Abstract
Transition metal complexes offer great potential as diagnostic and therapeutic agents, and a growing number of biological applications have been explored. To be effective, these complexes must reach their intended target inside the cell. Here we review the cellular accumulation of metal complexes, including their uptake, localization, and efflux. Metal complexes are taken up inside cells through various mechanisms, including passive diffusion and entry through organic and metal transporters. Emphasis is placed on the methods used to examine cellular accumulation, to identify the mechanism(s) of uptake, and to monitor possible efflux. Conjugation strategies that have been employed to improve the cellular uptake characteristics of metal complexes are also described.
Introduction
Transition metal complexes are appealing candidates in the search for new diagnostic and therapeutic agents. They represent a uniquely modular system, wherein the metal center holds its ligands in a precisely defined three-dimensional structure. These ligands can be varied relatively easily, in order to change the characteristics of the complex in either subtle or dramatic fashion. Transition metal complexes also offer rich photophysical and photochemical properties, expanding their utility beyond chemical recognition.
Biological applications of transition metal complexes are increasingly being explored (1–5). To be effective, these compounds must reach the desired location inside the cell. There are several routes across the cell membrane, the identity of which affects the rate of uptake and the intracellular distribution. Also of interest is the possibility of export by the multidrug efflux pumps, such as Pgp and MRP1, which recognize a multitude of structurally diverse substrates.
The current understanding of the cellular processing of transition metal complexes, outside of cisplatin, is relatively limited, though it continues to expand. The diversity in structure among metal complexes is at least as great as that for organic molecules. Therefore, one can expect the same diversity of uptake mechanisms and intracellular fate that has been characterized for organic drugs. Indeed, as we review here, this supposition holds true.
Methods to examine cellular accumulation
Metal complexes for diagnostic applications are frequently luminescent, allowing ready characterization of their uptake characteristics. They can be examined by fluorometry, confocal microscopy (6) and flow cytometry (7,8). For non-luminescent complexes, inductively coupled plasma mass spectrometry (ICP-MS) (9,10), atomic absorption spectroscopy (AAS) (11), and UV-visible absorption spectroscopy (12) are used. Analysis of exogenous transition metal complexes by ICP-MS or AAS is greatly enabled by the fact that no background exists within the cell. We have taken advantage of this within our own laboratory to study non-luminescent rhodium complexes.
While ICP-MS and AAS represent very sensitive methods to assay for metal content, these assays cannot be accomplished with monitoring in real time. Cell lysates are instead prepared from cells that have been incubated with metal complex, prior to assay for metal uptake. When adherent cells are used, they are either detached from the culture dish and then lysed, or lysed directly in the dish. Alternatively, the cells can be detached and treated with complex in suspension, though in this case, the cells are not in their normal growing environment. This cell lysate is analytically diluted, and the amount of metal in the solution is quantified. Amounts are typically reported versus cell number or total protein concentration. Independent of the quantification technique, attention must be paid to certain steps to ensure accurate results. Egger and colleagues have found that adsorption to the culture plates and sample storage conditions prior to analysis significantly influence recovery of the metal (9). Factors affecting adsorption include concentration of the complex, the amount of protein in the medium, the duration of contact of protein-containing medium before treatment with complex, and the lipophilicity of the complex. Adsorption-related artifacts are particularly an issue when lysis is performed directly in the culture dishes. To correct for these effects, adsorption blanks of cell-free samples treated with metal complex should be performed. A second major issue is the time that the sample is stored prior to measurement, as the recovery of analyte decreases with time. Consequently, samples should be quantified immediately after preparation. When these considerations are taken into account, reliable measurements of metal complex uptake can be performed.
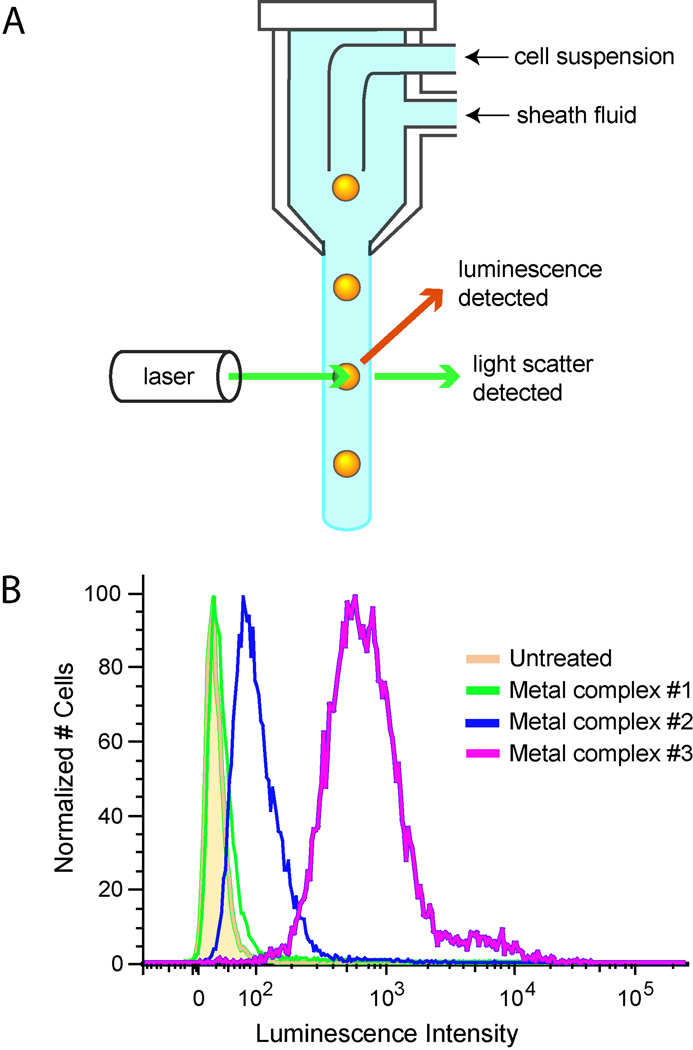
The cellular uptake of luminescent metal complexes is primarily examined using two complementary methods, flow cytometry and confocal microscopy; fluorometry of cell lysates can also be performed. For flow cytometry, cells are detached from culture either before or after incubation with the metal complex to produce a cell suspension. Untreated cells are used for as a control for autofluorescence. To exclude dead cells from analysis, a membrane-impermeable dead cell dye, such as propidium iodide, can be added (13). The cells are inspected individually as they pass single file through the laser beam(s) and the instrument records their light scatter and luminescence (Figure 1). Optical band pass filters separately collect the emission from multiple fluorophores. The result is a distribution of luminescence for the cell population, which can be depicted as a histogram of the number of cells versus luminescence intensity. The luminescence intensity of different cell populations, e.g. treated with different complexes or different incubation conditions, is easily compared.
Figure 1. Flow cytometry analysis of cellular uptake.
(A) The sample stream containing the cells are injected into a flowing stream of sheath fluid, which focuses the sample stream to roughly one cell in diameter. The luminescence and light scatter are recorded for each cell as it passes through the laser beam. (B) Histograms are shown representing cells treated with different metal complexes. Untreated cells display some background luminescence. Complexes with greater uptake exhibit increased luminescence intensity.
Flow cytometry is faster and less labor intensive than preparation of samples for ICP-MS, but still does not allow real-time monitoring. It also provides a distribution of cellular uptake, rather than only the mean uptake of all the cells. Samples prepared for flow cytometry will have the same adsorption issues described above, though they may be less significant, as the cells are detached from the culture dish after incubation with the metal compound, rather than lysed in the dish, or incubated in suspension following detachment. Flow cytometry distinguishes live from dead cells by uptake of a dead cell dye, whereas with ICP-MS, dead cells are eliminated from analysis if they have lost adherence to the culture dish and are washed away before the lysis step. Both techniques have their purpose, as ICP-MS provides absolute values for uptake, while flow cytometry is limited to luminescent compounds and is better suited for comparing the amount of uptake under different conditions.
Flow cytometry and analysis of cell lysates by ICP-MS and other methods only provide a measurement of the total amount of metal complex associated with the cell; they do not distinguish between membrane-bound and intracellular material. Localization is difficult to discern by these techniques, where cellular components, such as nuclei, must be physically isolated before the metal content can be determined (14).
Confocal microscopy, on the other hand, reveals the spatial distribution of luminescent metal complexes inside the cell. Co-staining with organelle dyes can be performed to further pinpoint their intracellular location. For non-luminescent complexes, displacement of dyes can provide indirect evidence of localization. For example, competitive displacement of the DNA stain Hoechst 33258 has been used to demonstrate nuclear accumulation (15). In fact confocal microscopy even offers the opportunity of real-time monitoring of luminescent complexes in situ. Another notable advantage of microscopy over ICP-MS is that lesser amounts of metal complex are typically required, as the incubations can be performed in small wells (e.g. those of a 96-well plate). To acquire better quality images, adherent cells are preferable over suspension cells, and the cells should not be confluent. Importantly, cells should be imaged live rather than fixed, as fixation can cause artifactual redistribution of compounds (16). In all uptake experiments, attention should be paid to the number of cells incubated with the metal complex, since the amount of uptake may be dependent on it. This has been shown to be the case for cell-penetrating peptides (CPP)(17). The merits of the different methods for measuring cellular accumulation are summarized in Table 1.
Table 1.
Comparison of methods for examining cellular accumulation
| Method | Advantages | Disadvantages |
|---|---|---|
| ICP-MS, AAS | Applicable to non-luminescent complexes Quantitative (mean metal content per cell or per mg protein) |
Low throughput Cannot distinguish surface-bound vs. internalized Sample is degraded |
| Flow cytometry | High throughput Semi-quantitative Provides population distribution of luminescence intensity Can distinguish live vs. dead cells |
Limited to luminescent complexes Cannot distinguish surface-bound vs. internalized |
| Confocal microscopy | Provides subcellular localization Real-time monitoring in situ Can distinguish live vs. dead cells |
Limited to luminescent complexes Low throughput |
Finally, in the absence of direct studies of uptake, biological activity may serve as a proxy. Here, of course, the lack of activity does not necessarily indicate the lack of cellular accumulation. However, biological read-outs of cytotoxicity, cellular proliferation, or other biochemical processes can be performed sensitively, and necessarily reflect uptake of the complex.
Identification of the uptake mechanism
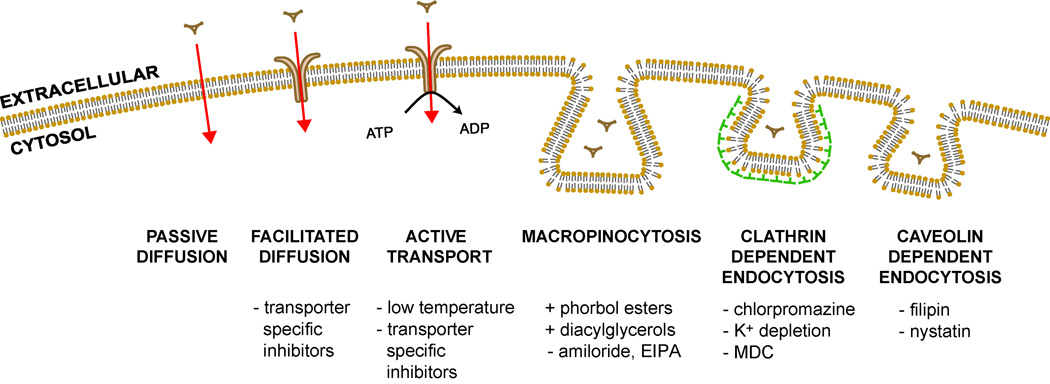
Understanding how metal-based probes and therapeutics gain entry to the cell is important for their practical development, as the import mechanism has implications for cell-type specificity, the rate of uptake, and their intracellular fate. The pathways into the cell include passive diffusion, transport proteins, and endocytosis (Figure 2). Passive diffusion involves the movement of molecules directly through the lipid bilayer down their concentration gradient. Transport proteins each move a particular class of cargo, and expression varies by tissue type. They can be energy-independent, as for channels and passive carriers, or be energy-dependent, as for ATP-powered pumps. Endocytosis is the uptake of macromolecules and solutes by vesicles derived from the plasma membrane. Diverse techniques including chemical, molecular biology, and imaging tools can be employed to reveal the mechanism of uptake.
Figure 2. Routes into the cell.
Passive diffusion, facilitated diffusion, active protein transporters, and several endocytic pathways are illustrated. Inhibitors and activators of each pathway are shown.
One clear dividing line between different mechanisms is whether uptake requires energy, as for endocytosis and active transport proteins, or is energy-independent, as is the case for passive diffusion through the membrane and diffusion facilitated by channels and carriers. Processes that require energy can be blocked by incubating cells at low temperature (4 °C) or by ATP-depletion with metabolic inhibitors, such as 2-deoxyglucose (competitively inhibits glycolysis)(18) and oligomycin (blocks oxidative phosphorylation)(19).
Passive diffusion is the pathway least susceptible to modulation. Uptake is energy-independent, is not saturable, and cannot be hindered by structural analogues. However, changes in the membrane fluidity, such as by cholesterol depletion or low temperature, can alter uptake. Cholesterol can be extracted from the plasma membrane using methyl-β-cyclodextrin (20,21). Also, the plasma membranes of most cells exhibit a membrane potential, with the inside of the cell negative with respect to the outside, which can serve as a driving force for the entry of positively charged compounds. In animal cells, the membrane potential depends primarily on the potassium concentration gradient. Consequently, the potential can be reduced by incubating the cells in a buffer with a potassium concentration equivalent to that found intracellularly. Gramicidin, a pore-forming peptide, also reduces the membrane potential by facilitating K+ equilibration (22). Conversely, hyperpolarization can be achieved using valinomycin, a cyclic peptide that selectively shuttles potassium ions across the membrane (22). Cationic dyes such as 3,3′-dihexyloxacarbocyanine are useful for verifying changes in the membrane potential (23). One should note that maintenance of the membrane potential requires ATP, and experiments involving metabolic inhibition may also affect the membrane potential, depending on the severity and duration of the metabolic blockade.
Protein-mediated transport can be active or passive. Examples of passive carriers include the organic cation transporters (OCTs)(24). Other proteins couple downhill transport of an ion to the uphill movement, such as the peptide transporters (PEPTs)(25), or take advantage of ATP hydrolysis. In contrast to passive diffusion, uptake is susceptible to inhibitors and can saturate. To study transport proteins, known inhibitors of suspected transporters can be used, and then cells observed for decreased uptake of the test compound. Alternatively, a specific transporter can be overexpressed, and the level of uptake compared to cells with low transporter expression.
Endocytosis encompasses several distinct pathways, which can be divided into two broad categories, phagocytosis and pinocytosis. Phagocytosis, the uptake of large particles, occurs only in specialized cells. Pinocytosis occurs in all cells by at least four distinct pathways: macropinocytosis, clathrin-mediated endocytosis, caveolae-dependent endocytosis, and clathrin- and caveolin-independent mechanisms (26,27). Uptake can be fluid-phase, adsorptive (nonspecific binding of solutes to the cell membrane), or receptor-mediated.
There are pharmacological treatments that can aid in distinguishing endocytic pathways, though the degree of their specificity varies substantially. Clathrin-mediated endocytosis is inhibited by hypertonic sucrose, potassium depletion, cytosolic acidification, chlorpromazine, monodansylcadaverine (MDC), and phenylarsine oxide. As all of these inhibitors have been shown to block uptake of fluid phase markers, they cannot be used to distinguish between clathrin-mediated endocytosis and macropinocytosis. Of these techniques, potassium depletion, chlorpromazine, and MDC appear to have the fewest side-effects (28).
Most inhibitors of caveolae-mediated endocytosis target cholesterol. Methyl-β-cyclodextrin (MβCD) forms soluble inclusion complexes with cholesterol, depleting it from the membrane. However, MβCD can also inhibit clathrin-mediated endocytosis and macropinocytosis, particularly at higher concentrations (29, 30). Filipin and nystatin sequester cholesterol in the form of membrane aggregates. These drugs disrupt caveolar structure and function but do not seem to impair clathrin-mediated endocytosis or macropinocytosis (31, 32). Another approach to deplete cholesterol uses cholesterol oxidase to convert cholesterol into 4-cholesten-3-one; this method has been shown to attenuate internalization of caveolae ligands (33).
For inhibition of macropinocytosis, drugs targeting actin polymerization, phosphoinositide metabolism, and sodium-proton exchange have been used. Agents that disrupt actin, such as cytochalasin D, are not specific for macropinocytosis as actin is required in multiple endocytic pathways (34). Wortmannin, a phosphoinositide 3-kinase (PI3K) inhibitor, blocks not only macropinocytosis and phagocytosis but also clathrin- and caveolae-mediated pathways (35,36). Amiloride and its derivatives, 5-(N-ethyl-N-isopropyl) amiloride (EIPA) and dimethyl amiloride (DMA), inhibitors of the Na+/H+ exchanger, block macropinocytosis and phagocytosis (37). Notably, the amilorides appear to have fewer side effects than the other available inhibitors of macropinocytosis. Stimulation of macropinocytosis can be achieved using phorbol esters (38) and diacylglycerols (39). Therefore, both suppression with amiloride and an increased response with the aforementioned stimulants can be used to define macropinocytosis.
The tools of molecular biology allow further characterization of the endocytic mechanism through identification of relevant proteins. The machinery driving clathrin-mediated endocytosis includes the clathrin triskelion, composed of three clathrin heavy chains and three light chains, which assemble into a polygonal lattice that molds the plasma membrane into a coated pit (27). Clathrin function can be modulated by overexpression of the heavy chain hub domain, which has a dominant negative effect on clathrin coat assembly, or by downregulation of clathrin heavy chain expression with siRNA (40). Caveolae contain members of the caveolin protein family, and they can be disrupted by overexpression of dominant-negative caveolin mutants and knockout of caveolin genes (27). A critical regulator of clathrin-mediated, caveolin-mediated and some clathrin- and caveolae-independent pathways is dynamin (41). The recruitment and assembly of dynamin at the neck of forming vesicles catalyzes their fission. Dominant-negative dynamin mutants can be used to study its involvement as well as the small molecule inhibitor dynasore (42, 43). As the mode of endocytosis is narrowed, the involvement of other proteins can be studied in a similar manner.
Colocalization of a luminescent metal complex with an endocytic marker supplies further evidence of entry via endocytosis. Numerous fluorescent tracers are commercially available (44). Clathrin-mediated endocytosis can be followed using fluorescently-tagged transferrin or low density lipoprotein (LDL) particles (45). After transferrin binds to its receptor on the plasma membrane, it is internalized in clathrin-coated vesicles, traffics through early and recycling endosomes, then returns to the plasma membrane. Similarly, LDL is recognized by the LDL receptor and enters by clathrin-mediated endocytosis. The LDL particle dissociates from its receptor in late endosomes, allowing its receptor to recycle back to the cell surface while LDL is transported into lysosomes (46). Labeled dextran is a fluid phase marker often used to follow macropinocytosis, though fluid phase markers have access to other types of vesicles. Cholera toxin B subunit has been used to track caveolae-mediated endocytosis, but it can enter by multiple mechanisms depending on the cell type, including clathrin-dependent and caveolae- and clathrin-independent endocytosis (47–50). The fluorescent glycosphingolipid analogue, BODIPY-LacCer, has also been used to track caveolae internalization (51).
Routes for Cell Entry by Metal Complexes
How do metal complexes exploit the various pathways into the cell? Though metal complexes are being investigated as probes and therapeutics, there are a relatively few studies on their mechanism of uptake. The cellular accumulation of cisplatin has received the most scrutiny and has been recently reviewed (52). Platinum drugs can enter the cell by passive diffusion, by organic cation transporters, by the copper transporter Ctr1, and possibly by endocytosis.
The mechanism of uptake for several luminescent Eu(III) and Tb(III) complexes has been characterized by Parker and coworkers (Figure 3)(53). These complexes enter cells by macropinocytosis and not via clathrin-mediated endocytosis or caveolae. Both wortmannin and amiloride suppress their uptake, while a phorbol ester and a diacylglycerol enhance uptake. The complexes also colocalize with fluorescein-labeled dextran.
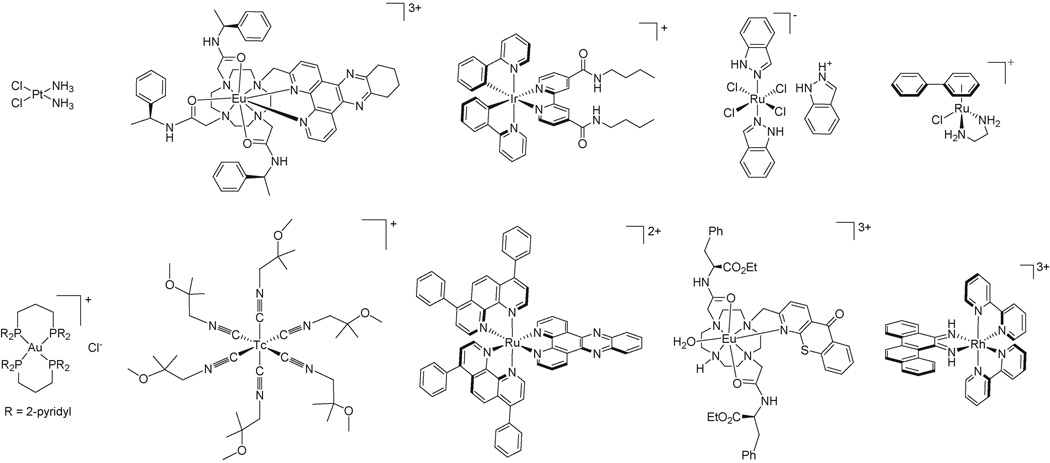
Figure 3. Metal complexes whose cellular accumulation have been explored.
Top row: cisplatin, a Eu(III) complex that enters cells by macropinocytosis, an Ir(III) polypyridine complex, the drug candidate KP1019, and a Ru(II) arene complex. Bottom row: a Au(I) phosphine complex that accumulates in mitochondria, the radiopharmaceutical [99mTc]Sestamibi, a Ru(II) dipyridophenazine complex, a Eu(III) complex that stains nucleoli, and a Rh(III) metalloinsertor that binds DNA mismatches.
Studies by Lo and colleagues on bis(cyclometalated) iridium(III) polypyridine complexes suggest their entry by more than one mechanism (54). The uptake of [Ir(ppy)2(bpyC4)]+, where ppy = 2-phenylpyridine and bpyC4 = 4,4′-bis(n-butyl-aminocarbonyl)-2,2′-bipyridine (Figure 3), into cytoplasmic foci was diminished at 4 °C, while the diffuse cytoplasmic staining was maintained. A similar effect was observed upon treatment with the colchicine, which interferes with microtubule assembly and inhibits endocytosis. Thus, uptake occurs both by an energy-dependent pathway (possibly endocytosis) and an energy-independent pathway.
For some Ru(III) complexes, cellular entry is mediated by the iron transport protein transferrin. The drug candidate KP1019, indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (Figure 3), binds transferrin with displacement of a chloride ligand, then enters the cell by clathrin-mediated endocytosis (55). Transferrin can bind two ruthenium moieties, one at each iron binding site, but this results in lower accumulation of ruthenium in the cell. Occupation of both iron binding sites by ruthenium probably causes a structural change in the protein that prevents it from binding to its receptor. In contrast, transferrin equilibrated with iron before binding of one ruthenium moiety results in substantially increased uptake. Following cellular entry, transferrin localizes in endosomes; therefore the release of the ruthenium complex from transferrin and subsequent endosomal escape likely occurs.
Metal complexes that are lipophilic cations may passively diffuse across the plasma membrane in response to the membrane potential. The imaging agent hexakis (2-methoxyisobutyl isonitrile) technetium(I), known as [99mTc]Sestamibi (Figure 3), accumulates inside cells driven by both the plasma and mitochondrial membrane potentials (56). Uptake is reduced by depolarization of the plasma membrane with high potassium buffer and depolarization of the mitochondrial membranes with the proton ionophores 2,4-dinitrophenol and carbonyl cyanide m-chlorophenylhydrazone. Similarly, a Ru(II) complex containing the lipophilic 4,7-diphenyl-1,10-phenanthroline ligand (Figure 3) enters by passive diffusion in a membrane-potential dependent manner (57).
Localization
Nuclear Targeting
Cellular accumulation alone is insufficient to enable activity; any potential therapeutic agent must also be able to reach its target within the cell. As many metal complexes are targeted to DNA, they must furthermore gain nuclear access, which is tightly regulated by the nuclear pore complex (NPC) (58). In addition to actively transporting cargo such as RNAs and large proteins, the NPC also facilitates the diffusion of small molecules, ions, and proteins. Although some fraction of the cytoplasmic concentration of a metal complex will certainly diffuse passively through the NPC, the efficiency of nuclear entry warrants consideration beyond this supposition. It is reasonable to assume that efficient nuclear localization would reduce off-target effects. Conversely, poor nuclear localization would pose a serious obstacle to the utility of any DNA-targeted agent.
To this end, Parker and coworkers have developed a class of luminescent probes based on cationic lanthanide complexes (59). The initial europium and terbium complexes were shown to reversibly localize within the nucleus above a 1 mM concentration threshold; subsequently, a europium complex has been developed that selectively targets nucleoli at lower concentrations (60). The long lifetime, large Stokes shift, and relatively high quantum yield of this complex (shown in Figure 3) make it an interesting tool for the further study of nucleoli, whose role is not yet fully understood (61).
Seymour’s group exploited the intrinsic fluorescence of zinc bis(thiosemicarbazone) (TSC) complexes to study their cellular uptake and localization through confocal fluorescent microscopy (62). Their work clearly showed that nuclear uptake varied with subtle structural differences in the complex as well as with cell line. In addition to their inherent potential as anti-tumor agents, these zinc TSC complexes may also provide insight into the uptake of analogous copper complexes, as the two classes are structurally very similar.
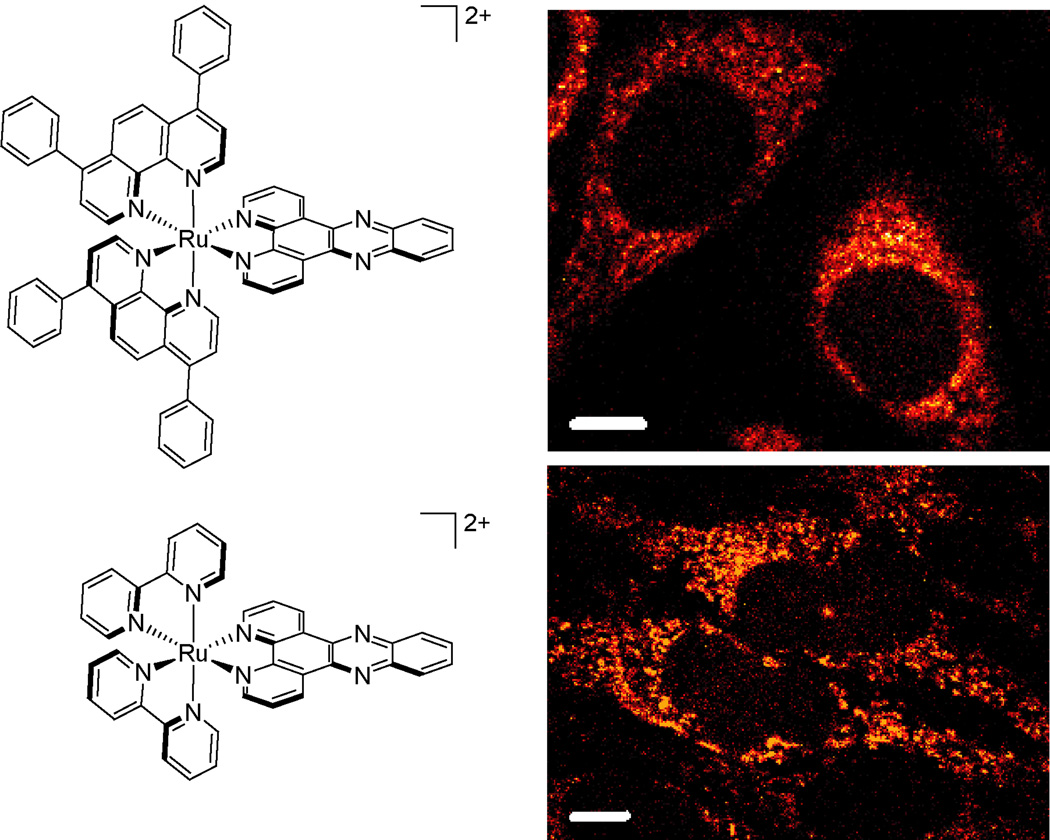
In our laboratory, we have used luminescent ruthenium analogues to study cellular uptake and localization (8, 57). Given the brightness and robust characteristics of polypyridyl complexes of Ru(II), we could make systematic changes in the ligand set and examine how uptake varied using flow cytometry and confocal microscopy. We observed, for example, that hydrophobicity of the complex was more of a driver for cell entry than size; [Ru(DIP)2dppz]2+ is taken up more readily inside cells versus [Ru(bpy)2dppz]2+ despite its large bulk (20.4 Å in diameter)(Figure 4).
Figure 4. Cellular uptake of dipyridophenazine (dppz) complexes of Ru(II).
The lipophilic complex [Ru(DIP)2dppz]2+ is observed inside HeLa cells at lower incubation concentrations and shorter times (top, 5 µM for 2 h) than [Ru(bpy)2dppz]2+ (bottom, 20 µM for 72 h). Structures of complexes are shown at left. Scale bars are 10 µm.
Importantly, the intrinsic luminescence of the ruthenium complex was also critical in revealing that fluorescent tagging with an organic fluorophore could alter subcellular localization, yielding nuclear accumulation that was not seen in the absence of the fluorescent tag (63). This result served as a striking example of the pitfalls of fluorescent labeling, how the fluorophore may provide more than just a “label”. This result also underscored the advantages of utilizing luminescent metal complexes that do require an extra “label”.
For non-luminescent complexes, ICP-MS and AAS can also yield semi-quantitative information about relative metal levels in isolated nuclear or mitochondrial fractions. These techniques are particularly amenable to analysis of exogenous transition metals. In addition, measurements of biological activity have served as a proxy for uptake for a class of rhodium metalloinsertors developed within our group (Figure 3) (8, 64). HCT116 +ch3/+ch2 cells were treated with [Rh(L)2chrysi]3+ complexes where chrysi = chrysenequinone and L = (NH3)2, 2,2′-bipyridine (bpy), 2,2′-dipyridylamine (HDPA), 1,10-phenanthroline (phen), or 4,7-diphenyl-1,10-phenanthroline (DIP) for various incubation times. Optimal activity for the DIP complex is observed with 12 hour incubation and a lower concentration than the other complexes, as would be expected from confocal microscopy and flow cytometry studies of [Ru(L)2dppz]2+ analogues. The 48–72 hours required for optimal activity of the remaining complexes also correlates well with ruthenium uptake times. Unexpectedly, the HDPA complex exhibits activity after only a short 12 hour incubation, suggesting accelerated uptake relative to that expected based upon hydrophobicity; subsequent ICP-MS uptake assays of the rhodium complexes confirms that the HDPA ligand modestly improves cellular uptake.
Mitochondrial Targeting
Mitochondria have emerged as an interesting alternative target for many applications (65, 66). Mitochondria use the energy released by the oxidation of substrates such as glucose to establish a proton gradient across their inner membrane. This proton gradient drives ATP synthetase, which produces the majority of the ATP required for active processes within the cell. Since mitochondria maintain their own genome, which codes for the enzymes of the electron transport chain, it is possible that DNA-targeting metal complexes could effectively compromise mitochondrial function. Thus mitochondrial targeting also presents an interesting strategy for new chemotherapies.
Indeed, any metal complex that disrupts mitochondrial function could hold potential as a chemotherapeutic agent, regardless of whether or not the complex targets DNA. To this end, Berners-Price and colleagues have developed gold(I) phosphine complexes that accumulate in mitochondria and trigger apoptosis through the caspase-3 and caspase-9 dependent mitochondrial pathway (Figure 3)(67). These complexes were shown to have selective activity in breast cancer cells at submicromolar concentrations. This approach has been expanded to include silver(I) phosphine complexes that also accumulate in the mitochondria in a membrane potential-dependent fashion and show cytotoxicities that correlate with their lipophilicities (68).
Compared to nuclear targeting, mitochondrial targeting is relatively facile, with a clearly understood chemical basis; lipophilic cations have been shown to accumulate in mitochondria due to the negative potential difference across the mitochondrial membrane (69, 70). Furthermore, carcinoma cells have been shown to have higher plasma and/or mitochondrial membrane potentials, providing a means of selectively targeting them (71, 72). The reasons for the increased membrane potential have yet to be fully understood, but work in this area holds the promise of further increasing selectivity.
In addition to the use of lipophilic cations as anti-cancer agents, Murphy and colleagues have employed triphenylphosphonium (TPP) salts as delivery vehicles for several other applications (73, 74). As a potential treatment for a broad range of diseases associated with oxidative damage in the mitochondria, an antioxidant based on the active moiety of vitamin E was conjugated to a TPP cation (75). The resulting conjugate was concentrated in the mitochondria by the membrane potential, reaching levels approximately 80-fold greater than those of vitamin E alone. Although most of the work in mitochondrial targeting to date has employed organic cations, organometallic ions potentially could deliver cargo to the mitochondria in a similar manner.
Efflux
Efflux mediated by protein export pumps is a major contributor to drug resistance in cancer cells, in which therapeutics are removed before they can exert their intended effect. Efflux transport proteins are also expressed in normal tissues, where they limit the absorption (intestine), block the distribution (blood-brain barrier), and facilitate the elimination (kidney and liver) of drugs. In human cancers, the major drug efflux pumps belong to the ATP-binding cassette (ABC) protein family. These include P-glycoprotein (Pgp, MDR1, ABCB1), members of the multidrug resistance-associated protein family (MRP1–9, ABCC1–9), and ABCG2 (MXR, BCRP). These proteins transport a remarkable range of substrates, with their transport profiles overlapping to some extent. Compounds recognized by Pgp are often large, amphipathic molecules that are neutral or have a net positive charge, though not all Pgp substrates conform to this generalization (76). The MRP family transports neutral and anionic drugs, as well as glutathione, glucuronate, and sulfate conjugates (77, 78). ABCG2 also has broad substrate specificity, and is able to transport both positively and negatively charged compounds (79).
Several in vitro systems can be used to assess whether a compound is susceptible to transport by an efflux pump. They measure cytotoxicity, accumulation/efflux, transport, or ATPase activity. These methods are summarized below, and further details are available in recent reviews (80, 81).
For drugs that show antiproliferative activity, cytotoxicity (resistance) assays are commonly used. Typically, cytotoxicity is expressed as the IC50 value, the concentration of drug required to kill half of the cells, calculated from the dose-response curve. To study protein-mediated export, cell survival following drug treatment is compared between parental cells and cells that overexpress a particular efflux protein. Transported compounds should exhibit decreased toxicity, and efflux pump inhibitors should restore activity in transporter-expressing cells. Numerous inhibitors are available for the major ABC efflux pumps (82).
Accumulation assays (vide supra) offer a more direct measurement of efflux transporter activity. They can be performed on cells or membrane vesicle preparations. Similar to the resistance assays, cell lines with differential transporter expression are used. If a compound is an efflux pump substrate, accumulation will be lower in the transporter-expressing cell line versus the parental line, and transporter inhibitors should increase accumulation. For compounds lacking an easy handle for monitoring accumulation, indirect competition methods using a fluorescent reference substrate are an option. Such probes include calcein-AM for Pgp and MRP1 and Hoechst 33342 for ABCG2 (76). Here, the effect of the test compound on the accumulation of the fluorescent dye is observed. This assay can show if a compound interacts with the transporter, but it does not distinguish between substrates and inhibitors. Alternatively, accumulation assays can be performed using inside-out membrane vesicles. In this case, the transporter pumps the test compound into the vesicle. Inhibition of the transporters would decrease accumulation of the compound in the vesicles. Inside-out vesicles each containing different efflux transporters are available from several commerical suppliers.
Measurement of transcellular transport, the passage of molecules across a cell layer, is another method for studying export. Polarized cells that express efflux transporters are grown to confluence on a permeable surface (a transwell). The cells must form tight extracellular junctions, such that movement of the test compound from one side of the monolayer to the other can only occur by passage through the cells. The transporters may be localized on the apical side of the polarized monolayer, such as for Pgp and ABCG2, or the basolateral side, the case for MRP1. Apical transporters will increase movement in the basal to apical direction; basal transporters will do the opposite. The test compound is added to one side of the cell monolayer, and over time its concentrations in the basal and apical compartments are measured. Transported compounds will exhibit a directional bias that can be altered by inhibition of the efflux transporter.
ABC efflux transporters are powered by ATP hydrolysis, and the presence of substrates usually stimulates this process. Hence, quantification of ATPase activity is another method for identifying transported compounds (83). For this assay, cell membrane preparations containing the transporter are incubated with ATP and the test compound. ATP hydrolysis releases inorganic phosphate, which can be detected by various analytical methods, such as reaction with a colorimetric indicator (84). ATP hydrolysis indicates that the test compound affects ATPase activity, but does not directly demonstrate efflux, and slowly transported substrates may not induce detectable ATPase activity.
Many metal complexes have been shown to be actively exported from cells. [99mTc]Sestamibi (Figure 3), a radiopharmaceutical used for imaging the blood flow of the heart, was the first metal complex shown to be transported out of the cell by Pgp (85). Similar to many other Pgp substrates, the complex is positively charged and hydrophobic. In cells expressing Pgp, the net accumulation of the [99mTc]Sestamibi is inversely proportional to the amount of Pgp expression, and export could be reversed by Pgp inhibitors such as verapamil, cyclosporin A, and quinidine. [99mTc]Sestamibi is also recognized by MRP1 (86, 87). Numerous other technetium complexes have been characterized regarding their Pgp transport activity, due to their promise in diagnostic imaging of Pgp-mediated transport in tumors (88).
The cellular efflux of cisplatin is mediated by multiple mechanisms (52, 89). Evidence suggests that the copper transporters ATP7A and ATP7B, which are responsible for the removal of excess copper ions from the cell, play a role in cisplatin export. Their expression can reduce cisplatin accumulation and/or efficacy, though not all cisplatin-resistant cell lines show increased expression. The MRP transporters may also export cisplatin, most likely as a glutathione conjugate. Pt(II) and glutathione (GSH) form a stable Pt(GS)2 complex, in which glutathione chelates Pt via cysteinyl sulfur and nitrogen atoms (90). Some cell lines with elevated MRP2 levels show increased cisplatin resistance, reduced accumulation, and decreased DNA platination. Cisplatin does not appear to be a Pgp substrate (91, 92).
The active export of two classes of ruthenium complexes has also been examined. The Ru(III) complex KP1019 (Figure 3) is a substrate and inhibitor of Pgp (93). Pgp expression reduced KP1019 cytotoxicity, and this effect could be reversed by Pgp inhibitors. KP1019 also restored accumulation of rhodamine 123, a fluorescent substrate of Pgp, in cells overexpresing P-glycoprotein. In contrast, activity of the complex was not diminished by overexpression of MRP1 or BCRP. Certain Ru(II) arene complexes may also be effluxed by Pgp (an example is shown in Figure 3)(94). A series of Ru(II) complexes exhibited reduced toxicity in a multi-drug resistant cell line, which has elevated levels of both Pgp and MRP2. Verapamil, a Pgp inhibitor, restored sensitivity to one of these Ru(II) complexes. In common with many other Pgp substrates, these complexes are hydrophobic and positively charged.
Conjugates
The most widely used strategy to accelerate the uptake or to increase the accumulation of metal complexes within the cell is to attach a biomolecule covalently that directs trafficking of the conjugated complex. Synthetic polymers and several different types of biomolecules have been utilized to this end, including peptides, B vitamins, carbohydrates, and hormones. Cisplatin and its derivatives are by far the most popular choice of cargo (Table 2).
Table 2.
Biocarriers and their metal cargoes
| Biocarrier | Metal Cargo | Comment | Reference |
|---|---|---|---|
| Peptides | |||
| RGD/NGR | platinum | integrin substrates | 95 |
| PKKKRKV | cobalt, technetium/rhenium | SV40 large tumor antigen NLS | 109–111, 113 |
| GRKKRRQRRRAP | zinc, technetium/rhenium | HIV-1 TAT peptide NLS/CPP | 106, 115–117 |
| rrrrrrrr | rhodium, ruthenium | synthetic NLS | 63, 108 |
| YGGFL | cobalt | enkephalin NLS | 112 |
| RKRKRK | gadolinium | acute lymphatic leukemia-1 NLS | 114 |
| B Vitamins | |||
| B12 | platinum | endocytosis and metabolism in lysosomes releases Pt | 118 |
| Carbohydrates | |||
| dextran | platinum | branched galactose clusters target cell surface receptors | 119–121 |
| Hormones | |||
| LH-RH | platinum | luteinizing hormone-releasing hormone peptide analogues | 122 |
| 1,7-β-estradiol | platinum | primary substrate for nuclear estrogen receptor | 123–126 |
| Synthetics | |||
| polymeric micelles | platinum | 28 nm micelles from poly(ethylene glycol)-poly(glutamic acid) | 127, 128 |
| single-walled carbon nanotubes | platinum | clathrin-mediated endocytosis | 129 |
| silica-encased coordination polymers | platinum | degradable shell provides controlled release | 130 |
| poly(ethylene glycol) | platinum | macromolecular PEG complexes, some conjugated to albumin | 131 |
Many groups have used short peptide sequences to direct uptake of a bioinorganic conjugate by targeting a particular receptor or cell compartment. Lippard and coworkers have combined platinum (IV) complexes with peptides containing an RGD or NGR motif (95). These sequences serve as substrates of the integrins αν/β3 and αν/β5 or aminopeptidase N, respectively, which are overexpressed during tumor-associated angiogenesis, providing an avenue to selectively target cancerous tissue. Indeed, conjugates of the substrate peptides were found to have roughly ten-fold greater activity than either conjugates of nonspecific sequences or the functionalized platinum (IV) complex without any conjugated peptide. No activity was observed for the peptide sequences themselves. Nils Metzler-Nolte and coworkers have synthesized and extensively characterized a wide variety of bioconjugates of amino acids, peptides, and peptide nucleic acids with various transition metals, including iron, copper, zinc, and cobalt with the intention of targeting specific organelles (96, 97). Their work examines metallocene and carbonyl alkyne complexes, and includes fluorescence microscopy and cytotoxicity studies that demonstrate the potential of a class of ferrocene-peptide conjugates to act as anti-microbial agents (98, 99).
Kelley and coworkers have shown that rationally designed peptide sequences containing cationic and lipophilic residues, e.g. FrFKFrFK (lower case denotes the D-isomer, used for enhanced biostability), can effectively and specifically deliver cargo to the mitochondria, but these sequences have not been used with metal complexes (100). This work is in accordance with findings that delocalized lipophilic cations accumulate in mitochondria (see Localization). While similar efforts to access selectively each organelle will undoubtedly remain a sought-after goal for those studying cellular uptake, the nucleus is by far the most prominently studied and therapeutically relevant target of peptide localization sequences and their conjugates.
To that end, short peptide sequences rich in positively charged residues such as arginine or lysine have been shown to function either as cell-penetrating peptides (CPPs), nuclear localization sequences (NLSs), or both. CPPs promote cellular uptake by translocation across the cell membrane or by endocytosis, whereas NLSs promote active transport through the nuclear pore complex. Early demonstrations of peptide-directed nuclear import involved the sequence PKKKRKV, from simian virus SV40 large tumor antigen, and the sequence GRKKRRQRRRAP, from the HIV-1 Tat peptide, which has also been shown to mediate cellular uptake (101–104). Since then, several other sequences, including synthetic sequences such as octaarginine, have been identified that share similar chemical characteristics and uptake properties (104, 105).
In an attempt to exploit the uptake properties that these peptide sequences confer, Vicente and coworkers conjugated photodynamic therapy (PDT) agents based on zinc(II) phthalocyanines to a bifunctional peptide sequence containing the Tat sequence and the NLS from nucleoplasmin (106). The peptide conjugates were shown to have slightly better cellular uptake and phototoxicity profiles than the unconjugated metal complexes. Fluorescent microscopy was performed on live HEp2 cells treated with the conjugates; in order to examine subcellular localization, the cells were simultaneously treated with organelle-specific fluorescent stains such as LysoSensor and MitoTracker. Significantly, image overlays showed that the conjugates were localizing mainly within lysosomes, consistent with cellular entry through endocytosis and subsequent failure to escape from the internalized endosomes. These experiments, then, reveal an obstacle to the widespread application of NLS peptides as delivery vehicles: endosomal trapping.
Interestingly, the extent of cellular uptake was also shown to be a function of the structure of the linker region; the conjugate containing a longer polyethylene glycol (PEG) linker region accumulated at higher levels. PEG linkers are commonly used in conjugate development due to the pharmacological benefits that they confer, including increased solubility and bioavailability and reduced systemic toxicity. Gibson and coworkers also employed PEG linkers to conjugate the anti-cancer agent carboplatin with a variety of NLS peptides (107). Based on the results of confocal microscopy studies with live M109FR cells and fluorescently labeled conjugates, the NLS successfully promoted rapid accumulation in the nucleus. However, the NLS-PEG-Pt conjugate formed less Pt-DNA adducts and was less cytotoxic than PEG-Pt or carboplatin alone. Gibson’s group interpreted these results as support for the notion that carboplatin requires cytosolic activation for its anti-cancer activity. Accordingly, bioconjugates of other metal complexes that require activation might encounter similar problems.
Within our own laboratory, octaarginine has been used to target rhodium mismatch-recognition agents to the nucleus. Early fluorescent microscopy experiments on live HeLa cells were very promising, showing dramatic nuclear accumulation at reduced incubation times (108). However, further investigations with fluorescent ruthenium complexes revealed yet another pitfall in conjugate development. The intrinsic luminescence of the ruthenium complexes allowed us to examine the distribution of the conjugates in the absence of a fluorescent organic tag, revealing that the organic fluorophore itself redirected the rhodium-peptide conjugate, which otherwise would not accumulate in the nucleus under the same conditions (63). This further emphasized that the uptake, and presumably inhibitory, properties of any conjugate are extremely sensitive to even subtle structural changes, such as addition of a fluorophore.
Metzler-Nolte and co-workers addressed this issue very elegantly by using atomic absorption spectroscopy (AAS) to study the cellular and nuclear uptake of cobalt bis(picolyl)amine complexes in live HT29 cells (109). This method allowed for the direct detection of unlabeled metal complex, eliminating any possibility of redirection; as mentioned in this report, inductively-coupled plasma mass spectrometry (ICP-MS) provides another alternative for direct detection. The complexes were conjugated with the SV40 antigen NLS, a PNA sequence (TGTTATCC), or both. Both the NLS and the PNA sequence were found to increase nuclear levels of cobalt, although the PNA sequence was slightly more effective in this case. The group had previously used the SV40 NLS to direct a fluorescently labeled cobaltocenium conjugate to the nucleus, as shown by fluorescent microscopy (110). Co-staining experiments also showed localization within the endosomes, consistent with endocytic entry into the cell. More recently, the group systematically investigated the cellular and nuclear uptake of an expanded class of conjugates, containing different metal complexes and peptide sequences, in the HepG2 cell line (111). Again, fluorescent microscopy and co-staining experiments were performed on live cells. Interestingly, scrambled versions of the SV40 NLS did not afford rapid cellular and nuclear uptake. In addition, the group has utilized the enkephalin peptide (YGGFL) as a vector for a cobalt carbonyl alkyne complex, although the cellular uptake properties of the conjugate were not determined (112).
The SV40 NLS has also been used to direct a trifunctional conjugate to the nucleus of mouse melanoma cells (113). The DNA intercalator pyrene was linked to an aliphatic triamine ligand which chelates either technetium-99m, an Auger electron emitter that exhibits strong radiotoxicity when in close contact with DNA, or rhenium, which provides a nonradioactive analogue. Appealingly, the pyrene moiety provided an intrinsic fluorescence for all conjugates. The SV40 NLS was used to complete the trifunctional conjugate and direct it to the nucleus. Both fluorescent micscropy and radiometry of extracts from 99mTc-treated cells confirmed that the conjugate accumulated in the nucleus, leading to radiotoxicity that was absent in the Re-treated cells.
Access to the nucleus for contrast agents such as gadolinium would also unlock a plethora of new applications in diagnostic imaging. With this goal, a combination of the SV40 and the acute lymphatic leukemia-1 (RKRKRK) NLS sequences was used to develop a gadolinium contrast agent that was found to localize in the nuclei of a human glioma xenograft in nude mice (114). Also technetium-99m and rhenium conjugates with the Tat peptide have been developed for diagnostic and radiotherapeutic applications, using radiometric and fluorescent microscopy techniques to study the effects of peptide chirality and sequence on cellular uptake (115–117).
Although peptides are the most common biomolecular carrier for metal complexes, there are many others. Alberto’s group is also developing conjugates to use vitamin B12 as a vehicle for platinum (II) complexes (118). Moreover, even dextran and galactose conjugates have been harnassed to deliver platinum prodrugs (119–121).
Hormone conjugates have garnered more attention due to the promise of tissue specificity. The first hormone conjugates came from Schally’s laboratory, where a variety of cytotoxic agents, including a cisplatin-like complex, were linked with luteinizing hormone-releasing hormone in 1992 (122). More recently, estrogen conjugates in particular have been developed as a means of targeting breast cancer cells, which express estrogen receptors. Hannon and coworkers have created and extensively characterized an estrogen-platinum complex that binds serum albumin and DNA (123). They demonstrated that it was capable of binding the estrogen recepetor in isogenic MCF-7 ER+/ER− cell lines, human breast adenocarcinoma lines that either express or do not express the nuclear estrogen receptor, by a competitive radioligand binding assay (124). Schobert, Berhardt, and Hammond have collaborated to develop cisplatin conjugates of estradiol and various other steroidal alcohols and have shown that these conjugates bind sex hormone binding globulin (SHBG), a transport protein for estrogens and androgens, and the nuclear estrogen receptor (ERα) (125). Curiously, the complex that displayed the greatest cytotoxicity failed to distort plasmid DNA in a gel shift assay, suggesting an unexpected mechanism of action. Lippard’s laboratory has also developed a similar complex with platinum (IV) metal centers and demonstrated enhanced cytotoxicity against the MCF-7 line (126). This approach represents an additional level of sophistication, as the platinum (IV) metal center is reduced to platinum (II) within the cell, leading to dissociation of the axial ligands, shedding of the covalent linkage, and intracellular release of unmodified cisplatin. This eliminates any possibility of interference with the mechanism of the drug’s action.
In addition to biomolecules, several groups have used synthetic polymers as drug vectors, either to improve pharmacological properties at the systemic level, or to improve uptake at the cellular level. Kataoka’s laboratory at the University of Tokyo has led the development of cisplatin-incorporated polymeric micelles, with microarray expression profiling suggesting that the conjugates would have additional therapeutic benefits over free cisplatin (127, 128). Lippard’s laboratory has tethered their platinum (IV) prodrug analogues of cisplatin to single-walled carbon nanotubes (SWNT) (129). Confocal microscopy experiments with fluorescently labeled conjugates showed punctate staining, consistent with cellular entry through endocytosis. Furthermore, AAS confirmed the release of free cisplatin throughout the cell. Intracellular concentration of the conjugates was increased six-fold with respect to the unconjugated platinum complexes, and the conjugates displayed enhanced toxicity versus cisplatin in the NTera-2 cell line, even after normalization of platinum concentrations for the polynuclear conjugates. Lin’s laboratory has developed nanoscale coordination polymers of platinum (IV) prodrugs encased in silica to provide a controlled release of cisplatin; cytotoxicity in HT-29 cells was similar to that of cisplatin alone (130). Jaehde and coworkers attempted to combat cisplatin resistance in A2780 ovarian carcinoma cells, which exhibit reduced cellular uptake, by conjugating cisplatin to albumin and to PEG (131).
Like any design effort, the rational development of bioconjugates is met by risk and opportunity. Active uptake has the promise to increase intracellular concentrations dramatically and to provide a basis of cellular specificity, but to remain effective, the conjugate must escape endosomal trapping. Nuclear localization signals may be able to direct conjugates more efficiently to their targets, but the consequences of conjugation may alter the functionality of the active moiety. Even seemingly innocent structural changes, such as the length of the linker region or the inclusion of a fluorophore can have dramatic effects on uptake and localization that must be considered in order to deliver active complexes successfully to their targets.
Conclusions
A variety of methods are useful in studying the cellular accumulation of metal complexes. Flow cytometry and confocal microscopy are complementary methods appropriate for luminescent complexes, whereas ICP-MS or AAS must be used to study the overall uptake of non-luminescent complexes. Metal complexes can enter cells by passive diffusion, with the help of transport proteins, or by endocytosis. Experiments with metabolic inhibitors can differentiate between active and passive uptake mechanisms, while channel inhibitors can differentiate between passive and facilitated diffusion. Identification of the uptake mechanism can greatly aid in the development of new diagnostic and therapeutic applications. Importantly, studies of cellular accumulation should not neglect the issue of cellular efflux, which can render a diagnostic agent useless or lead to clinical resistance in the therapeutic case.
Although a balance of uptake and efflux that leads to cellular accumulation is necessary for therapeutic activity, it is not sufficient. Metal complexes must also be able to reach their target within the cell, and presumably for DNA-targeted complexes, within the nucleus. Several conjugation strategies have been developed to improve the cellular accumulation and nuclear targeting of metal-based agents; peptides, B vitamins, carbohydrates, and hormones have all been covalently attached to metal moieties, most notably cisplatin and its derivatives. These strategies also introduce new challenges. For example, the covalent linkage must not interfere with the mechanism of activity. The most sophisticated designs are metabolized within the cell to release an active subunit. While this approach offers promise, new strategies will be needed to control specific organelle targeting. Nevertheless, transition metal complexes offer rich opportunities in the design of completely new diagnostics and therapeutics, and thus various routes to cell entry and organelle delivery need to be explored and exploited for a new generation of versatile design.
Acknowledgments
† Financial support for this work from the National Institutes of Health (Grant GM33309 to J.K.B.) is acknowledged.
References
- 1.Erkkila KE, Odom DT, Barton JK. Chem. Rev. 1999;99:2777–2796. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Lippard SJ. Nat. Rev. Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 3.Zeglis BM, Pierre VC, Barton JK. Chem. Commun. 2007:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruijnincx PCA, Sadler PJ. Curr. Opin. Chem. Biol. 2008;12:197–206. doi: 10.1016/j.cbpa.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakupec MA, Galanski M, Arion VB, Hartinger CG, Keppler BH. Dalton Trans. 2008:183–194. doi: 10.1039/b712656p. [DOI] [PubMed] [Google Scholar]
- 6.Montgomery P, Murray BS, New EJ, Pal R, Parker D. Acc. Chem. Res. 2009;42:925–937. doi: 10.1021/ar800174z. [DOI] [PubMed] [Google Scholar]
- 7.Jiménez-Hernández ME, Orellana G, Montero F, Portolés MT. Photochem. Photobiol. 2000;72:28–34. doi: 10.1562/0031-8655(2000)072<0028:arpfcv>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Puckett CA, Barton JK. J. Am. Chem. Soc. 2007;129:46–47. doi: 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egger E, Rappel C, Jakupec MA, Hartinger CG, Heffeter P, Keppler BK. J. Anal. At. Spectrom. 2009;24:51–61. doi: 10.1039/B810481F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghezzi, Aceto M, Cassino C, Gabano E, Osella D. J. Inorg. Biochem. 2004;98:73–78. doi: 10.1016/j.jinorgbio.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Kirin SI, Ott I, Gust R, Mier W, Weyhermüller T, Metzler-Nolte N. Angew. Chem. Int. Ed. 2008;47:955–959. doi: 10.1002/anie.200703994. [DOI] [PubMed] [Google Scholar]
- 12.Jonas SK, Riley PA. Cell Biochem. Funct. 1991;9:245–253. doi: 10.1002/cbf.290090406. [DOI] [PubMed] [Google Scholar]
- 13.Davies D, Hughes C. Dead cell discrimination. In: Diamond RA, DeMaggio S S, editors. In Living Color: Protocols in Flow Cytometry and Cell Sorting. 1st edition. Berlin: Springer; 2000. [Google Scholar]
- 14.Liu JJ, Galettis P, Farr A, Maharaj L, Samarasinha H, McGechan AC, Baguley BC, Bowen RJ, Berners-Price SJ, McKeage MJ. J. Inorg. Biochem. 2008;102:303–310. doi: 10.1016/j.jinorgbio.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Hotze ACG, Hodges NJ, Hayden RE, Sanchez-Cano C, Paines C, Male N, Tse M-K, Bunce CM, Chipman JK, Hannon MJ. Chemistry and Biology. 2008;15:1258–1267. doi: 10.1016/j.chembiol.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 17.Hällbrink M, Oehlke J, Papsdorf G, Bienert M. Biochim. Biophys. Acta. 2004;1667:222–228. doi: 10.1016/j.bbamem.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Barban S, Schulze H. J. Biol. Chem. 1961;236:1887–1890. [PubMed] [Google Scholar]
- 19.Slater C. Methods Enzymol. 1967;10:48–57. [Google Scholar]
- 20.Ohtani Y, Irie T, Uekama K, Fukunaga K, Pitha J. Eur. J. Biochem. 1989;186:17–22. doi: 10.1111/j.1432-1033.1989.tb15171.x. [DOI] [PubMed] [Google Scholar]
- 21.Kilsdonk EPC, Yancey PG, Stoudt GW, Wen Bangerter F, Johnson WJ, Phillips MC, Rothblat GH. J. Biol. Chem. 1995;270:17250–17256. doi: 10.1074/jbc.270.29.17250. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro HM. Methods. 2000;21:271–279. doi: 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
- 23.Tanner MK, Wellhausen SR. Flow cytometric detection of fluorescent redistributional dyes for measurement of cell transmembrane potential. In: Jaroszeski MJ, Heller R, editors. Methods in Molecular Biology, vol 91: Flow cytometry protocols. Totowa, NJ: Humana Press Inc; 1997. [DOI] [PubMed] [Google Scholar]
- 24.Koepsell H, Lips K, Volk C. Pharmaceutical Research. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 25.Rubio-Aliaga, Daniel H. Trends Pharmacol. Sci. 2002;23:434–440. doi: 10.1016/s0165-6147(02)02072-2. [DOI] [PubMed] [Google Scholar]
- 26.Doherty GJ, McMahon HT. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 27.Conner SD, Schmid SL. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov AI, editor. Exocytosis and Endocytosis. Totowa, NJ: Humana Press; 2008. Ivanov. Pharmacological inhibition of endocytic pathways: Is it specific enough to be useful? [DOI] [PubMed] [Google Scholar]
- 29.Rodal SK, Skretting G, Garred Ø, Vilhardt F, van Deurs B, Sandvig K. Mol. Biol. Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimmer, van Deurs B, Sandvig K. J. Cell Sci. 2002;115:2953–2962. doi: 10.1242/jcs.115.14.2953. [DOI] [PubMed] [Google Scholar]
- 31.Smart J, Anderson RG. Methods Enzymol. 2002;353:131–139. doi: 10.1016/s0076-6879(02)53043-3. [DOI] [PubMed] [Google Scholar]
- 32.Ros-Baró, López-Iglesias C, Peiró S, Bellido D, Palacín M, Zorzano A, Camps M. Proc. Natl. Acad. Sci. USA. 2001;98:12050–12055. doi: 10.1073/pnas.211341698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov, Nusrat A, Parkos CA. Mol. Biol. Cell. 2004;15:176–188. doi: 10.1091/mbc.E03-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girao H, Geli M-I, Idrissi F-Z. FEBS Letters. 2008;582:2112–2119. doi: 10.1016/j.febslet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Jess TJ, Belham CM, Thomson FJ. Cell Signal. 1996;8:297–304. doi: 10.1016/0898-6568(96)00054-x. [DOI] [PubMed] [Google Scholar]
- 36.Amyere M, Payrastre B, Krause U, Van Der Smissen P, Veithen V, Courtoy PJ. Mol. Biol. Cell. 2000;11:3453–3467. doi: 10.1091/mbc.11.10.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West MA, Bretscher MS, Watts C. J. Cell Biol. 1989;109:2731–2739. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamaze, Schmid SL. Curr. Opin. Cell Biol. 1995;7:573–580. doi: 10.1016/0955-0674(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 39.Keller HU. J. Cell. Physiol. 1990;145:465–471. doi: 10.1002/jcp.1041450311. [DOI] [PubMed] [Google Scholar]
- 40.Vassilieva EV, Nusrat A. Vesicular trafficking: molecular tools and targets. In: Ivanov AI, Walker JM, editors. Exocytosis and Endocytosis. Totowa, NJ: Humana Press; 2008. [DOI] [PubMed] [Google Scholar]
- 41.Hinshaw JE. Annu. Rev. Cell Dev. Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Damke H, Gossen M, Freundlieb S, Bujard H, Schmid SL. Methods Enzymol. 1995;257:209–220. doi: 10.1016/s0076-6879(95)57026-8. [DOI] [PubMed] [Google Scholar]
- 43.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dev. Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Haugland RP. In: The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. 10th ed. Spence MTZ, editor. Eugene, OR: Molecular Probes; 2005. [Google Scholar]
- 45.Ghosh RN, Gelman DL, Maxfield FR. J Cell Sci. 1994;107:2177–2189. doi: 10.1242/jcs.107.8.2177. [DOI] [PubMed] [Google Scholar]
- 46.Lodish H, Berk A, Zipursky LS, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. 4th edition. New York: W. H. Freeman; 2000. [Google Scholar]
- 47.Orlandi PA, Fishman PH. J. Cell Biol. 1998;141:905–915. doi: 10.1083/jcb.141.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torgersen ML, Skretting G, van Deurs B, Sandvig K. J. Cell Sci. 2001;114:3737–3747. doi: 10.1242/jcs.114.20.3737. [DOI] [PubMed] [Google Scholar]
- 49.Nichols J. Nat. Cell Biol. 2002;4:374–378. doi: 10.1038/ncb787. [DOI] [PubMed] [Google Scholar]
- 50.Kirkham M, Fujita A, Chadda R, Nixon SJ, Kurzchalia TV, Sharma DK, Pagano RE, Hancock JF, Mayor S, Parton RG. J. Cell Biol. 2005;168:465–476. doi: 10.1083/jcb.200407078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng Z-J, Singh RD, Sharma DK, Holicky EL, Hanada K, Marks DL, Pagano RE. Mol. Biol. Cell. 2006;17:3197–3210. doi: 10.1091/mbc.E05-12-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. Annu. Rev. Pharmacol. Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 53.New J, Parker D. Org. Biomol. Chem. 2009;7:851–855. doi: 10.1039/b822145f. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Lo KK-W. Inorg. Chem. 2009;48:6011–6025. doi: 10.1021/ic900412n. [DOI] [PubMed] [Google Scholar]
- 55.Hartinger G, Zorbas-Seifried S, Jakupec MA, Kynast B, Zorbas H, Keppler BK. J. Inorg. Biochem. 2006;100:891–904. doi: 10.1016/j.jinorgbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 56.Piwnica-Worms, Kronauge JF, Chiu ML. Circulation. 1990;82:1826–1838. doi: 10.1161/01.cir.82.5.1826. [DOI] [PubMed] [Google Scholar]
- 57.Puckett CA, Barton JK. Biochemistry. 2008;47:11711–11716. doi: 10.1021/bi800856t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nigg EA. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 59.Poole R, Bobba G, Cann M, Frias J, Parker D, Peacock R. Org. Biomol. Chem. 2005;3:1013–1024. doi: 10.1039/b418964g. [DOI] [PubMed] [Google Scholar]
- 60.Pandya S, Yu J, Parker D. Dalton Trans. 2006:2757–2766. doi: 10.1039/b514637b. [DOI] [PubMed] [Google Scholar]
- 61.Yu J, Parker D, Pal R, Poole RA, Cann MJ. J. Am. Chem. Soc. 2006;128:2294–2299. doi: 10.1021/ja056303g. [DOI] [PubMed] [Google Scholar]
- 62.Cowley AR, Davis J, Dilworth JR, Donnelly PS, Dobson R, Nightingale A, Peach JM, Shore B, Kerr D, Seymour L. Chem. Commun. 2005:845–847. doi: 10.1039/b417206j. [DOI] [PubMed] [Google Scholar]
- 63.Puckett CA, Barton JK. J. Am. Chem. Soc. 2009;131:8738–8739. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ernst RJ, Song H, Barton JK. J. Am. Chem. Soc. 2009;131:2359–2366. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Napolitano SM, Aprille JR. Adv. Drug Delivery Rev. 2001;49:63–70. doi: 10.1016/s0169-409x(01)00125-9. [DOI] [PubMed] [Google Scholar]
- 66.Murphy MP, Smith RAJ. Adv. Drug Delivery Rev. 2000;41:235–250. doi: 10.1016/s0169-409x(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 67.Rackham O, Nichols SJ, Leedman PJ, Berners-Price SJ, Filipovska A. Biochem. Pharmacol. 2007;74:992–1002. doi: 10.1016/j.bcp.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 68.Liu JJ, Galettis P, Farr A, Maharaj L, Samarasinha H, McGechan AC, Baguley BC, Bowen RJ, Berners-Price SJ, McKeage MJ. J. Inorg. Biochem. 2008;102:303–310. doi: 10.1016/j.jinorgbio.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 69.Liberman EA, Topali VP, Tsofina LM, Jasaitis AA, Skulachev VP. Nature. 1969;222:1076–1078. doi: 10.1038/2221076a0. [DOI] [PubMed] [Google Scholar]
- 70.Johnson LV, Walsh ML, Chen LB. Proc. Natl. Acad. Sci. U. S. A. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis S, Weiss MJ, Wong JR, Lampidis TJ, Chen LB. J. Biol. Chem. 1985;260:13844–13850. [PubMed] [Google Scholar]
- 72.Dairkee SH, Hackett AJ. Breast Cancer Res. Treat. 1991;18:57–61. doi: 10.1007/BF01975444. [DOI] [PubMed] [Google Scholar]
- 73.Burns RJ, Smith RAJ, Murphy MP. Arch. Biochem. Biophys. 1995;322:60–68. doi: 10.1006/abbi.1995.1436. [DOI] [PubMed] [Google Scholar]
- 74.Burns RJ, Murphy MP. Arch. Biochem. Biophys. 1997;339:33–39. doi: 10.1006/abbi.1996.9861. [DOI] [PubMed] [Google Scholar]
- 75.Smith RAJ, Porteous CM, Coulter CV, Murphy MP. Eur. J. Biochem. 1999;263:709–716. doi: 10.1046/j.1432-1327.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- 76.Eckford PDW, Sharom FJ. Chem. Rev. 2009;109:2989–3011. doi: 10.1021/cr9000226. [DOI] [PubMed] [Google Scholar]
- 77.Borst P, Evers R, Kool M, Wijnholds J. J. Nat. Cancer Inst. 2000;92:1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 78.Kruh GD, Belinsky MG. Oncogene. 2003;22:7537–7552. doi: 10.1038/sj.onc.1206953. [DOI] [PubMed] [Google Scholar]
- 79.Krishnamurthy P, Schuetz JD. Annu. Rev. Pharmacol. Toxicol. 2006;46:381–410. doi: 10.1146/annurev.pharmtox.46.120604.141238. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, Bachmeier C, Miller DW. Adv. Drug Del. Rev. 2003;55:31–51. doi: 10.1016/s0169-409x(02)00170-9. [DOI] [PubMed] [Google Scholar]
- 81.Szakacs, Varadi A, Ozvegy-Laczka C, Sarkadi B. Drug Disc. Today. 2008;13:379–393. doi: 10.1016/j.drudis.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 82.Matsson P, Pedersen JM, Norinder U, Bergström CAS, Artursson P. Pharm. Res. 2009;26:1816–1831. doi: 10.1007/s11095-009-9896-0. [DOI] [PubMed] [Google Scholar]
- 83.Sarkadi, Price EM, Boucher RC, Germann UA, Scarborough GA. J. Biol. Chem. 1992;267:4854–4858. [PubMed] [Google Scholar]
- 84.Drueckes P, Schinzel R, Palm D. Anal. Biochem. 1995;230:173–177. doi: 10.1006/abio.1995.1453. [DOI] [PubMed] [Google Scholar]
- 85.Piwnica-Worms, Chiu ML, Budding M, Kronauge JF, Kramer RA, Croop JM. Cancer Res. 1993;53:977–984. [PubMed] [Google Scholar]
- 86.Hendrikse N, Franssen E, Van der Graaf W, Meijer C, Piers D, Vaalbrug W, De Vries E. Br. J. Cancer. 1998;77:353–358. doi: 10.1038/bjc.1998.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moretti J-L, Cordobes M, Starzec A, de Beco V, Vergote J, Benazzouz F, Boissier B, Cohen H, Safi N, Piperno-Neumann S, Kouyoumdjian J-C. J. Nucl. Med. 1998;39:1214–1218. [PubMed] [Google Scholar]
- 88.Sharma V, Piwnica-Worms D. Top. Curr. Chem. 2005;252:155–178. [Google Scholar]
- 89.Kuo MT, Chen HHW, Song I-S, Savaraj N, Ishikawa T. Cancer Metastatis Rev. 2007;26:71–83. doi: 10.1007/s10555-007-9045-3. [DOI] [PubMed] [Google Scholar]
- 90.Ishikawa T, Ali-Osman F. J. Biol. Chem. 1993;268:20116–20125. [PubMed] [Google Scholar]
- 91.Piwnica-Worms, Rao VV, Kronauge JF, Croop JM. Biochemistry. 1995;34:12210–12220. doi: 10.1021/bi00038a015. [DOI] [PubMed] [Google Scholar]
- 92.Müller H, Klinkhammer W, Globisch C, Kassack MU, Pajeva IK, Wiese M. Bioorg. Med. Chem. 2007;15:7470–7479. doi: 10.1016/j.bmc.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 93.Heffeter P, Pongratz M, Steiner E, Chiba P, Jakupec MA, Elbling L, Marian B, Körner W, Sevelda F, Micksche M, Keppler BK, Berger W. J. Pharmacol. Exp. Ther. 2005;312:281–289. doi: 10.1124/jpet.104.073395. [DOI] [PubMed] [Google Scholar]
- 94.Aird RE, Cummings J, Ritchie AA, Muir M, Morris RE, Chen H, Sadler PJ, Jodrell DI. Br. J. Cancer. 2002;86:1652–1657. doi: 10.1038/sj.bjc.6600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mukhopadhyay S, Barnés CM, Haskel A, Short SM, Barnes KR, Lippard SJ. Bioconjugate Chem. 2008;19:39–49. doi: 10.1021/bc070031k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kirin SI, Dübon P, Weyhermüller T, Bill E, Metzler-Nolte N. Inorg. Chem. 2005;44:5405–5415. doi: 10.1021/ic048343b. [DOI] [PubMed] [Google Scholar]
- 97.Gasser G, Neukamm MA, Ewers A, Brosch O, Weyhermüller T, Metzler-Nolte N. Inorg. Chem. 2009;48:3157–3166. doi: 10.1021/ic900013r. [DOI] [PubMed] [Google Scholar]
- 98.Chantson J, Varga Falzacappa MV, Crovella S, Metzler-Nolte N. ChemMedChem. 2006;1:1268–1274. doi: 10.1002/cmdc.200600117. [DOI] [PubMed] [Google Scholar]
- 99.Chantson J, Varga Falzacappa MV, Crovella S, Metzler-Nolte N. J. Organomet. Chem. 2005;690:4564–4572. [Google Scholar]
- 100.Horton KL, Stewart KM, Fonseca SB, Guo Q, Kelley SO. Chemistry & Biology. 2008;15:375–382. doi: 10.1016/j.chembiol.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 101.Kalderon D, Roberts BL, Richardson WD, Smith AE. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 102.Dang CV, Lee WM. J. Biol. Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- 103.Siomi H, Shida H, Maki M, Hatanaka M. J. of Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pouton CW. Adv. Drug Del. Rev. 1998;34:51–64. doi: 10.1016/s0169-409x(98)00050-7. [DOI] [PubMed] [Google Scholar]
- 105.Stewart KM, Horton KL, Kelley SO. Org. Biomol. Chem. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- 106.Sibrian-Vazquez M, Ortiz J, Nesterova IV, Fernández-Lázaro F, Sastre-Santos A, Soper SA, Vicente MGH. Bioconjugate Chem. 2007;18:410–420. doi: 10.1021/bc060297b. [DOI] [PubMed] [Google Scholar]
- 107.Aronov O, Horowitz AT, Gabizon A, Fuertes MA, Pérez JM, Gibson D. Bioconjugate Chem. 2004;15:814–823. doi: 10.1021/bc0499331. [DOI] [PubMed] [Google Scholar]
- 108.Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 109.Kirin SI, Ott I, Gust R, Mier W, Weyhermüller T, Metzler-Nolte N. Angewandte Chemie. 2008;47:955–959. doi: 10.1002/anie.200703994. [DOI] [PubMed] [Google Scholar]
- 110.Noor F, Wüstholz A, Kinscherf R, Metzler-Nolte N. Angew. Chem. Int. Ed. 2005;44:2429–2432. doi: 10.1002/anie.200462519. [DOI] [PubMed] [Google Scholar]
- 111.Noor F, Kinscherf R, Bonaterra GA, Walczak S, Wölfl S, Metzler-Nolte N. Chembiochem. 2009;10:493–502. doi: 10.1002/cbic.200800469. [DOI] [PubMed] [Google Scholar]
- 112.Neukamm MA, Pinto A, Metzler-Nolte N. Chem. Commun. 2008:232–234. doi: 10.1039/b712886j. [DOI] [PubMed] [Google Scholar]
- 113.Haefliger P, Agorastos N, Renard A, Giambonini-Brugnoli G, Marty C, Alberto R. Bioconjugate Chemistry. 2005;16:582–587. doi: 10.1021/bc0500084. [DOI] [PubMed] [Google Scholar]
- 114.Heckl S, Vogel U. J. Pharmacol. Exp. Ther. 2006;319:657–662. doi: 10.1124/jpet.106.109868. [DOI] [PubMed] [Google Scholar]
- 115.Polyakov V, Sharma V, Dahlheimer JL, Pica CM, Luker GD, Piwnica-Worms D. Bioconjugate Chem. 2000;11:762–771. doi: 10.1021/bc000008y. [DOI] [PubMed] [Google Scholar]
- 116.Bullok KE, Dyszlewski M, Prior JL, Pica CM, Sharma V, Piwnica-Worms D. Bioconjugate Chem. 2002;13:1226–1237. doi: 10.1021/bc025573a. [DOI] [PubMed] [Google Scholar]
- 117.Gammon ST, Villalobos VM, Prior JL, Sharma V, Piwnica-Worms D. Bioconjugate Chem. 2003;14:368–376. doi: 10.1021/bc0256291. [DOI] [PubMed] [Google Scholar]
- 118.Ruiz-Sánchez P, Mundwiler S, Spingler B, Buan NR, Escalante-Semerena JC, Alberto R. J. Biol. Inorg. Chem. 2008;13:335–347. doi: 10.1007/s00775-007-0329-4. [DOI] [PubMed] [Google Scholar]
- 119.Ohya Y, Oue H, Nagatomi K, Ouchi T. Biomacromolecules. 2001;2:927–933. doi: 10.1021/bm010053o. [DOI] [PubMed] [Google Scholar]
- 120.Ohya Y, Nagatomi K, Ouchi T. Macromol. Biosci. 2001;1:355–363. [Google Scholar]
- 121.Ichinose K, Tomiyama N, Nakashima M, Ohya Y, Ichikawa M, Ouchi T, Kanematsu T. Anticancer Drugs. 2000;11:33–38. doi: 10.1097/00001813-200001000-00006. [DOI] [PubMed] [Google Scholar]
- 122.Janáky T, Juhász A, Rékási Z, Serfõzõ P, Pinski J, Bokser L, Srkalovic G, Milovanovic S, Redding TW, Halmos G, Nagy A, Schally AV. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10203–10207. doi: 10.1073/pnas.89.21.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jackson, Davis J, Pither RJ, Rodger A, Hannon MJ. Inorg. Chem. 2001;40:3964–3973. doi: 10.1021/ic010152a. [DOI] [PubMed] [Google Scholar]
- 124.Hannon MJ, Green PS, Fisher DM, Derrick PJ, Beck JL, Watt SJ, Ralph SF, Sheil MM, Barker PR, Alcock NW, Price RJ, Sanders KJ, Pither R, Davis J, Rodger A. Chemistry. 2006;12:8000–8013. doi: 10.1002/chem.200501012. [DOI] [PubMed] [Google Scholar]
- 125.Schobert R, Bernhardt G, Biersack B, Bollwein S, Fallahi M, Grotemeier A, Hammond GL. Chem. Med. Chem. 2007;2:333–342. doi: 10.1002/cmdc.200600173. [DOI] [PubMed] [Google Scholar]
- 126.Barnes KR, Kutikov A, Lippard SJ. Chem. Biol. 2004;11:557–564. doi: 10.1016/j.chembiol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 127.Nishiyama N, Okazaki S, Cabral H, Miyamoto M, Kato Y, Sugiyama Y, Nishio K, Matsumura Y, Kataoka K. Cancer Research. 2003;63:8977–8983. [PubMed] [Google Scholar]
- 128.Nishiyama N, Koizumi F, Okazaki S, Matsumura Y, Nishio K, Kataoka K. Bioconjugate Chem. 2003;14:449–457. doi: 10.1021/bc025555t. [DOI] [PubMed] [Google Scholar]
- 129.Feazell RP, Nakayama-Ratchford N, Dai H, Lippard SJ. J. Am. Chem. Soc. 2007;129:8438–8439. doi: 10.1021/ja073231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rieter WJ, Pott KM, Taylor KML, Lin W. J. Am. Chem. Soc. 2008;130:11584–11585. doi: 10.1021/ja803383k. [DOI] [PubMed] [Google Scholar]
- 131.Garmann D, Warnecke A, Kalayda GV, Kratz F, Jaehde U. J. Controlled Release. 2008;131:100–106. doi: 10.1016/j.jconrel.2008.07.017. [DOI] [PubMed] [Google Scholar]