Abstract
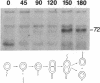
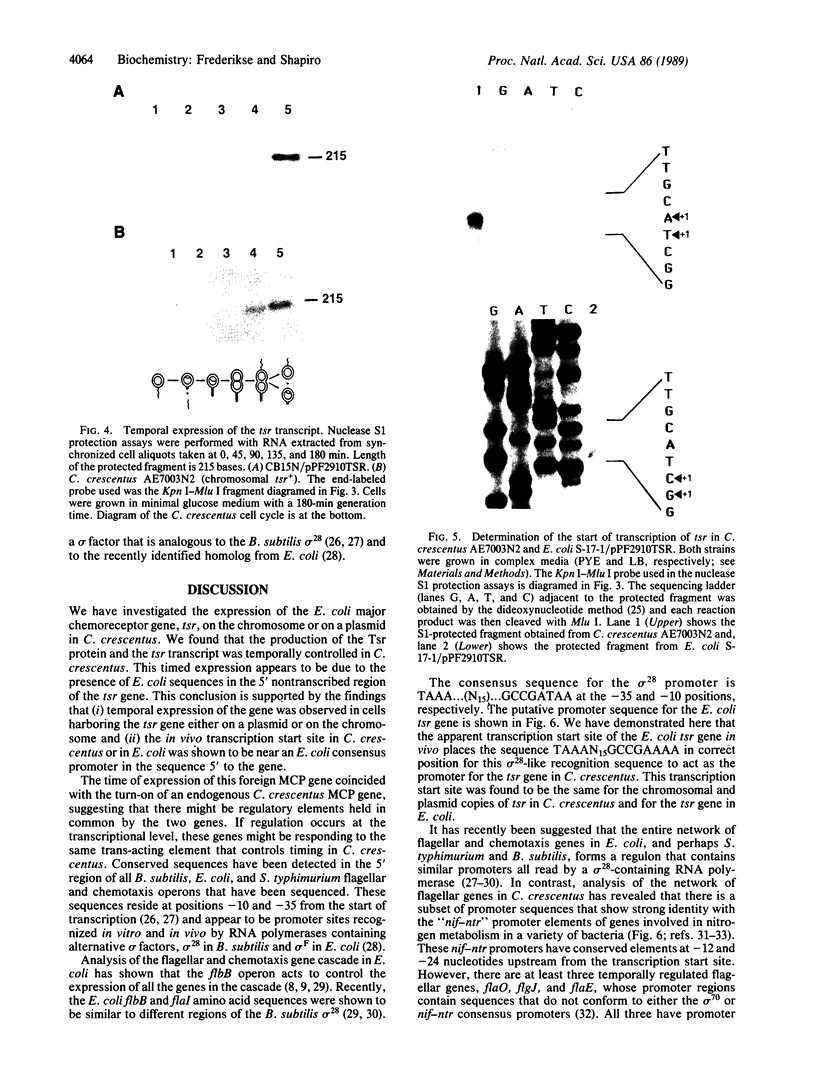
Flagellar and chemotaxis genes are transcribed at a discrete time in the Caulobacter cell cycle. We demonstrate here that the expression of the Escherichia coli chemoreceptor gene tsr, with 2.6 kilobases of its upstream sequence, is temporally controlled in Caulobacter crescentus. The tsr gene was placed on the chromosome in single copy or on a low-copy-number plasmid. It was found that the Tsr protein appeared at the same point in the cell cycle as an endogenous C. crescentus methyl-accepting chemotaxis protein. Nuclease S1 mapping experiments showed that the tsr transcript was also controlled by the cell cycle, suggesting that the E. coli tsr gene is regulated by C. crescentus factors that mediate the timing of transcription initiation. The apparent transcription start site of the E. coli tsr gene was determined in both E. coli and C. crescentus, and we found that in both backgrounds the promoter used conforms to the consensus sequence for the promoters of the flagellar and chemosensory genes of Bacillus subtilis and E. coli. The use of this promoter suggests that C. crescentus has a cognate sigma factor and predicts that other C. crescentus genes are expressed from this consensus promoter.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L. N., Hanson R. S. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985 Mar;161(3):955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya K., Bellofatto V., Shapiro L., Feingold J. Transcription initiation in vitro and in vivo at a highly conserved promoter within a 16 S ribosomal RNA gene. J Mol Biol. 1986 Jan 5;187(1):1–14. doi: 10.1016/0022-2836(86)90401-8. [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Chamberlin M. J. Secondary sigma factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Feb;86(3):830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. H., Frantz B. B., Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5' consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988 Apr;170(4):1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Beynon J., Cannon M., Buchanan-Wollaston V., Cannon F. The nif promoters of Klebsiella pneumoniae have a characteristic primary structure. Cell. 1983 Sep;34(2):665–671. doi: 10.1016/0092-8674(83)90399-9. [DOI] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Champer R., Bryan R., Gomes S. L., Purucker M., Shapiro L. Temporal and spatial control of flagellar and chemotaxis gene expression during Caulobacter cell differentiation. Cold Spring Harb Symp Quant Biol. 1985;50:831–840. doi: 10.1101/sqb.1985.050.01.101. [DOI] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- Contreras I., Shapiro L., Henry S. Membrane phospholipid composition of Caulobacter crescentus. J Bacteriol. 1978 Sep;135(3):1130–1136. doi: 10.1128/jb.135.3.1130-1136.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H., Gerardot C. J. Genetic mapping of genes required for motility in Caulobacter crescentus. Genetics. 1984 Nov;108(3):523–532. doi: 10.1093/genetics/108.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J., Fleming D. L., Gomes S. L., Frederikse P., Shapiro L. General nonchemotactic mutants of Caulobacter crescentus. Genetics. 1986 Nov;114(3):717–730. doi: 10.1093/genetics/114.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977 Oct;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes S. L., Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984 Sep 25;178(3):551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6422–6424. doi: 10.1073/pnas.84.18.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Márquez L. M., Chamberlin M. J. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J Bacteriol. 1988 Apr;170(4):1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B., Dingwall A., Bryan R., Champer R., Shapiro L. Temporal regulation and overlap organization of two Caulobacter flagellar genes. J Mol Biol. 1989 Jan 5;205(1):71–83. doi: 10.1016/0022-2836(89)90365-3. [DOI] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., DeRosier D. J. Bacterial flagellar structure and function. Can J Microbiol. 1988 Apr;34(4):442–451. doi: 10.1139/m88-077. [DOI] [PubMed] [Google Scholar]
- Mansour J. D., Henry S., Shapiro L. Differential membrane phospholipid synthesis during the cell cycle of Caulobacter crescentus. J Bacteriol. 1980 Jan;141(1):262–269. doi: 10.1128/jb.141.1.262-269.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich S. A., Newton A. Promoter mapping and cell cycle regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin D., Minnich S., Chen L. S., Newton A. A set of positively regulated flagellar gene promoters in Caulobacter crescentus with sequence homology to the nif gene promoters of Klebsiella pneumoniae. J Mol Biol. 1987 Jun 20;195(4):939–943. doi: 10.1016/0022-2836(87)90497-9. [DOI] [PubMed] [Google Scholar]
- Nowlin D. M., Nettleton D. O., Ordal G. W., Hazelbauer G. L. Chemotactic transducer proteins of Escherichia coli exhibit homology with methyl-accepting proteins from distantly related bacteria. J Bacteriol. 1985 Jul;163(1):262–266. doi: 10.1128/jb.163.1.262-266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Swanson E., Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985 Nov 5;186(1):107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]