The introduction of fluorescent probes into proteins provides a powerful tool to study protein structure and function either in vitro or in vivo.1 One particularly useful method involves genetically fusing the protein of interest to fluorescent proteins.2 However, the size of naturally occurring fluorescent proteins (>20 kDa) and the typical requirement for C- or N-terminal fusions limit their utility as a local probe of structure, and in many cases can perturb the structure or function of a protein. A variety of other methods have been developed to introduce fluorescent probes into protein in vitro and in vivo including the selective chemical3 or enzymetic4 modification of specific recognition motifs, the use of intein-based semisynthetic methods5 and in vitro protein translation6 with chemically aminoacylated tRNAs. Although very useful, each approach has its own limitations including protein size and yields, modifications to the native protein sequence, or reagents/protocols that are not compatible with live cells. To augment and extend these methods, we have attempted to genetically encode small, fluorescent amino acids directly in living cells.7 In this study, we report the site-specific incorporation of an environmentally sensitive fluorescent amino acid into proteins in response to the amber nonsense codon (TAG) with good yield and high fidelity in yeast. We demonstrate that the method can be used as a local probe of conformational changes in protein structure induced by ligand binding.
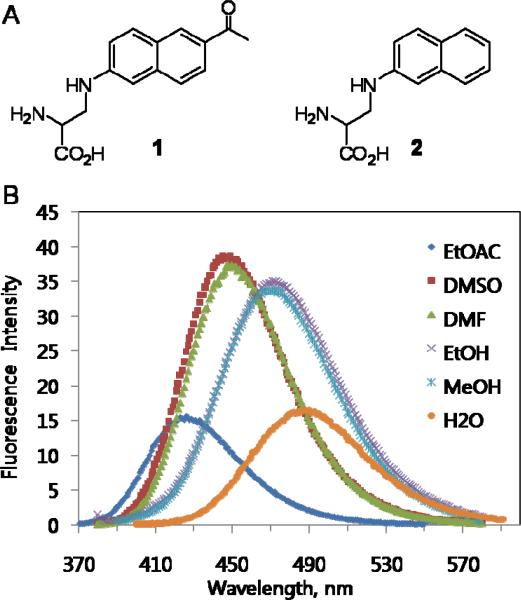
6-Propionyl-2-(N,N-dimethyl)aminonaphthalene, prodan8, is a highly environmentally sensitive fluorophore that is widely used in biochemistry and cell biology.9 To genetically encode the amino acid analog of prodan, 3-(6-acetylnaphthalen-2-ylamino)-2-aminopropanoic acid (Anap, 1), it was synthesized in six steps starting with 1-(6-hydroxynaphthalen-2-yl)ethanone (Figure S1). The extinction coefficient (17 500 cm−1 M−1) and quantum yield (0.48, Figure S2) of Anap in EtOH (360 nm) are comparable to those of prodan (18 400 cm−1M−1 and 0.71)8,10. The fluorescence spectra in various solvents also exhibit similar sensitivity to solvent polarity as other prodan probes with a significant shift in emission maximum on transitioning from water to ethyl acetate (Figure 1). Compared with dansyl- or coumarin-containing amino acids9, Anap has enhanced environmental sensitivity with comparable or increased brightness.9c,11
Figure 1.
(a) Chemical structures of Anap (1) and Nap (2). (b) Fluorescence spectra of 1 in various solvents; samples were excited at 350 nm.
To genetically encode this amino acid in S. cerevisiae, an orthogonal Escherichia coli amber suppressor tRNA/leucyl-tRNA synthetase pair12 was used on the basis of previous experiments7a,13 which showed that LeuRS mutants can accommodate unnatural amino acids with large hydrophobic side chains. To alter the specificity of LeuRS so that it aminoacylates with Anap and no endogenous amino acids, a LeuRS library was constructed by randomizing 5 residues (M40, L41, Y499, Y527 and H537) in the leucine-binding site.7a,12 The library was then subjected to rounds of alternating positive and negative selections as previously described14, but unfortunately no mutant LeuRS with the desired selectivity was isolated. We next attempted a stepwise strategy15 in which the same selection scheme was used first to evolve an aminoacyl-tRNA synthetase specific for the Anap analogue, 3-(naphthalen-2-ylamino)-2-aminopropanoic acid (Nap, 2). This amino acid lacks the acyl substituent of Anap, and therefore requires a lower degree of hydrogen bonding complementarity in the amino acid binding site of LeuRS. Five rounds of selection generated 92 clones that survived only in the presence of Nap in uracil dropout medium; DNA sequencing of 10 clones revealed three unique mutants (Table S1). The ability of one of these clones, Nap-1 (G40, P41, G499, A527, T537), to selectively incorporate Nap into proteins was tested by suppression of an amber mutation in C-terminal His6-tagged human superoxide dismutase (hSOD-33TAG) in the presence of Nap-1 and 1 mM Nap. SDS/PAGE analysis and MALDI-TOF MS showed selective incorporation of Nap with little detectable incorporation of endogenous yeast amino acids (Figure S3). The mutation of the Met, Leu and Tyr residues of wild-type LeuRS to small hydrophobic residues in Nap-1 is consistent with the generation of a larger binding pocket to accommodate the naphthyl side chain of 2.
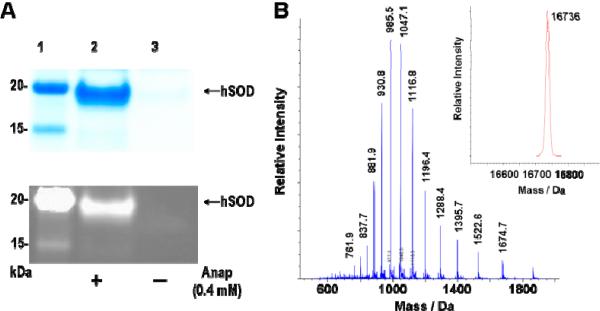
Next, a LeuRS library was generated based on the sequences of the LeuRS mutants evolved for Nap and the crystal structure of Thermus thermophilus LeuRS (which is homologous to E. coli LeuRS).16 Residues 40, 41, 499, 527 and 537 were fixed as Gly, Pro, Gly, Ala and Thr, respectively, and five additional residues (L38, Y500, L538, F541 and A560) in the leucine- binding site were randomized (Lib. 1, Table S2). Five rounds of selection for Anap-specific synthetases afforded 61 clones that survived only in the presence of Anap in uracil dropout medium; DNA sequencing of 10 clones revealed only one unique mutant (Anap-1, Table 1). This mutant LeuRS was used to express hSOD-33TAG in order to test its ability to selectively incorporate Anap into proteins. SDS/PAGE analysis and MALDI-TOF MS showed that the incorporation efficiency of Anap into hSOD-33TAG by the mutant LeuRS was poor, and that the LeuRS incorporated natural amino acids (Leu or Ile) to a significant degree (data not shown). To improve the efficiency and selectivity of Anap-1, another library of LeuRS mutants was generated by randomizing eight residues (L38, M40, Y499, Y500, H537, L538, F541, and A560) (Lib. 2, Table S2) based on the sequences of the LeuRS mutants from the previous selections. After five rounds of selection, three unique mutants with the desired phenotype were identified (Table 1) and the ability of one of these clones (Anap-2C: F38, G40, P41, V499, L500, A527, G537, S538, C541 and V560) to incorporate Anap into hSOD-33TAG was tested. SDS/PAGE analysis and ESI-MS showed that the mutant LeuRS incorporated Anap into the protein with good efficiency (1.2 mg/L in shake flasks) and selectivity (Figure 2).
Table 1.
Selected Anap-specific LeuRS mutants.
| LeuRS | frequency | Leu 38 | Met 40 | Leu 41 | Tyr 499 | Tyr 500 | Tyr 527 | His 537 | Leu 538 | Phe 541 | Ala 560 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anap-1 |
10/10 |
Phe |
Gly |
Pro |
Gly |
Tyr |
Ala |
Thr |
Cys |
Ser |
Val |
| Anap-2A | 5/10 | Phe | Gly | Pro | Val | His | Ala | Glu | Ser | Cys | Val |
| Aanp-2B | 3/10 | Phe | Gly | Pro | Ile | His | Ala | Glu | Ser | Cys | Val |
| Anap-2C | 2/10 | Phe | Gly | Pro | Val | Leu | Ala | Glu | Ser | Cys | Val |
Figure 2.
Expression of hSOD with an amber codon at position 33 in the presence of the pair and 0.4 mM Anap. (a) SDS-PAGE analysis by GelCode Blue straining (top) and fluorescence (bottom); Lane 1: protein marker; Lane 2: hSOD expressed in the presence of the pair and 0.4 mM Anap; Lane 3: hSOD expressed in the presence of the pair, but in the absence of Anap. (b) ESI-MS analysis. The insert shows the deconvoluted spectum, calculated 16738, observed 16736.
We next asked whether our ability to site-specifically incorporate Anap into proteins could be used to measure ligand-induced local conformational changes in proteins directly without the need for a FRET pair, using glutamine-binding proteinah (QBP) from E. coli. QBP is a monomeric protein composed of 224 amino acid residues that is responsible for the first step in the active transport of L-glutamine across the cytoplasmic membrane.17 QBP consists of two similar globular domains which are linked by two peptide hinges.17 The X-ray crystal structure of QBP shows that the deep cleft formed between the two domains contains the glutamine binding site and glutamine binding causes a significant conformational change.17 To incorporate Anap into QBP, an amber nonsense codon was substituted for Asn16018, which lies in the cleft between the two domains. Expression and purification of the amber mutant of QBP containing a C-terminal His6 tag (QBP-160TAG) was carried out in the presence of the evolved tRNA Leu CUA /Anap-2C pair and 0.4 mM Anap in S. cerevisiae using the reported method.19 The selective incorporation of Anap into QBP was confirmed by ESI-MS (Figure S4).
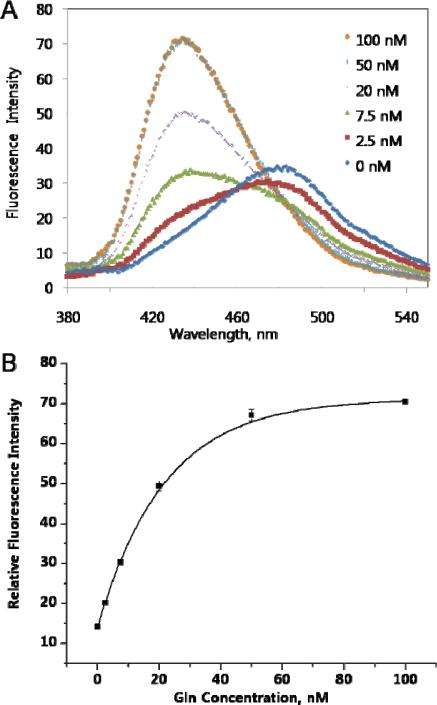
To examine the ability of Anap to detect local structural changes, the mutant N160Anap QBP (QBP-N160Anap) was titrated with glutamine and fluorescence was measured. With increasing concentrations of glutamine, of QBP-N160Anap undergoes a large shift from 480 nm to 430 nm (Figure 3); the fluorescence intensity at 430 nm also increased roughly 5-fold. We then measured the fluorescence anisotropy before (0.20 ± 0.01) and after (0.23 ± 0.01) glutamine binding to determine if differences in the rotational mobility of the Anap side chain contribute to the spectral changes. However, the anisotropy difference between the two states is not significant suggesting that the fluorescence changes are mainly caused by differences in the polarity of the environment around Anap in the two conformational states. The Kd of the mutant protein for glutamine, which was measured directly from the change in Anap fluorescence, was found to be 10 nM (Figure 3), close to that of the wild-type protein (6.4 nM, Figure S5), suggesting that the relatively small size of Anap does not cause significant structural perturbations to QBP. Since the genetic incorporation of Anap into proteins by simple mutagenesis is quantitative and site-specific, and no additional components or steps are required (such as a reactive linker to conjugate the protein and fluorophore, or a specific targeting sequence), this method represents a straightforward approach to interrogate changes at specific sites in protein structure versus global changes in overall protein structure.20
Figure 3.
Fluorescence measurements of QBP-Anap with different concentrations of Gln. Protein samples (10 nM) in D-PBS were excited at 350 nm. Fluorescence spectra were recorded after adding Gln in the same buffer to the final indicated concentrations without further incubation. (a) Fluorescence spectra. (b) Fluorescence intensity at 430 nm.
In summary, a fluorescent amino acid was incorporated into proteins in yeast with high efficiency and specificity in response to the amber codon. The amino acid was site-specifically incorporated into E. coli glutamine binding protein and used to directly probe local structural changes caused by ligand binding. The small size of Anap and ability to introduce it by simple mutagenesis at defined sites in the proteome should make it a useful local probe of protein structure, molecular interactions, protein folding and localization. We are currently applying this method to imaging protein localization in both yeast and mammalian cells.21
Supplementary Material
Acknowledgments
We thank Dr. Lei Wang for providing plasmids for protein expression. We are grateful to the NIH GM62159, and the Skaggs Institute for Chemical Biology for support of this work.
Footnotes
Supporting Information Available: Materials and methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]; b Zhang J, Campbell RE, Ting AY, Tsien RY. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]; c Ting AY, Kain AH, Klemke RL, Tsien RY. Proc. Natl. Acad. Sci. U.S.A. 2002;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]; e MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 2.a Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]; b Hu CD, Kerppola TK. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 3.a Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]; b Griffin BA, Adams SR, Jones J, Tsien RY. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]; c Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 4.a Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]; b Chen I, Howarth M, Lin W, Ting AY. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]; c Carrico IS, Carlson BL, Bertozzi CR. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 5.Cotton GJ, Muir TW. Chem. Biol. 2000;7:253–261. doi: 10.1016/s1074-5521(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 6.Hamada H, Kameshima N, Szymanska A, Wegner K, Lankiewicz L, Shinohara H, Taki M, Sisido M. Bioorg. Med. Chem. 2005;13:3379–3384. doi: 10.1016/j.bmc.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. Proc. Natl. Acad. Sci. USA. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang J, Xie J, Schultz PG. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 8.Weber G, Farris FJ. Biochemistry. 1979;18:3075–3078. doi: 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- 9.a Macgregor RB, Weber G. Nature. 1986;319:70–73. doi: 10.1038/319070a0. [DOI] [PubMed] [Google Scholar]; b Cohen BE, McAnaney TB, Park ES, Jan YN, Boxer SG, Jan LY. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]; c Lakowicz JR. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Press; New York: 1999. [Google Scholar]
- 10.Davis BN, Abelt CJ. J. Phys. Chem. A. 2005;109:1295–1298. doi: 10.1021/jp046050y. [DOI] [PubMed] [Google Scholar]
- 11.a Jones G, II, Jackson WR, Choi C. J. Phys. Chem. 1985;89:294–300. [Google Scholar]; b Bramhall J. Biochemistry. 1986;25:3479–3486. doi: 10.1021/bi00359a057. [DOI] [PubMed] [Google Scholar]
- 12.Wu N, Deiters A, Cropp TA, King D, Schultz PG. J. Am. Chem. Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- 13.Tippmann EM, Schultz PG. Tetrahedron. 2007;63:6182–6184. [Google Scholar]
- 14.a Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]; b Chin JW, Cropp TA, Chu S, Meggers E, Schultz PG. Chem. Biol. 2003;10:511–519. doi: 10.1016/s1074-5521(03)00123-6. [DOI] [PubMed] [Google Scholar]
- 15.Xie J, Liu W, Schultz PG. Angew. Chem. Int. Ed. 2007;46:9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- 16.Cusack S, Yaremchuk A, Tukalo M. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun YJ, Rose J, Wang B, Hsiao C. J. Mol. Biol. 1998;278:219–229. doi: 10.1006/jmbi.1998.1675. [DOI] [PubMed] [Google Scholar]
- 18.Wada A, Mie M, Aizawa M, Lahoud P, Cass AEG, Kobatake E. J. Am. Chem. Soc. 2003;125:16228–16234. doi: 10.1021/ja036459l. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Wang L. J. Am. Chem. Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 20.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Brock A, Chen S, Chen S, Schultz PG. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.