Abstract
The photo-crosslinking amino acid, p-benzoyl-L-phenylalanine (pBpa), was genetically incorporated into E. coli catabolite activator protein (CAP) in bacteria in response to an amber nonsense codon using an orthogonal tRNA/aminoacyl-tRNA synthetase pair. The mutant CAP (CAP-K26Bpa) containing pBpa formed a covalent complex with a DNA fragment containing the consensus operator sequence upon UV irradiation. Because this amino acid can be genetically incorporated into any DNA-binding protein in E. coli, yeast or mammalian cells with minimal perturbation of protein structure, this method should be generally useful for investigating DNA-protein interactions.
Sequence-specific DNA binding proteins control a large number of cellular processes including replication, transcription, epigenetic modifications and DNA repair. Photochemical crosslinking methods have long been used to identify and characterize protein-DNA interactions, and can be carried out by introducing a photo-reactive group into the DNA or the protein.1-4 In the former case, one can use either solid phase synthesis or DNA polymerases to incorporate a photo-reactive moiety, but one must typically know the DNA binding site for the protein of interest. Alternatively, photocrosslinkers can be introduced into DNA binding proteins by chemical modification or semisynthesis, or by in vitro protein translation systems using chemically aminoacylated tRNAs. Recently, we developed a method that allows one to genetically incorporate unnatural amino acids, including those with chemically and photo-chemically reactive groups, directly into proteins in both prokaryotic or eukaryotic cells in response to an amber nonsense codon (TAG).5-7 Here, we demonstrate that this method can be used to site-specifically introduce a photo-reactive probe into a DNA binding protein which upon irradiation forms a stable complex with the cognate DNA-binding site in a sequence-specific fashion.
Benzophenones have been used extensively as photophysical probes to identify and map interactions between biomolecules. They are chemically more stable than other photo-crosslinking groups such as diazo esters, aryl azides and diazirines; insert relatively nonspecifically into C—H bonds upon irradiation at < 365 nm (hydrogen abstraction by oxygen in the excited state followed by carbon-centered radical recombination); and efficiently repopulate the ground state in the absence of photo-reaction.8 Recently, the photo-crosslinking amino acid, p-benzoyl-L-phenylalanine (pBpa, Fig. 1A) was genetically encoded by means of orthogonal tRNA/aminoacyl-tRNA synthetase (aaRS) pairs evolved in E. coli and yeast, and used to investigate protein-protein interactions both in vitro and in living cells.5,9-13 The evolved tRNA/aaRS pairs site-specifically incorporate pBpa into proteins at sites encoded by the amber nonsense codon, and the pBpa-containing proteins efficiently form stable complexes with their partners upon UV irradiation. Similarly, we expected that the genetic incorporation of pBpa into nucleic acid-binding proteins would enable them to selectively form covalent complexes with their cognate DNA or RNA binding site upon irradiation.
Figure 1.
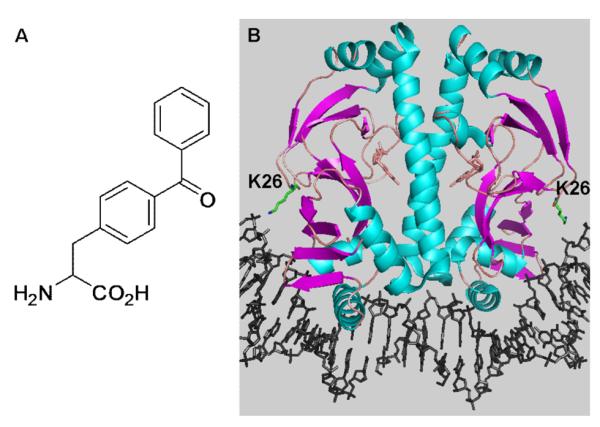
(A) Chemical structure of pBpa. (B) Crystal structure of the CAP-DNA complex showing the position of Lys26.
To demonstrate the feasibility of this approach, pBpa was incorporated into the E. coli catabolite activator protein (CAP). CAP is a well-characterized transcriptional activator which regulates a number of catabolite-sensitive operons in E. coli.14 CAP functions as a homodimer by binding a 2-fold symmetric 22-bp consensus binding site located near CAP-dependent promoters in the presence of the allosteric effector, cAMP.14 Based on the crystal structure of the CAP-DNA complex,15,16 Lys26 was substituted by pBpa. Lys26 comes into close contact with DNA when CAP binds its operator site as a result of an approximate 90° bend induced in the double helix (Fig. 1B).15,17 Earlier studies showed that replacement of Lys26 in CAP with metal-chelating groups afforded a selective affinity cleaving agent with minimal perturbation of DNA binding affinity.17,18
A CAP derivative containing a C-terminal strep-tag (WSHPQFEK) was generated by amplifying the CAP gene by PCR with primers containing the strep-tag sequence and inserting it into the pBAD vector. To incorporate pBpa, the codon corresponding to Lys26 was mutated to the amber codon (TAG) by site-directed mutagenesis. Expression of the mutant CAP was carried out in the presence of an evolved Methanococcus jannashii tRNA/aaRS pair5 and 1 mM pBpa in E. coli strain DH10B grown in glycerol minimum medium according to the previously reported protocol.18 Wildtype (WT) and mutant proteins were isolated by strep-tag affinity purification19, and the yield of the mutant CAP (CAP-K26Bpa) was 3–5 mg/L (WT CAP yield was 6–10 mg/L). SDS-PAGE analysis of the purified protein showed a band for the amber mutant in the presence of 1 mM pBpa whose size approximated that of WT CAP; in contrast no full length protein was detected in the absence of pBpa (Fig. 2A). ESI-mass spectrometric analysis of CAP-K26Bpa gave an observed average mass of 25284 Da, in close agreement with the calculated mass of 25283 Da (we also observed a peak corresponding to the mass of the acetylated protein) (Fig. 2B). These results confirm the efficient and selective incorporation of pBpa into CAP.
Figure 2.
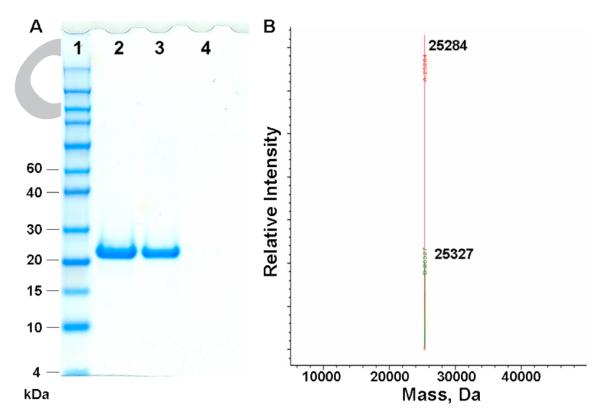
(A) SDS-PAGE analysis of CAP expression after His6-tag affinity purification. Lane 1, protein marker; lane 2, WT CAP; lane 3, CAP-K26Bpa expressed in the presence of 1 mM pBpa; lane 4, CAP-K26Bpa expressed in the absence of pBpa. (B) ESI-mass spectrometric analysis of CAP-K26Bpa after His6-tag affinity purification. Full-length protein (−Met) 25284 (calculated 25283); N-terminal acetylated protein 25327 (calculated 25325).
The dissociation constant for the binding of the CAP-K26Bpa mutant to its operator site was measured to determine whether incorporation of pBpa at position 26 affects binding affinity. Gel-retardation assays were carried out with a 50-bp 5′-32P-end labeled double-stranded DNA fragment containing the 22-bp DNA recognition site for CAP.18 The dissociation constants for WT CAP and CAP-K26Bpa were determined to be 1.9 nM and 3.0 nM, respectively (Fig. 3) indicating that the pBpa group does not significantly perturb the interaction of CAP with its operator sequence.
Figure 3.
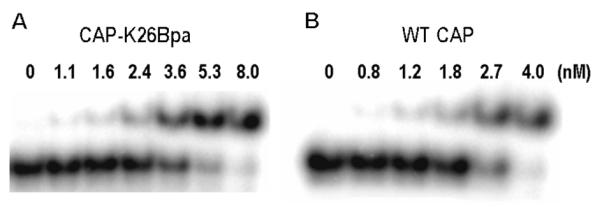
Gel-retardation assay of CAP-K26Bpa (A) and WT CAP (B) with 50-bp 5′-32P-end labeled DNA fragment. Conditions: 2 nM DNA, CAP, 50 μg/mL BSA, 0.2 mM cAMP, 10 mM Mops (pH 7.3), 200 mM NaCl, 10 μL total volume, 37 °C, 10 min.
Next, photo-crosslinking of CAP-K26Bpa with DNA was performed using the same 50-bp DNA fragment that was used in the gel-retardation assay. We also used a 50-bp DNA fragment with six mutations (AAATGTGATCTAGATCACATTT → AATTCTCATCTACATGACAATT) in the CAP-binding sequence (which abrogate DNA binding) in order to determine whether the crosslinking reaction is sequence-specific. After 10 min incubation of CAP with the DNA fragment in phosphate buffer (pH 8.0) at room temperature, samples were exposed to UV (300–320 nm) for 5 min20 and the crosslinked product was then analyzed.21 Denaturing SDS-PAGE analysis revealed one new band whose electrophoretic mobility is substantially less than that of the free DNA, as expected for the crosslinked adduct.22 No corresponding band was observed either for the DNA/CAP-K26Bpa complex in the absence of irradiation, or for irradiated samples containing the DNA/WT CAP complex (Fig. 4). In addition, the DNA fragment with six mutations did not give any crosslinked product with CAP-K26Bpa under the same conditions, demonstrating that the photo-crosslinking reaction is sequence-specific. Analysis of band intensities with Image Quant software indicated that approximately 5% of DNA was crosslinked by CAP-K26Bpa. This is ∼5 times more efficient than a previous experiment1 in which an azidophenyl group was chemically introduced at position 178 in CAP, resulting in ∼1% of a DNA-protein photo-adduct. The low efficiency may be due to the instability of CAP under the photolysis conditions (in the absence of DNA CAP photodecomposes as determined by mass spectral analysis), competitive trapping by molecular oxygen which is typically present in such experiments, or a nonideal crosslinking geometry.
Figure 4.

SDS-PAGE analysis of CAP-DNA photo-crosslinking reaction. The CAP-binding sequence is shown in red with underlines. DNA 2 has 6 mutations in the CAP-binding sequence. Conditions: 2 nM DNA, 50 nM CAP, 50 μg/mL BSA, 0.2 mM cAMP, 35 mM NaH2PO4 (pH 8.0), 200 mM NaCl, 10 μL total volume, room temperature, 5 min
In summary, we have site-specifically incorporated pBpa into the DNA-binding protein, CAP, by means of an evolved Methanococcus jannashii tRNA/aaRS pair. The mutant CAP containing pBpa was able to form a covalent complex with a DNA fragment containing the consensus operator sequence upon UV irradiation. Because this photo-crosslinking amino acid can be genetically incorporated into any nucleic acid-binding protein in prokaryotic or eukaryotic organisms, this method should be generally useful for mapping DNA- or RNA-protein interactions. The fact that simple mutagenesis can be used to rapidly introduce pBpa at multiple sites within a protein suggests that this method will also prove useful in the absence of detailed structural information. Future efforts will focus on the incorporation of cleavable photocrosslinking amino acids in order to further facilitate analysis of DNA-binding sites in native nucleic acids.
Acknowledgements
We are grateful to the NIH GM62159 and the Skaggs Institute for Chemical Biology for support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Pendergrast PS, Chen Y, Ebright YW, Ebright RH. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10287. doi: 10.1073/pnas.89.21.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shigdel UK, Zhang J, He C. Angew. Chem. Int. Ed. 2008;47:90. doi: 10.1002/anie.200703625. [DOI] [PubMed] [Google Scholar]
- 3.Pingoud V, Thole H, Christ F, Grindl W, Wende W, Pingoud A. J. Biol. Chem. 1999;274:10235. doi: 10.1074/jbc.274.15.10235. [DOI] [PubMed] [Google Scholar]
- 4.Meisenheimer KM, Koch TH. Crit. Rev. Bio-chem. Mol. Biol. 1997;32:101. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]
- 5.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Proc. Natl. Acad. Sci. USA. 2002;99:11020. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. J. Am. Chem. Soc. 2002;124:9026. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 7.Tippmann EM, Liu W, Summerer D, Mack AV, Schultz PG. ChemBioChem. 2007;8:2210. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- 8.Dorman G, Prestwich GD. Biochemistry. 1994;33:5661. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 9.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. Science. 2003;301:964. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 10.Chin JW, Schultz PG. ChemBioChem. 2002;3:1135. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Nat. Methods. 2005;2:201. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 12.Okuda S, Tokuda H. Proc. Natl. Acad. Sci. USA. 2009;106:5877. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori H, Ito K. Proc. Natl. Acad. Sci. USA. 2006;103:16159. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Crombrugghe B, Busby S, Buc H. Science. 1984;224:831. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 15.Schultz SC, Shields GC, Steitz TA. Science. 1991;253:1001. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson G, Wilson C, Gunaseker A, Ebright YW, Ebright RH. J. Mol. Biol. 1996;260:395. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 17.Pendergrast PS, Ebright YW, Ebright RH. Science. 1994;265:959. doi: 10.1126/science.8052855. [DOI] [PubMed] [Google Scholar]
- 18.Lee HS, Schultz PG. J. Am. Chem. Soc. 2008;130:13194. doi: 10.1021/ja804653f. [DOI] [PubMed] [Google Scholar]
- 19.Proteins were purified according to the manufacturer’s protocol (IBA).
- 20.Photo-crosslinking efficiency was best at 5 min of UV irradiation and longer exposure time did not increase the efficiency (data not shown).
- 21.Photo-crosslinking samples containing 35 mM NaH2PO4 (pH 8.0), 200 mM NaCl, 2 nM DNA, 50 nM CAP, 50 μg/mL BSA and 0.2 mM cAMP were incubated in PCR-tubes for 10 min at room temperature, followed by UV irradiation to the bottom of the tubes at room temperature using a UV transilluminator (300–320 nm). Samples were directly analyzed by SDS-PAGE without further purification.
- 22.Small amount of background crosslinking was observed with CAP-K26Bpa (lane 3 and 4).