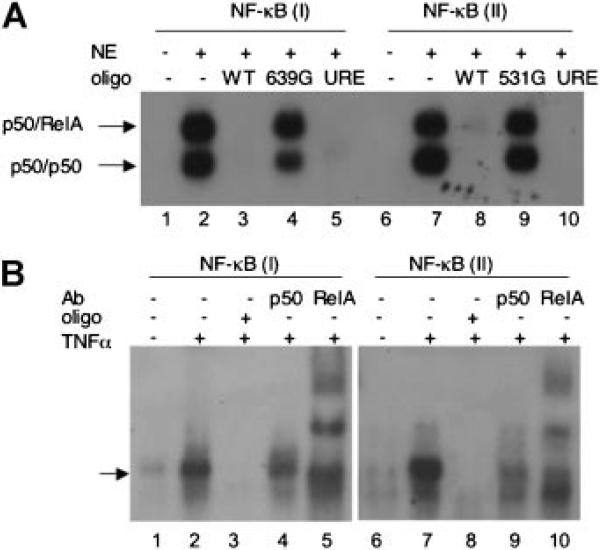
p50/RelA SPECIFICALLY BINDS TO NF-κB SITES IN THE TAp63 PROMOTER IN VITRO
Next, we performed electrophoretic mobility shift assay (EMSA) using nuclear extracts derived from NIH3T3 cells transiently transfected with p50/RelA. As shown in Figure 4A, the p50/RelA heterodimer and p50/p50 homodimer were readily detected using 32P-labeled oligonucleotides containing NF-κB (I) (lane 2) or NF-κB (II) (lane 7), in keeping with other reports [Duckett et al., 1993; Lin et al., 1998]. NF-κB-DNA complex formation was completely blocked by an excess of cold wild type NF-κB (I) oligonucleotides (WT, lane 3) or NF-κB (II) oligonucleotides (WT, lane 8), but not by their respective mutant derivatives (639G, Lane 4; and 531G, lane 9). In addition, cold oligonucleotides corresponding to the c-Myc promoter URE (upstream regulatory element), which has been shown to be an NF-κB responsive element [Duyao et al., 1990], completely blocked the NF-κB-DNA complex formation, indicating that both NF-κB (I) and NF-κB (II) elements in the TAp63 promoter are likely functional for NF-κB-binding.
Fig. 4.
NF-κB directly binds to the TAp63 promoter fragments by EMSA. A: Nuclear extract derived from NIH3T3 cells transiently transfected with p50-and RelA-expressing plasmids were incubated with either 32P-labeled NF-κB (I) oligonucleotides (lane1–5) or NF-κB (II) oligonucleotides (lane 6–10). No nuclear extract was used in the control lanes (lanes 1 and 6). Competition experiments were performed by adding a 30-fold excess of cold oligonucleotides as indicated prior to the addition of 32P-labled probe. URE: upstream NF-κB-responding element [Duyao et al., 1990]. B: Human embryonic kidney 293 cells were treated with TNFα (1 ng/ml) for 30 min and nuclear extracts were isolated. Equal amounts of nuclear protein was subjected to EMSA and supershift assay by using 32P-labeled oligonucleotides containing NF-κB (I) or NF-κB (II). Antibodies specific for p50 or RelA were used as indicated. Cold WT oligonucleotides were used in the competition assay (see Supplementary Table I). Nuclear extracts derived from cells without TNFα treatment were used as a control (Lanes 1 and 6).
We then examined the effects of TNFα, a well-defined NF-κB activator [Kaltschmidt et al., 2005], on NF-κB-TAp63 promoter complex formation by EMSA. To this purpose, we used nuclear extracts from 293 cells untreated or treated with TNFα. As shown in Figure 4B, treatment with TNFα significantly increased NF-κB binding to elements NF-κB (I) (lane 2) and NF-κB (II) (lane 7). As expected, cold oligonucleotides completely blocked complex formation (lanes 3 and 8). In addition, we performed EMSA- antibody supershift assay. Treatment with a p50-specific antibody led to a decrease in NF-κB-DNA complex formation (compare lane 4 to lane 2 and lane 9 to lane 7), whereas treatment with a RelA-specific antibody led to a supershift of the NF-κB-DNA complexes (lanes 5 and 10). Thus, these data indicate that activation of NF-κB by TNFα stimulates their binding to NF-κB elements in the TAp63 promoter.
NF-κB PLAYS AN IMPORTANT ROLE IN TAp63 EXPRESSION
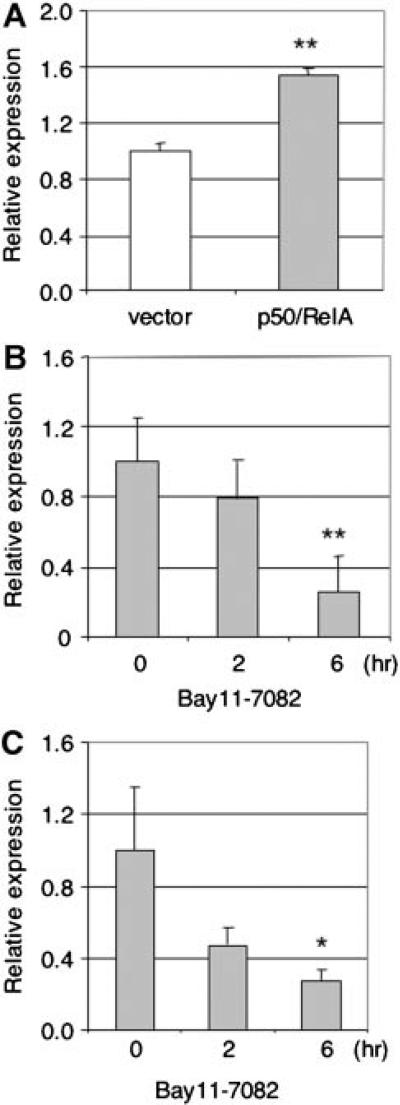
We next examined whether NF-κB is involved in endogenous expression of TAp63. We first ectopically expressed NF-κB and examined TAp63 mRNA levels in MCF10A cells, an immortalized human mammary epithelial cell line that expresses endogenous TAp63 [Li et al., 2006]. Our data show that ectopic expression of p50/RelA in MCF10A cells leads to a significant increase of TAp63 mRNA levels (P < 0.01) as assessed by Q-PCR (Fig. 5A). Since these cells have detectable levels of NF-κB, we next treated MCF10A cells with Bay 11-7082, a specific inhibitor of NF-κB [Pierce et al., 1997]. TAp63 mRNA levels were markedly decreased in a dose-dependent manner (Fig. 5B).
Fig. 5.
NF-κB regulates TAp63 mRNA expression. MCF10A cells were transiently transfected with p50- and RelA-expression plasmids for 24 h (A) or treated with a specific NF-κB inhibitor Bay 11-7082 (10 μM) for the indicated times (B). Total RNA was extracted and subjected to quantitative-PCR (Q-PCR) analysis. The amount of TAp63 mRNA was normalized to GAPDH in the same samples and presented as mean ± SE. Three independent experiments were performed in triplicate. ** P < 0.01. C: Primary cortical neuron cells isolated form E15.5 embryonic mice were treated with Bay 11-7082 (10 μM) for the indicated times followed by Q-PCR. Two independent experiments were performed in triplicate. * P < 0.05.
It has been reported that NF-κB is constitutively active in mouse cortical neurons [Bhakar et al., 2002], and that TAp63 is expressed in developing cortical neurons [Jacobs et al., 2005]. We therefore examined the effects of NF-κB inhibition on the expression of TAp63 in primary mouse embryonic day 15.5 cortical neurons. Consistent with the results from MCF10A cells, inhibition of NF-κB by Bay11-7082 decreased TAp63 mRNA levels in a dose-dependent manner (Fig. 5C). Taken together, these data indicate that NF-κB is important for TAp63 expression.
DISCUSSION
In this study, we demonstrated that NF-κB transcriptionally induces TAp63 expression. The responsible elements for NF-κB-mediated TAp63 induction are located within the region from –784 to –296 in the TAp63 promoter, which contains two NF-κB binding sites. Ectopic RelA expression stimulates TAp63 reporter activity and increases endogenous TAp63 mRNA levels, while inhibition of NF-κB by IκBα super-repressor or a chemical inhibitor leads to down regulation of TAp63 mRNA expression and activity. In addition, mutations in the critical NF-κB binding sites significantly abolish the effects of NF-κB on TAp63. Activation of NF-κB by TNFα enhances p50/RelA binding to the NF-κB binding sites. Furthermore, we show that the Sp1 site between the NF-κB binding elements plays a role in NF-κB-mediated upregulation of TAp63 expression.
The p63 gene can be transcribed from an upstream promoter, giving rise to TAp63, or from an intronic promoter, resulting in ΔNp63. Koster et al. [2004] observed that the TAp63 isoforms are primarily expressed in the mouse from E7.5 through E15 during embryogenesis, the interval for ectoderm commitment and initiation to epithelial lineage, followed by increasing and dominant expression of ΔNp63 isoforms. Other reports have shown that TAp63 expression can be detected by RT-PCR only until E13 [Laurikkala et al., 2006], or that TAp63 is undetectable from E10 to 3 days after birth by in situ hybridization [Laurikkala et al., 2006; Mikkola, 2007]. In general, TAp63 expression is at very low levels in adult cells and established cell lines. Interestingly, it has been shown that TAp63α is induced during oocytogenesis at E18.5, and that γ-irradiation induces TAp63α phosphorylation [Suh et al., 2006]. Notably, TAp63 is induced under certain stress conditions, such as withdrawal of growth factors [Jacobs et al., 2005], DNA damage [Katoh et al., 2000; Gressner et al., 2005], or upon wounding [Su et al., 2009]. In particular, TAp63 appears to be highly transcriptionally potent and is essential for maintaining adult epidermal stem cells by regulating cell proliferation, senescence and genomic stability [Su et al., 2009].
NF-κB has been shown to positively regulate p53 transcription [Kirch et al., 1999], although the physiological relevance is not clear. Here, we show that p50/RelA complexes bind to the NF-κB binding sites in the TAp63 promoter and upregulate TAp63 expression. In addition, we show that the Sp1 site adjacent to the NF-κB sites plays an important role in this regulation, consistent with reports that Sp1 can play an important role in NF-κB-induced promoter activity [Pazin et al., 1996; Boekhoudt et al., 2003]. Of note, it has been shown that activation of CD74 in murine mature follicular B cells upregulates TAp63 via the NF-κB pathway, which in turn transactivates Bcl-2 thereby increasing cell survival [Lantner et al., 2007]. Furthermore, it has been reported that IKKβ, which activates NF-κB by promoting IκBα degradation, can also phosphorylate and stabilize TAp63γ protein [MacPartlin et al., 2008]. Thus, activation of NF-κB by various stimuli can upregulate TAp63 at different levels.
p63 is required for developmental neuronal death [Jacobs et al., 2005]. NF-κB induction by cytokines, oxidative stress, viral products, glutamate and beta-amyloid has been implicated in the pathogenesis of several neurodegenerative and inflammatory disorders [Hunot et al., 1997; Kaltschmidt et al., 1997; Gveric et al., 1998; Mattson, 2005]. Although NF-κB complexes are inactive and sequestered in the cytoplasm in most of resting cells, they are constitutively active in cortical neurons [Bhakar et al., 2002]. It has also been reported that TAp63 is the predominant p63 isoform in the developing cortex, and that TAp63 can induce apoptosis of developing cortical neurons [Jacobs et al., 2005]. Our data show the presence of TAp63 mRNA expression in mouse embryonic cortical neuron (E15.5). Importantly, TAp63 mRNA levels are approximately 100-fold higher than that of ΔNp63 isoforms (data not shown). In addition, our data indicate that NF-κB plays a pivotal role in TAp63 expression since inhibition of NF-κB dramatically decreases TAp63 mRNA levels. Of note, TAp63 is also expressed in sympathetic neurons during the developmental death period, and protein levels are induced by NGF withdrawal [Jacobs et al., 2005]. Since activation of NF-κB by various stimuli can promote neuronal cell death [Grilli et al., 1996; Bian et al., 2001; Goffi et al., 2005; Sarnico et al., 2008], it is conceivable that TAp63 plays an important role in these processes.
p63 is essential for skin, limb and nervous system development [Celli et al., 1999; Mills et al., 1999; Yang et al., 1999; Jacobs et al., 2005], by regulation of cell proliferation, differentiation and maintenance of epithelial stem cells [Koster et al., 2007; Senoo et al., 2007]. The specific functions for different p63 isoforms are still under vigorous investigation. Ectopic expression of TAp63α, but not ΔNp63, induces K5 and K14, two well defined differentiation markers for keratinocytes and commitment to stratification [Koster et al., 2004, 2006]. Interestingly, studies of TAp63–/– mice suggest that the TAp63 isoforms are dispensable for epidermal development. However, these mice are defective in DNA-damage induced oocyte death [Suh et al., 2006]. A recent study shows that conditional TAp63 knockout mice display accelerated aging and develop blisters, skin ulcerations, and senescence of hair follicle-associated dermal and epidermal cells. These phenotypes are caused by the loss of TAp63 in dermal and epidermal precursors, since both cell types show defective proliferation, early senescence and genomic instability [Su et al., 2009]. Importantly, skin-derived precursors (SKP) lacking TAp63 proliferate 4-5 fold more than wild type counterparts. Intriguingly, inhibition of NF-κB function in the epidermis causes massive tissue hyperplasia, whereas in transgenic mouse models expression of constitutively active NF-κB subunits produces epithelial hypoplasia [Seitz et al., 1998]. Furthermore, increasing NF-κB activity in normal keratinocytes leads to growth arrest via induction of p21Cip independently of p53 [Seitz et al., 2000; Basile et al., 2003]. Thus, it is tempting to speculate that TAp63 negatively regulates cell growth of SKP and that activation of NF-κB by various stimuli leads to cell growth arrest via TAp63-mediated pathway. Whether NF-κB/TAp63 pathway is involved in epidermal differentiation remains to be investigated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. M. Dobbelstein for providing pGL3-TAp63, and we thank Dr. Hongwu Zheng for help of isolating mouse cortical neurons and Xiaobo Wang for technical assistance on EMSA. We are grateful to members of Xiao's laboratory for stimulating discussions. This work was supported in part by NIH grants CA129129 to G.E.S., CA79804 and GM70017 to Z.X.X.
Grant sponsor: NIH; Grant numbers: CA129129, CA79804, GM70017.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Akama KT, Albanese C, Pestell RG, Van Eldik LJ. Amyloid beta-peptide stimulates nitric oxide production in astrocytes through an NFkappaB-dependent mechanism. Proc Natl Acad Sci USA. 1998;95:5795–5800. doi: 10.1073/pnas.95.10.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleyasin H, Cregan SP, Iyirhiaro G, O'Hare MJ, Callaghan SM, Slack RS, Park DS. Nuclear factor-(kappa)B modulates the p53 response in neurons exposed to DNA damage. J Neurosci. 2004;24:2963–2973. doi: 10.1523/JNEUROSCI.0155-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile JR, Eichten A, Zacny V, Munger K. NF-kappaB-mediated induction of p21(Cip1/Waf1) by tumor necrosis factor alpha induces growth arrest and cytoprotection in normal human keratinocytes. Mol Cancer Res. 2003;1:262–270. [PubMed] [Google Scholar]
- Bhakar AL, Tannis LL, Zeindler C, Russo MP, Jobin C, Park DS, MacPherson S, Barker PA. Constitutive nuclear factor-kappa B activity is required for central neuron survival. J Neurosci. 2002;22:8466–8475. doi: 10.1523/JNEUROSCI.22-19-08466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, McAllister-Lucas LM, Shao F, Schumacher KR, Feng Z, Porter AG, Castle VP, Opipari AW., Jr. NF-kappa B activation mediates doxorubicin-induced cell death in N-type neuroblastoma cells. J Biol Chem. 2001;276:48921–48929. doi: 10.1074/jbc.M108674200. [DOI] [PubMed] [Google Scholar]
- Boekhoudt GH, Guo Z, Beresford GW, Boss JM. Communication between NF-kappa B and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J Immunol. 2003;170:4139–4147. doi: 10.4049/jimmunol.170.8.4139. [DOI] [PubMed] [Google Scholar]
- Bours V, Bentires-Alj M, Hellin AC, Viatour P, Robe P, Delhalle S, Benoit V, Merville MP. Nuclear factor-kappa B, cancer, and apoptosis. Biochem Pharmacol. 2000;60:1085–1089. doi: 10.1016/s0006-2952(00)00391-9. [DOI] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- Brunner HG, Hamel BC, von Bokhoven H. P63 gene mutations and human developmental syndromes. Am J Med Genet. 2002;112:284–290. doi: 10.1002/ajmg.10778. [DOI] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- Chan H, Bartos DP, Owen-Schaub LB. Activation-dependent transcriptional regulation of the human Fas promoter requires NF-kappaB p50-p65 recruitment. Mol Cell Biol. 1999;19:2098–2108. doi: 10.1128/mcb.19.3.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett CS, Perkins ND, Kowalik TF, Schmid RM, Huang ES, Baldwin AS, Jr., Nabel GJ. Dimerization of NF-KB2 with RelA(p65) regulates DNA binding, transcriptional activation, and inhibition by an I kappa B-alpha (MAD-3). Mol Cell Biol. 1993;13:1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao MP, Buckler AJ, Sonenshein GE. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc Natl Acad Sci USA. 1990;87:4727–4731. doi: 10.1073/pnas.87.12.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffi F, Boroni F, Benarese M, Sarnico I, Benetti A, Spano PF, Pizzi M. The inhibitor of I kappa B alpha phosphorylation BAY 11-7082 prevents NMDA neurotoxicity in mouse hippocampal slices. Neurosci Lett. 2005;377:147–151. doi: 10.1016/j.neulet.2004.11.088. [DOI] [PubMed] [Google Scholar]
- Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, Lena AM, Candi E, Terrinoni A, Catani MV, Oren M, Melino G, Krammer PH, Stremmel W, Muller M. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilli M, Pizzi M, Memo M, Spano P. Neuroprotection by aspirin and sodium salicylate through blockade of NF-kappaB activation. Science. 1996;274:1383–1385. doi: 10.1126/science.274.5291.1383. [DOI] [PubMed] [Google Scholar]
- Gveric D, Kaltschmidt C, Cuzner ML, Newcombe J. Transcription factor NF-kappaB and inhibitor I kappaBalpha are localized in macrophages in active multiple sclerosis lesions. J Neuropathol Exp Neurol. 1998;57:168–178. doi: 10.1097/00005072-199802000-00008. [DOI] [PubMed] [Google Scholar]
- Hunot S, Brugg B, Ricard D, Michel PP, Muriel MP, Ruberg M, Faucheux BA, Agid Y, Hirsch EC. Nuclear translocation of NF-kappaB is increased in dopaminergic neurons of patients with Parkinson disease. Proc Natl Acad Sci USA. 1997;94:7531–7536. doi: 10.1073/pnas.94.14.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs WB, Govoni G, Ho D, Atwal JK, Barnabe-Heider F, Keyes WM, Mills AA, Miller FD, Kaplan DR. p63 is an essential proapoptotic protein during neural development. Neuron. 2005;48:743–756. doi: 10.1016/j.neuron.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B, Uherek M, Volk B, Baeuerle PA, Kaltschmidt C. Transcription factor NF-kappaB is activated in primary neurons by amyloid beta peptides and in neurons surrounding early plaques from patients with Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:2642–2647. doi: 10.1073/pnas.94.6.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt B, Widera D, Kaltschmidt C. Signaling via NF-kappaB in the nervous system. Biochim Biophys Acta. 2005;1745:287–299. doi: 10.1016/j.bbamcr.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Katoh I, Aisaki KI, Kurata SI, Ikawa S, Ikawa Y. p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene. 2000;19:3126–3130. doi: 10.1038/sj.onc.1203644. [DOI] [PubMed] [Google Scholar]
- Kirch HC, Flaswinkel S, Rumpf H, Brockmann D, Esche H. Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-kappaB and Myc/Max. Oncogene. 1999;18:2728–2738. doi: 10.1038/sj.onc.1202626. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Lu SL, White LD, Wang XJ, Roop DR. Reactivation of developmentally expressed p63 isoforms predisposes to tumor development and progression. Cancer Res. 2006;66:3981–3986. doi: 10.1158/0008-5472.CAN-06-0027. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki R, Ikawa S, Maeda T, Nakazaki Y, Kurita R, Harata M, Shutoh Y, Bai YS, Soda Y, Tanabe T, Dohi T, Kato R, Ikawa Y, Asano S, Tani K. p51/p63, a novel p53 homologue, potentiates p53 activity and is a human cancer gene therapy candidate. J Gene Med. 2006;8:1121–1130. doi: 10.1002/jgm.945. [DOI] [PubMed] [Google Scholar]
- Lantner F, Starlets D, Gore Y, Flaishon L, Yamit-Hezi A, Dikstein R, Leng L, Bucala R, Machluf Y, Oren M, Shachar I. CD74 induces TAp63 expression leading to B cell survival. Blood. 2007;110:4303–4311. doi: 10.1182/blood-2007-04-087486. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Li N, Li H, Cherukuri P, Farzan S, Harmes DC, DiRenzo J. TA-p63-gamma regulates expression of DeltaN-p63 in a manner that is sensitive to p53. Oncogene. 2006;25:2349–2359. doi: 10.1038/sj.onc.1209270. [DOI] [PubMed] [Google Scholar]
- Lin SC, Wortis HH, Stavnezer J. The ability of CD40L, but not lipopolysaccharide, to initiate immunoglobulin switching to immunoglobulin G1 is explained by differential induction of NF-kappaB/Rel proteins. Mol Cell Biol. 1998;18:5523–5532. doi: 10.1128/mcb.18.9.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPartlin M, Zeng SX, Lu H. Phosphorylation and stabilization of TAp63gamma by IkappaB kinase-beta. J Biol Chem. 2008;283:15754–15761. doi: 10.1074/jbc.M801394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. NF-kappaB in the survival and plasticity of neurons. Neurochem Res. 2005;30:883–893. doi: 10.1007/s11064-005-6961-x. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. p63 in skin appendage development. Cell Cycle. 2007;6:285–290. doi: 10.4161/cc.6.3.3798. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, Jones KA. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- Sarnico I, Boroni F, Benarese M, Alghisi M, Valerio A, Battistin L, Spano P, Pizzi M. Targeting IKK2 by pharmacological inhibitor AS602868 prevents excitotoxic injury to neurons and oligodendrocytes. J Neural Transm. 2008;115:693–701. doi: 10.1007/s00702-007-0016-1. [DOI] [PubMed] [Google Scholar]
- Seitz CS, Lin Q, Deng H, Khavari PA. Alterations in NF-kappaB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-kappaB. Proc Natl Acad Sci USA. 1998;95:2307–2312. doi: 10.1073/pnas.95.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz CS, Deng H, Hinata K, Lin Q, Khavari PA. Nuclear factor kappaB subunits induce epithelial cell growth arrest. Cancer Res. 2000;60:4085–4092. [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Shin SR, Sanchez-Velar N, Sherr DH, Sonenshein GE. 7,12-dimethyl-benz(a)anthracene treatment of a c-rel mouse mammary tumor cell line induces epithelial to mesenchymal transition via activation of nuclear factor-kappaB. Cancer Res. 2006;66:2570–2575. doi: 10.1158/0008-5472.CAN-05-3056. [DOI] [PubMed] [Google Scholar]
- Sonenshein GE. Rel/NF-kappa B transcription factors and the control of apoptosis. Semin Cancer Biol. 1997;8:113–119. doi: 10.1006/scbi.1997.0062. [DOI] [PubMed] [Google Scholar]
- Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, Miller FD, Flores ER. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- Tamaki T, Ohnishi K, Hartl C, LeRoy EC, Trojanowska M. Characterization of a GC-rich region containing Sp1 binding site(s) as a constitutive responsive element of the alpha 2(I) collagen gene in human fibroblasts. J Biol Chem. 1995;270:4299–4304. doi: 10.1074/jbc.270.9.4299. [DOI] [PubMed] [Google Scholar]
- Waltermann A, Kartasheva NN, Dobbelstein M. Differential regulation of p63 and p73 expression. Oncogene. 2003;22:5686–5693. doi: 10.1038/sj.onc.1206859. [DOI] [PubMed] [Google Scholar]
- Wartiovaara K, Barnabe-Heider F, Miller FD, Kaplan DR. N-myc promotes survival and induces S-phase entry of postmitotic sympathetic neurons. J Neurosci. 2002;22:815–824. doi: 10.1523/JNEUROSCI.22-03-00815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JT, Kral JG. The NF-kappaB/IkappaB signaling system: A molecular target in breast cancer therapy. J Surg Res. 2005;123:158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Wu M, Lee H, Bellas RE, Schauer SL, Arsura M, Katz D, FitzGerald MJ, Rothstein TL, Sherr DH, Sonenshein GE. Inhibition of NF-kappaB/Rel induces apoptosis of murine B cells. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ouaaz F, Bruzzo P, Singh V, Gerondakis S, Beg AA. NF-kappa B RelA (p65) is essential for TNF-alpha-induced fas expression but dispensable for both TCR-induced expression and activation-induced cell death. J Immunol. 2001;166:4949–4957. doi: 10.4049/jimmunol.166.8.4949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.