Abstract
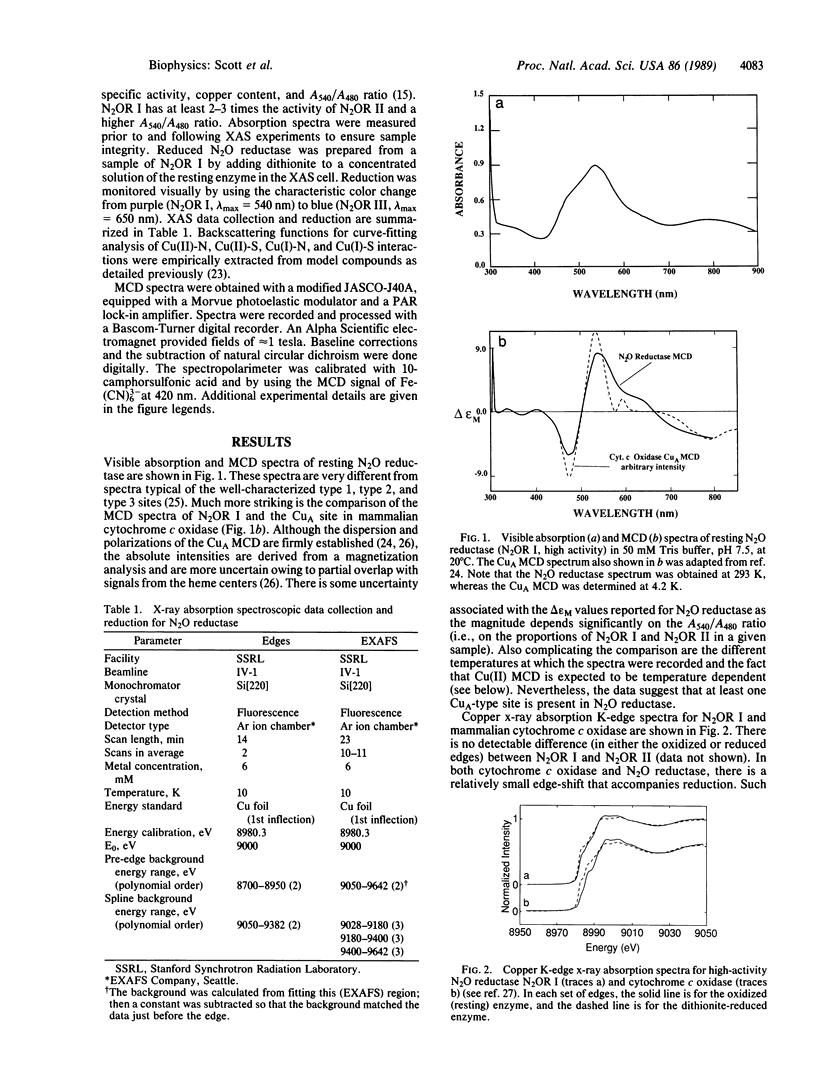
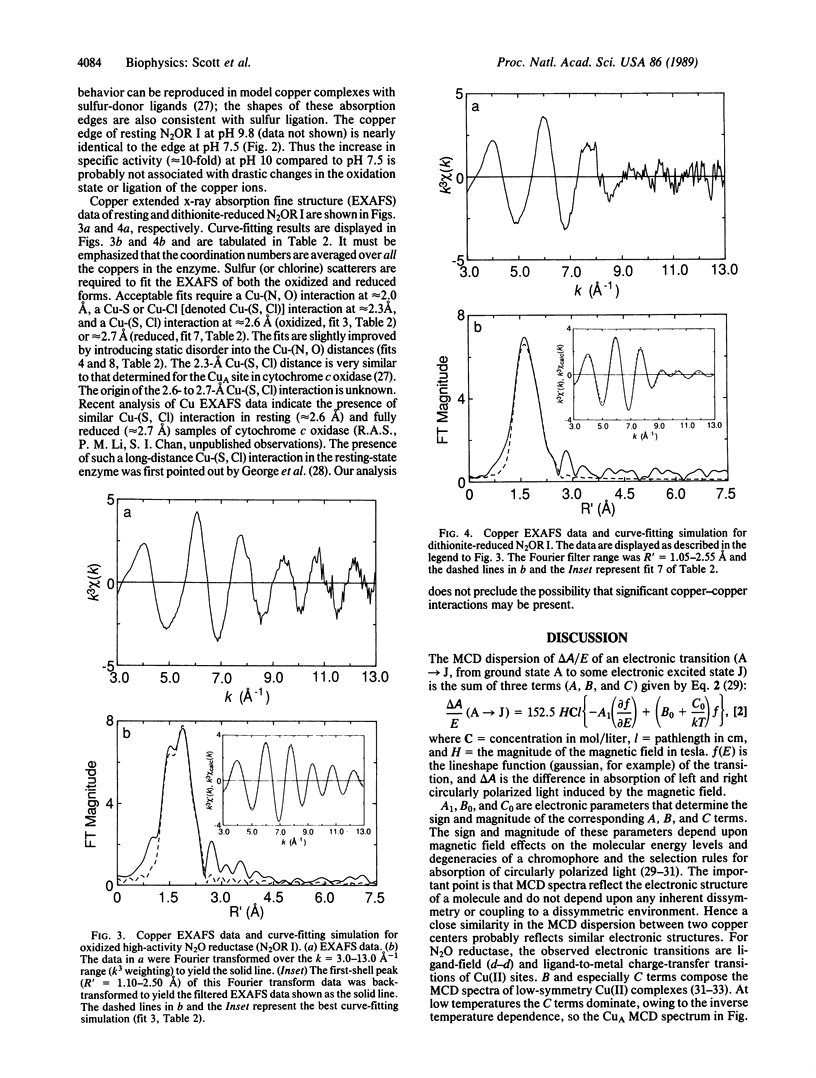
N2O reductase (N2O----N2) is the terminal enzyme in the energy-conserving denitrification pathway of soil and marine denitrifying bacteria. The protein is composed of two identical subunits and contains eight copper ions per enzyme molecule. The magnetic circular dichroism spectrum of resting (oxidized) N2O reductase is strikingly similar to the magnetic circular dichroism spectrum of the CuA site in mammalian cytochrome c oxidase [Greenwood, C., Hull, B. C., Barber, D., Eglinton, D. G. & Thomson, A. J. (1983) Biochem. J. 215, 303-316] and is unlike the magnetic circular dichroism spectra of all other biological copper chromophores obtained to date. Sulfur (or chlorine) scatterers are required to fit the copper extended x-ray absorption fine structure data of both the oxidized and reduced forms of N2O reductase. Satisfactory fits require a Cu-N or Cu-O [denoted Cu-(N, O)] interaction at 2.0 A, a Cu-(S, Cl) interaction at 2.3 A and an additional Cu(S, Cl) interaction at approximately 2.6 A (oxidized) or approximately 2.7 A (reduced). Approximately eight sulfur ions (per eight copper ions) at approximately 2.3 A are required to fit the extended x-ray absorption fine structure data for both the oxidized and reduced N2O reductase. The 2.3-A Cu-(S, Cl) distance is nearly identical to that previously determined for the CuA site in cytochrome c oxidase. A 2.6-2.7 A Cu-(S, Cl) interaction is also present in resting and fully reduced cytochrome c oxidase. Comparison of the N2O reductase sequence, determined by translating the structural NosZ gene, with cytochrome c oxidase subunit II sequences from several sources indicates that a Gly-Xaa-Xaa-Xaa-Xaa-Xaa-Cys-Ser-Xaa-Xaa-Cys-Xaa-Xaa-Xaa-His stretch is highly conserved. This sequence contains three of the probable ligands (two cysteines and one histidine) in a CuA-type site. Collectively these data establish that Pseudomonas stutzeri N2O reductase contains CuA-type sites.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALLEN M. B., VAN NIEL C. B. Experiments on bacterial denitrification. J Bacteriol. 1952 Sep;64(3):397–412. doi: 10.1128/jb.64.3.397-412.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boogerd F. C., Van Verseveld H. W., Stouthamer A. H. Respiration-driven proton translocation with nitrite and nitrous oxide in Paracoccus denitrificans. Biochim Biophys Acta. 1981 Dec 14;638(2):181–191. doi: 10.1016/0005-2728(81)90226-7. [DOI] [PubMed] [Google Scholar]
- Coyle C. L., Zumft W. G., Kroneck P. M., Körner H., Jakob W. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina. Purification and properties of a novel multicopper enzyme. Eur J Biochem. 1985 Dec 16;153(3):459–467. doi: 10.1111/j.1432-1033.1985.tb09324.x. [DOI] [PubMed] [Google Scholar]
- Gelles J., Blair D. F., Chan S. I. The proton-pumping site of cytochrome c oxidase: a model of its structure and mechanism. Biochim Biophys Acta. 1986;853(3-4):205–236. doi: 10.1016/0304-4173(87)90002-4. [DOI] [PubMed] [Google Scholar]
- Greenwood C., Hill B. C., Barber D., Eglinton D. G., Thomson A. J. The optical properties of CuA in bovine cytochrome c oxidase determined by low-temperature magnetic-circular-dichroism spectroscopy. Biochem J. 1983 Nov 1;215(2):303–316. doi: 10.1042/bj2150303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Moubarak A., O'Brien P., Pan L. P., Cho I., Millett F. Topological studies of monomeric and dimeric cytochrome c oxidase and identification of the copper A site using a fluorescence probe. J Biol Chem. 1988 Jun 15;263(17):8142–8149. [PubMed] [Google Scholar]
- Hoffman B. M., Roberts J. E., Swanson M., Speck S. H., Margoliash E. Copper electron-nuclear double resonance of cytochrome c oxidase. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1452–1456. doi: 10.1073/pnas.77.3.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Saraste M., Wikström M. Structural models of the redox centres in cytochrome oxidase. EMBO J. 1987 Sep;6(9):2819–2823. doi: 10.1002/j.1460-2075.1987.tb02578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike I., Hattori A. Energy yield of denitrification: an estimate from growth yield in continuous cultures of Pseudomonas denitrificans under nitrate-, nitrite- and oxide-limited conditions. J Gen Microbiol. 1975 May;88(1):11–19. doi: 10.1099/00221287-88-1-11. [DOI] [PubMed] [Google Scholar]
- Kroneck P. M., Antholine W. A., Riester J., Zumft W. G. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II),Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 1988 Dec 19;242(1):70–74. doi: 10.1016/0014-5793(88)80987-6. [DOI] [PubMed] [Google Scholar]
- Körner H., Frunzke K., Döhler K., Zumft W. G. Immunochemical patterns of distribution of nitrous oxide reductase and nitrite reductase (cytochrome cd1) among denitrifying pseudomonads. Arch Microbiol. 1987 Jun;148(1):20–24. doi: 10.1007/BF00429641. [DOI] [PubMed] [Google Scholar]
- Li P. M., Gelles J., Chan S. I., Sullivan R. J., Scott R. A. Extended X-ray absorption fine structure of copper in CuA-depleted, p-(hydroxymercuri)benzoate-modified, and native cytochrome c oxidase. Biochemistry. 1987 Apr 21;26(8):2091–2095. doi: 10.1021/bi00382a005. [DOI] [PubMed] [Google Scholar]
- Maret W., Dietrich H., Ruf H. H., Zeppezauer M. Active site-specific reconstituted copper(II) horse liver alcohol dehydrogenase: a biological model for type 1 Cu2+ and its changes upon ligand binding and conformational transitions. J Inorg Biochem. 1980 Jun;12(3):241–252. doi: 10.1016/s0162-0134(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Scholes C. P., Chan S. I. On the nature of cysteine coordination to CuA in cytochrome c oxidase. J Biol Chem. 1988 Jun 15;263(17):8420–8429. [PubMed] [Google Scholar]
- McCarthy J. E., Ferguson S. J., Kell D. B. Estimation with an ion-selective electrode of the membrane potential in cells of Paracoccus denitrificans from the uptake of the butyltriphenylphosphonium cation during aerobic and anaerobic respiration. Biochem J. 1981 Apr 15;196(1):311–321. doi: 10.1042/bj1960311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhele K., Tang Y. J., Clark M. A., Ingraham J. L. A Pseudomonas stutzeri outer membrane protein inserts copper into N2O reductase. J Bacteriol. 1987 Dec;169(12):5721–5726. doi: 10.1128/jb.169.12.5721-5726.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Gelles J., Li P. M., Chan S. I. Chemical modification of the CuA site affects the proton pumping activity of cytochrome c oxidase. Biochemistry. 1988 Jan 12;27(1):296–301. doi: 10.1021/bi00401a045. [DOI] [PubMed] [Google Scholar]
- Rich P. R., West I. C., Mitchell P. The location of CuA in mammalian cytochrome c oxidase. FEBS Lett. 1988 Jun 6;233(1):25–30. doi: 10.1016/0014-5793(88)81349-8. [DOI] [PubMed] [Google Scholar]
- Riester J., Zumft W. G., Kroneck P. M. Nitrous oxide reductase from Pseudomonas stutzeri. Redox properties and spectroscopic characterization of different forms of the multicopper enzyme. Eur J Biochem. 1989 Jan 2;178(3):751–762. doi: 10.1111/j.1432-1033.1989.tb14506.x. [DOI] [PubMed] [Google Scholar]
- Scott R. A., Schwartz J. R., Cramer S. P. Structural aspects of the copper sites in cytochrome c oxidase. An X-ray absorption spectroscopic investigation of the resting-state enzyme. Biochemistry. 1986 Sep 23;25(19):5546–5555. doi: 10.1021/bi00367a030. [DOI] [PubMed] [Google Scholar]
- Scott R. A., Sullivan R. J., DeWolf W. E., Jr, Dolle R. E., Kruse L. I. The copper sites of dopamine beta-hydroxylase: an X-ray absorption spectroscopic study. Biochemistry. 1988 Jul 26;27(15):5411–5417. doi: 10.1021/bi00415a005. [DOI] [PubMed] [Google Scholar]
- Snyder S. W., Hollocher T. C. Purification and some characteristics of nitrous oxide reductase from Paracoccus denitrificans. J Biol Chem. 1987 May 15;262(14):6515–6525. [PubMed] [Google Scholar]
- Steinrücke P., Steffens G. C., Panskus G., Buse G., Ludwig B. Subunit II of cytochrome c oxidase from Paracoccus denitrificans. DNA sequence, gene expression and the protein. Eur J Biochem. 1987 Sep 15;167(3):431–439. doi: 10.1111/j.1432-1033.1987.tb13356.x. [DOI] [PubMed] [Google Scholar]
- Stevens T. H., Martin C. T., Wang H., Brudvig G. W., Scholes C. P., Chan S. I. The nature of CuA in cytochrome c oxidase. J Biol Chem. 1982 Oct 25;257(20):12106–12113. [PubMed] [Google Scholar]
- Thomson A. J., Greenwood C., Peterson J., Barrett C. P. Determination of the optical properties of CuA(II) in bovine cytochrome c oxidase using magnetic circular dichroism as an optical detector of paramagnetic resonance. J Inorg Biochem. 1986 Oct-Nov;28(2-3):195–205. doi: 10.1016/0162-0134(86)80083-6. [DOI] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988 Oct;170(10):4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Physical mapping of transposon Tn5 insertions defines a gene cluster functional in nitrous oxide respiration by Pseudomonas stutzeri. J Bacteriol. 1987 Oct;169(10):4577–4580. doi: 10.1128/jb.169.10.4577-4580.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G., Döhler K., Körner H. Isolation and characterization of transposon Tn5-induced mutants of Pseudomonas perfectomarina defective in nitrous oxide respiration. J Bacteriol. 1985 Sep;163(3):918–924. doi: 10.1128/jb.163.3.918-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


