Abstract
Human α-defensins (HNPs) are immune defense mini-proteins that act by disrupting microbial cell membranes. Elucidating the three-dimensional structures of HNPs in lipid membranes is important for understanding their mechanisms of action. Using solid-state NMR, we have determined the three-dimensional structure of HNP-1 in a microcrystalline state outside the lipid membrane, which provides benchmarks for structure determination and comparison with the membrane-bound state. From a suite of 2D and 3D magic-angle spinning experiments, 13C and 15N chemical shifts were obtained that yielded torsion angle constraints while inter-residue distances were obtained to restrain the three-dimensional fold. Together, these constraints led to the first high-resolution SSNMR structure of a human defensin. The SSNMR structure has close similarity to the crystal structures of the HNP family, with the exception of the loop region between the first and second β-strands. The difference, which is partially validated by direct torsion angle measurements of selected loop residues, suggests possible conformational variation and flexibility of this segment of the protein, which may regulate HNP interaction with the phospholipid membrane of microbial cells.
Keywords: Human α-defensin, solid-state NMR, resonance assignment, structure determination, antimicrobial peptides
Introduction
Antimicrobial peptides (AMPs) are small cationic peptides that constitute part of the innate immune system of many plants and animals to rapidly kill invading microbial pathogens. In mammals the two main classes of antimicrobial peptides are cathelicidins and defensins 1. While most cathelicidins are α-helical in structure, defensins contain β-strands stabilized by three disulfide bonds 2-4. Based on the disulfide-linkage patterns, three classes of defensins, α-, β-, and θ-defensins, are identified in vertebrates. Humans have six α-defensins that contain 29-32 residues. Four of these proteins are present in the azurophilic granules of neutrophils and are called human neutrophil peptides 1-4 (HNP 1-4) 2; 5, and two are expressed in intestinal Paneth cells and are called HD-5 and HD-6 6; 7. Except for HD-6, all five human α-defensins have wide-spectrum antimicrobial activities with LD50 in the μg/ml range 8; 9. Similar to most other AMPs, the main mechanism of action of HNPs is believed to be permeabilization of the microbial cell membrane 10; 11.
The presence of three antiparallel disulfide-stabilized β-strands makes defensins larger and more complex proteins than most other AMPs. In particular, they are natural extensions of the two-stranded β-hairpin AMPs such as protegrins from porcine leukocytes and tachyplesins from horseshoe crabs, whose membrane-bound structure and lipid interaction have been characterized in detail by solid-state NMR recently 12-21. Compared to these smaller β-sheet AMPs, HNPs have weaker antimicrobial activities. Moreover, although HNP 1-3 have nearly identical sequences, differing only in the N-terminus residue, their activities differ by as much as 4-fold 8. Understanding the activity differences among human defensins requires structural investigations in the lipid membrane. However, so far most high-resolution structures of human defensins come from X-ray crystallography in the absence of any membrane-mimetic solvents 3; 4. A few biophysical studies that investigated the interactions of defensins with lipid membranes have also been reported, but did not contain high-resolution structural information 22-24.
Solid-state NMR magic-angle spinning (MAS) NMR has been recently shown to be able to elucidate the atomic-resolution structure of 13C, 15N-labeled proteins in microcrystalline states 25-28, in fibrillar forms 29-31, and in lipid membranes 32; 33. A wide variety of 2D and 3D correlation techniques have been developed to obtain resonance assignment 34; 35, torsion angles 36-38 and inter-atomic distances 39. Therefore, the technology is now in place to determine the full structures of human α-defensins in lipid membranes to elucidate the mechanism of action of this important class of immune defense molecules.
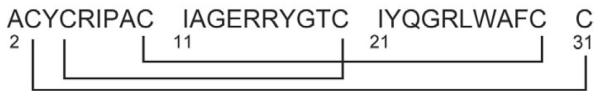
In this work, we report the three-dimensional structure determination of recombinant HNP-1, whose sequence is shown in Figure 1, by SSNMR. We have carried out resonance assignment and internuclear distance measurements of HNP-1 in an ordered microcrystalline state, in preparation for structure determination of the protein bound to the lipid membrane, since membrane-bound proteins usually give lower-resolution spectra due to the disorder created by the lipid bilayer. The conformation of microcrystalline HNP-1 also represents the protein structure before binding to the lipid membrane, and is thus necessary for understanding potential conformational changes of HNPs induced by the lipid bilayer. We compare the SSNMR structure with various published crystal structures of HNPs 3; 4; 40.
Figure 1.
Amino acid sequence and disulfide bond connectivities of HNP-1.
Results
Preparation of microcrystalline HNP-1 samples for solid-state NMR experiments
The solid-state NMR data presented here were obtained from microcrystalline samples of HNP-1 precipitated from a polyethylene glycol (PEG) solution. Two samples were prepared using different concentrations of the protein stock solution. The high-concentration stock solution (36 mg/ml) yielded microcrystals one hour after mixing with PEG, while the low-concentration stock solution (30 mg/ml) produced microcrystals gradually in a 4-day period, with ~80% of the protein eventually precipitating from the supernatant. The slower precipitation produced larger crystals of ~20 μm diameter compared to 2 μm for the rapidly precipitated sample. Figure 2 shows an image of HNP-1 microcrystals obtained from the slower precipitation procedure.
Figure 2.
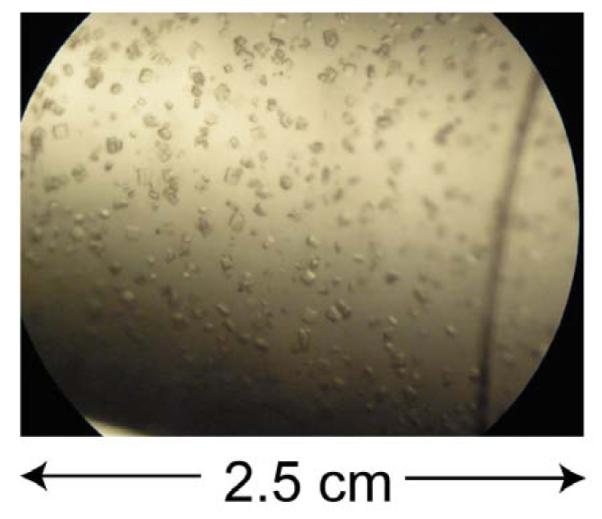
An image of microcrystalline HNP-1. The average size of the crystals is ~20 μm.
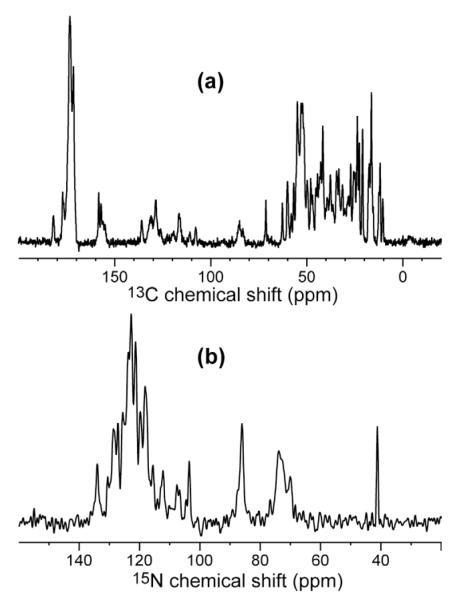
Both microcrystalline HNP-1 samples yielded well-resolved 13C and 15N MAS spectra, shown in Figure 3. For the rapidly precipitated sample in the 4 mm rotor, the 13C full widths at half maxima were 0.6 – 1.0 ppm and the 15N linewidths were 1.3 – 1.7 ppm. For the slowly precipitated sample in the 2.5 mm rotor, the linewidths improved to 0.4 – 0.8 ppm for 13C and 1.0 – 1.5 ppm for 15N. Faster spinning frequencies and stronger 1H decoupling fields further contributed to the narrower linewidths of the 2.5 mm rotor sample.
Figure 3.
1D CP-MAS spectra of microcrystalline U-13C, 15N-labeled HNP-1. (a) 13C spectrum, measured at 258 K at a 13C Larmor frequency of 225 MHz. (b) 15N spectrum, measured at 268 K at a 15N Larmor frequency of 60 MHz.
Resonance assignment of HNP-1 by 2D and 3D SSNMR
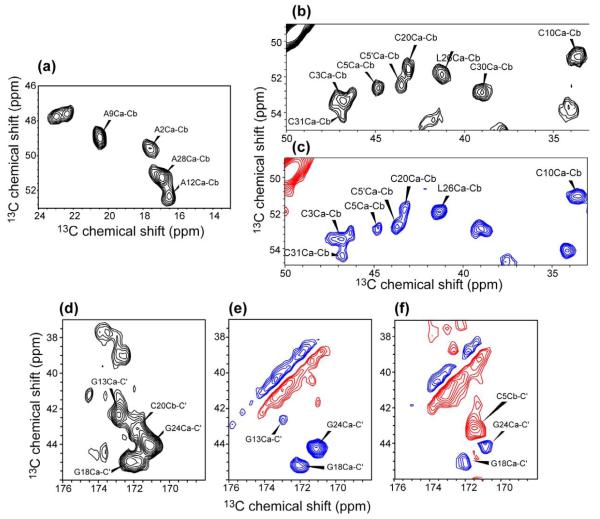
We first assigned the amino acid spin systems of HNP-1 using 2D 13C-13C correlation experiments, including a DARR experiment with 40 ms mixing, and double-quantum (DQ)-filtered experiments using SPC5 41 and CM5RR sequences 42. The 40 ms DARR spectrum allowed the assignment of most amino acid types (Figure 4a, b, d), including Cys, Ala, Ile, Arg, Leu, Pro, and Thr, which have distinct cross peak patterns. The aromatic residues were assigned based on the Cα/Cβ cross peaks to the aromatic carbons. For example, the Cγ of the unique tryptophan W27 resonates at 107.8 ppm (Figure S1), which allowed the rest of the W27 spin system to be identified. For the unique glutamate E14 and glutamine Q23, the sidechain Cδ chemical shifts (181 ppm for E14 and 176 ppm for Q23) served as useful guides for identifying the chemical shifts of the other sidechain carbons.
Figure 4.
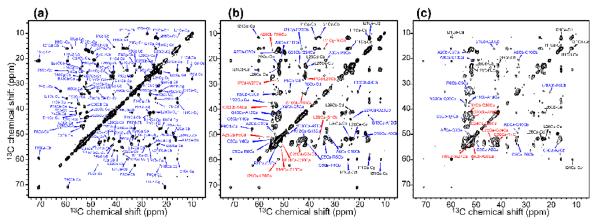
Representative regions of 2D 13C-13C correlation spectra of HNP-1. (a) Ala Cα-Cβ region from a 40 ms DARR spectrum. (b) Cys Cα-Cβ region from a 40 ms DARR spectrum. (c) Cys Cα-Cβ region from a 0.8 ms CM5RR spectrum. Blue: positive intensities. Red: negative intensities. (d-f) CO-Cα/Cβ region. (d) 40 ms DARR spectrum, (e) 0.8 ms CM5RR spectrum. (f) 1.5 ms CM5RR. The sign of the cross peak intensities distinguishes the one-bond Gly CO-Cα peaks (red, indicating negative) from the two-bond Cys CO-Cβ peaks (blue, indicating positive).
The DQ-filtered 2D spectra confirmed the DARR assignment by exhibiting opposite intensities for one-bond and two-bond cross peaks. For example, Gly Cα and Cys Cβ, which both resonate at 40-45 ppm, are distinguished based on the negative intensities of the one-bond Gly Cα-CO peaks and the positive intensities of the two-bond Cys Cβ-CO peaks in the CM5RR spectra (Figure 4c, e, f). Similarly, while Q23 Cβ has similar chemical shifts to Arg Cγ in the DARR spectrum, in the DQ filtered spectra Q23 Cβ exhibits a positive one-bond Cα-Cβ cross peak, which is distinct from the negative two-bond Arg Cα-Cγ cross peaks. By comparing various 2D 13C-13C spectra, we assigned all amino acid spin systems except for one Arg.
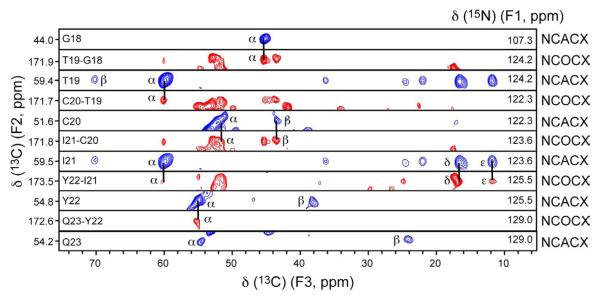
Next, we obtained sequence-specific assignment from inter-residue NCOCX and intra-residue NCACX 3D experiments 34; 35. Figure 5 shows a representative strip of F2-F3 planes of the two 3D experiments for residues G18-Q23. We first identified the 15N chemical shift of residue i in the NCACX spectrum by comparing the 13C chemical shifts with those obtained from the 2D CC spectra. The 15N chemical shift of residue i+1 was then obtained from the NCOCX spectrum through its cross peaks with the 13C chemical shifts of residue i. The 15N plane of residue i+1 was then searched in the NCACX spectrum to identify the 13C chemical shifts of residue i+1.
Figure 5.
F2-F3 strips of 3D NCACX (blue) and NCOCX (red) spectra for residues G18 to Q23, illustrating sequential resonance assignment. The strip width for the F2 dimension is 2.3 ppm. The F1 15N chemical shifts for the strips are indicated on the right end of each strip.
A2, A9, T19 and C30 are clear starting points for assigning a consecutive segment of residues. The N-terminal residue A2 has a distinct 15N chemical shift of 41.7 ppm, which is folded to 106.9 ppm. Residue A9 has cross peaks with P8, which has characteristic 13C chemical shifts. Residue T19 has characteristic Cα and Cβ chemical shifts.
The four Ala residues, which are well resolved in the 2D CC spectra, were sequentially assigned in the 3D spectra. For example, A9 was assigned based on its correlation with the unique P8 and with C10, and A12 was assigned based on its correlation with the well defined I11 and G13. For the three Gly residues, G24 was identified by inter-residue correlations Q23N-G24Cα and R25N-G24Cα in the 3D spectra and in the 100 ms DARR spectrum, while G18 was assigned based on the T19N-G18Cα correlations in the 3D spectrum.
The six Cys residues were resolved and assigned based on their cross peaks with their neighboring residues in the NCOCX spectrum. For example, C3 has characteristic cross peaks with A2 Cβ and Cα, and C5 has cross peaks with the Y4 spin system. Interestingly, C5 exhibits two Cβ chemical shifts differing by 0.9 ppm, suggesting the presence of two conformations at this site. C10 has a Cβ chemical shift of 33.4 ppm (on the TMS scale), which is slightly low for an oxidized cysteine. However, its assignment was corroborated by multiple sequential C10 – A9 cross peaks and I11 – C10 cross peaks. For example, C10N-A9C’-A9C /C cross peaks were identified in the 3D NCOCX spectrum, and sequential C10C’- A9CÛ, C10Cÿ- A9CÛ, and A9C - C10C cross peaks were also detected in the 100 ms DARR spectrum (Figure S2).
The complete 13C and 15N isotropic chemical shifts of HNP-1 are summarized in Table 1. Among the six cysteines, the C10 – C30 pair has relatively upfield Cβ chemical shifts. To further verify that C10 and C30 are indeed disulfide-bonded, we compared the Cα and Cβ chemical shifts of all HNP-1 cysteines with literature statistical analyses of the dependence of cysteine 13C chemical shifts on the oxidation state and conformation 43; 44. On the DSS scale, the oxidized cysteines have Cβ chemical shifts of 33 - 51 ppm and Cα chemical shifts of 50 - 61 ppm. When the HNP-1 cysteine chemical shifts are also referenced to DSS (thus increasing the 13C chemical shifts in Table 1 by 1.7 ppm), we find that all six cysteines, including C10 and C30, lie in the oxidized range (Figure S3), thus confirming the disulfide-bonded nature of all three pairs.
Table 1.
13C and 15N chemical shifts of microcrystalline HNP-1 without lipids a.
| N | Cα | Cβ | Cγ | Cδ | Cε | CO | |
|---|---|---|---|---|---|---|---|
| A2 | 41.7 | 49.5 | 17.5 | 171.0 | |||
| C3(C31) b | 117.8 | 53.0 | 46.6 | 171.4 | |||
| Y4 | 117.6 | 54.8 | 172.6 | ||||
| C5 (C20) | 130.6 | 52.7 | 44.5/43.4 | 171.4 | |||
| R6 | 123.1 | 53.9 | 33.7 | 26.7 | 41.5 | 173.6 | |
| I7 | 122.3 | 56.5 | 41.2 | 28.0 | 15.3 | 10.1 | 172.6 |
| P8 | 134.4 | 62.2 | 32.9 | 23.1/22.1 | 47.5 | 172.7 | |
| A9 | 122.6 | 48.8 | 20.6 | 172.9 | |||
| C10(C30) | 115.6 | 50.8 | 33.4 | 173.0 | |||
| I11 | 115.4 | 57.9 | 37.5 | 23.8 | 15.9 | 12.2 | 173.0 |
| A12 | 121.9 | 51.8 | 16.6 | 175.2 | |||
| G13 | 109.6 | 42.6 | |||||
| E14 | 121.3 | 52.2 | 31.4 | 34.4 | 181.8 | 173.6 | |
| R15 | 130.6 | 52.7 | 171.4 | ||||
| R16 | 122.8 | 54.5 | 28.1 | 27.0 | 42.4 | 157.1 | 175.3 |
| Y17 | 128.1 | 56.3 | 38.5 | 131.5 | 173.5 | ||
| G18 | 107.3 | 45.0 | 171.9 | ||||
| T19 | 124.2 | 59.4 | 69.6 | 21.7 | 171.7 | ||
| C20(C5) | 122.3 | 51.6 | 43.0 | 171.8 | |||
| I21 | 123.6 | 59.5 | 35.8 | 24.3 | 16.2 | 11.4 | 173.5 |
| Y22 | 125.5 | 54.8 | 37.7 | 172.6 | |||
| Q23 | 129.0 | 54.2 | 23.8 | 31.9 | 176.5 | 173.9 | |
| G24 | 103.4 | 44.0 | 170.3 | ||||
| R25 | 118.4 | 51.6 | 32.2 | 25.5 | 41.2 | 172.0 | |
| L26 | 120.2 | 51.6 | 41.2 | 25.4 | 23.2 | 22.4 | 174.4 |
| W27 | 120.7 | 52.0 | 29.0 | 175.3 | |||
| A28 | 127.1 | 51.0 | 16.9 | 172.8 | |||
| F29 | 123.8 | 56.6 | 39.8 | 129.2 | |||
| C30(C10) | 121.9 | 53.0 | 38.6 | 172.7 | |||
| C31(C3) | 121.1 | 55.3 | 47.0 | 177.5 |
The 13C chemical shifts are referenced to TMS. To convert to DSS-referenced chemical shifts, the 13C frequencies in this table should be increased by 1.7 ppm. The 15N chemical shifts are referenced to liquid ammonia.
The partner cysteine involved in the disulfide bond is indicated in the bracket.
Backbone dihedral angles and verification by dipolar correlation experiments
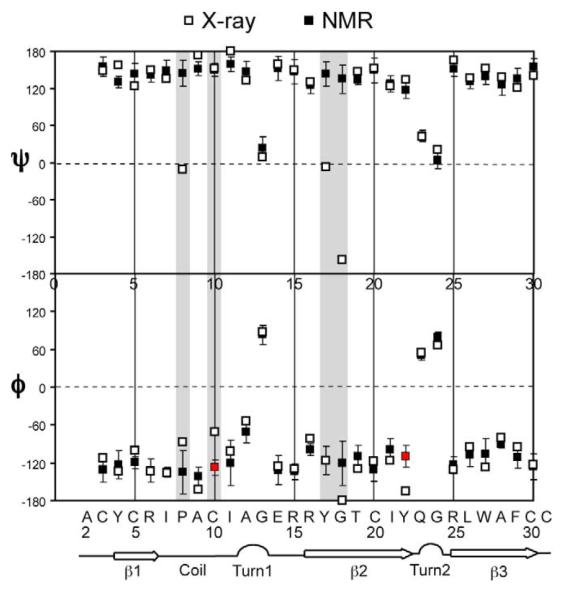
The 13C and 15N chemical shifts allowed us to determine the backbone (φ, ψ) torsion angles of HNP-1 using the well-known empirical relation between N, Cα, Cβ, CO chemical shifts and protein secondary structure 45. We used the TALOS program 46 to predict the (φ, ψ) angles and compared them with the values found from the crystal structure of HNP-3 3 (PDB code: 1DFN). The chemical-shift constrained torsion angles indicate three well-defined β-strands, β1, β2 and β3, separated by a type-II β-turn at A12-G13 and a type I’ β-turn at Q23-G24 (Figure 6). These secondary structure motifs are consistent with those of the HNP-3 structure, although the boundaries of the loop or turn segments between β-strands differ slightly between HNP-3 and HNP-1.
Figure 6.
HNP-1 (φ, ψ) torsion angles determined from solid-state NMR 13C and 15N chemical shifts (filled squares). The torsion angles of HNP-3 are shown for comparison (open squares and ribbons) (PDB accession code: 1DFN). Residues with significantly different torsion angles from the crystal structure values of HNP-3 are shaded.
Among the 28 (φ, ψ) pairs predicted by TALOS, excluding the terminal A2 and C31, 24 φ angles and 24 ψ angles agree well with the crystal structure values within the experimental uncertainty. Residues with large dihedral angle discrepancies are P8, C10, Y17, G18 and Y22 (Figure 6 and Table S1). P8 and C10 both lie in the loop between β1 and β2 strands and might be expected to be dynamically disordered. However, the HNP-3 crystal structure shows well-defined positions for these residues, with B factors not higher than the average 3. Direct measurement of C-H order parameters of the HNP-1 sample by NMR (see Figure 11 below) also indicates that the loop region in HNP-1 is not dynamic.
Figure 11.
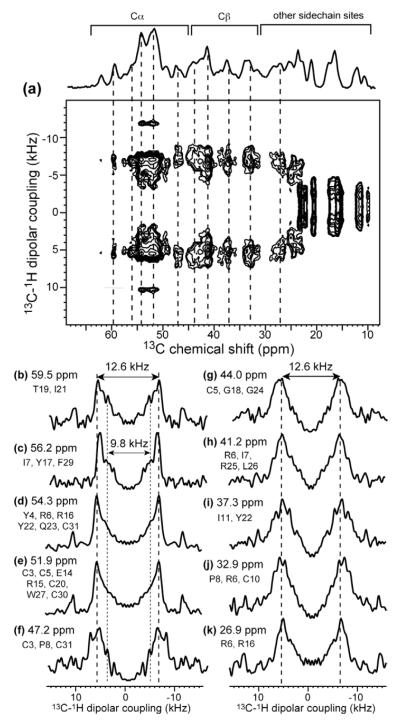
2D 13C-1H LG-CP spectrum to determine the mobility of microcrystalline HNP-1. (a) 2D spectrum, measured under 11 kHz MAS and 293 K. (b-k) 1D dipolar cross sections at selected chemical shifts. Most Cα and Cβ cross sections show rigid-limit values (after taking into account the LG scaling factor of 0.577), except for the 56.2-ppm cross section (c), which has a second coupling that is smaller than the rest.
To verify the torsion angle differences between TALOS and the crystal structure, we conducted the HNCH experiment to directly measure the φ angles of C10 and Y22. The experiment measures the φH angle (defined as HN-N-Cα-Hα, which is related to φ by φH+60°=φ) by correlating the N-H and Cα-Hα dipolar couplings 36; 47, and is particularly sensitive to the β-sheet conformation, which has φH angles around 180°. To maximize the angular resolution, we implemented the N-H doubled version of the experiment 47. In principle, a 3D experiment where two chemical shift dimensions provide site resolution while one dipolar dimension gives the angular information is the most desirable. However, the low sensitivity of the 3D experiment and the limited protein amount precluded its implementation. Thus, we used one 13C chemical shift dimension for site resolution and measured the dipolar evolution at time 0 and half a rotor period. Simulations have shown that the HNCH dipolar dephasing at the middle of the rotor period is the most sensitive to φH angle differences 48.
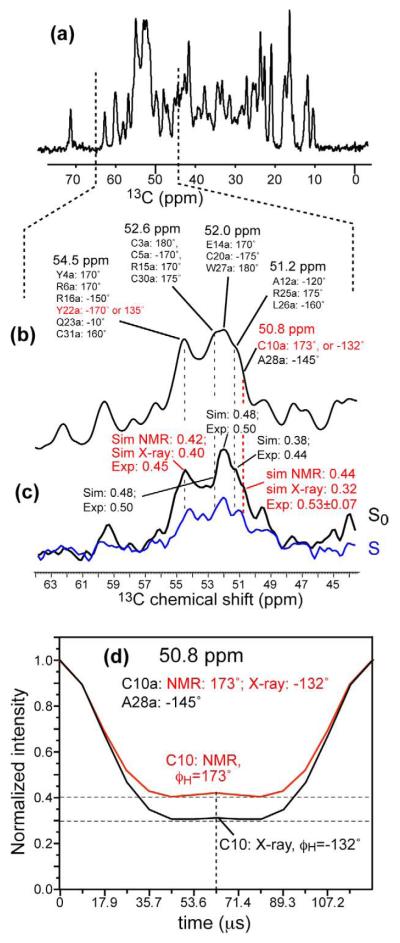
Figure 7c shows the Cα region of the 13C spectra with 0 and half a rotor period of HNCH evolution. The peaks were assigned based on the 2D and 3D spectra above. Although single-site resolution is not available, most sites have consistent φ angles between the TALOS prediction and the crystal structure within ±10° (Table S1). The only uncertain residues are C10 and Y22, thus their φH angles can be extracted from the intensities. Specifically, the C10 Cα signal at 50.8 ppm overlaps with only one residue, A28, whose φH angle is −145°±5°, as indicated by both TALOS and the crystal structure (Figure 7b). Thus, the calculated HNCH curve for 50.8 ppm is very sensitive to the C10 φH angle (Figure 7d). The experimental intensity (0.53) is much more consistent with the TALOS prediction of a β-sheet like φH angle of about 180° rather than a low value of about −130° (Figure S4). Similarly, the measured HNCH intensity for the 54.5-ppm peak, which is a composite of Y22 Cα with five other Cα sites, is more consistent with the TALOS prediction than with the crystal structure, although the distinction between the two possibilities is smaller for this peak due to more resonances overlapping at this position.
Figure 7.
Direct measurement of φ torsion angles by HNCH. (a) Aliphatic region of the 1D 13C spectrum. (b) Cα region (44-64 ppm) of the 13C spectrum, indicating the assignment of the resolved peaks and the φH torsion angles (φH+60°=φ) of each residue. For most residues the φH angles agree between TALOS and the crystal structure within ±10°. But for C10 and Y22, significant deviations exist. (c) Measured HNCH spectra with dipolar evolution time of 0 (S0) and half a rotor period (S, shown in blue). The measured S.S0 values are indicated along with calculated values. The 50.8-ppm peak has an S/S0 value that is consistent with the chemical shift derived φ angles and not the HNP-3 value. (d) Calculated HNCH curves for the 50.8 ppm C10/A28 Cα peak. The A28 φH angle is fixed while the C10 φH angle is either 173° or 132°. The measured HNCH intensity at the middle of the rotor period is consistent with the TALOS φH angle but not the crystal structure value.
For P8, both φ and ψ angles differ between the chemical shift prediction and the crystal structure (Table S1). The lack of HN at Pro precludes direct measurement of its φ angle. In principle, the ψ angle can be measured using the NCCN technique 37. At present the protein amount is insufficient for this experiment, which requires 13C double-quantum filtration in addition to 13C-15N REDOR. Thus, future experiments are necessary to clarify the conformational discrepancy at P8. Nevertheless, based on distance constraints involving this loop, we hypothesize that the difference may be real instead of due to inaccuracy of the NMR structure determination (see below).
The torsion angle difference for Y17-G18 is also noteworthy. TALOS predicted a relatively ideal β-strand ψ angle for Y17 whereas the crystal structure gives an unusual ψ angle near 0°. On the other hand, the TALOS Y17 torsion angles have relatively large uncertainties (ΔΦ=23°, ΔΨ=20°) (Table S1). Among the 10 pairs of torsion angles predicted, seven pairs lie in the β-sheet region while three pairs lie near the α-helical region, with ψ angles of about −20°, which are similar to the crystal structure ψ value. Thus, the ensemble of NMR torsion angles for Y17 encompasses the crystal structure value.
For G18, chemical shifts predicted a φ angle of −120° whereas the crystal structure shows a φ angle near 180°. Since glycine residues have two Hα protons, the HNCH experiment is not ideal for measuring their φ angles. The TALOS uncertainties for G18 are (Δφ=35°, Δψ=23°) (Table S1), and three of the predicted pairs of angles lie in the right-handed α-helix region of the Ramachandran diagram. Thus, the conformation of this residue is not well predicted from the chemical shifts. On the other hand, G18 is strictly conserved in all human defensins. Based on comparisons of HNP-4, HD5 and HD6 crystal structures, it has been suggested that G18 may act as a hinge for the G18-L29 β-hairpin 4, which has significantly different orientations in various HNPs. Thus, it remains a possibility that the G18 torsion angle difference between the NMR and crystal structures may reflect real conformational variations at this residue.
Inter-residue distances
To constrain the three-dimensional fold of HNP-1, we measured distances between sequential and non-sequential residues using DARR experiments with long mixing times. Two spin diffusion mixing times, 100 ms and 200 ms, were chosen, which resulted in larger numbers of cross peaks that deteriorated the spectral resolution (Figure 8). To bypass this problem, we used a comparative strategy to identify the long-distance correlation peaks. For each inter-residue cross peak, we first found all possible assignments that agree with the peak position to ±0.2 ppm. We then measured the distances of these inter-residue pairs in the HNP-3 crystal structure 3, and selected the assignment with the shortest distance. For these inter-residue cross peaks, since distances between sequential residues are usually the shortest, where ambiguity arises, we gave preferences to sequential distances over medium or long-range distances.
Figure 8.
2D 13C-13C DARR spectra of HNP-1 with mixing times (a) 40 ms, (b) 100 ms, and (c) 200 ms. In (a), all peaks are assigned. In (b) and (c), only inter-residue and intra-residue multiple-bond correlations not observed in (a) are assigned. Red: non-sequential inter-residue correlations; blue: sequential correlations; black: intra-residue multiple-bond correlations.
In this way, we identified 40 inter-residue CC correlations, among which 26 were sequential contacts, 1 was medium range (1< ∣i-j∣ <4), and 13 were long-range contacts (∣i-j∣ > 4). Non-sequential correlations were observed for residues A2, Y4, R6, I7, C10, E14, R16, I21, L26, W27, F29 and C30 (Table 2). Six long-range correlation peaks were detected between β1 and β3 strands, and six contacts between β2 and β3 strands. These inter-strand correlations provided important restraints to the three-dimensional fold of the protein.
Table 2.
Non-sequential inter-residue distance restraints.
| residues | spectrum | type of correlation | Distance (A) |
|---|---|---|---|
| C10Cα-E14Cβ | 40 ms DARR | medium range: i – (i+4) | 2.5 – 4.8 |
| E14Cγ-C30Cα | 40 ms DARR | long range | 2.5 – 4.8 |
| A2Cα-C30Cα | 100 ms DARR | long range: β1 – β3 | 2.5 – 5.4 |
| Y4Cα-C30Cβ | 100 ms DARR | long range: β1 – β3 | 2.5 – 5.4 |
| R6Cα-A28Cα | 100 ms DARR | long range: β1 – β3 | 2.5 – 5.4 |
| I7Cγ-L26Cβ | 100 ms DARR | long range | 2.5 – 5.4 |
| I7Cγ-W27Cα | 100 ms DARR | long range | 2.5 – 5.4 |
| C10Cβ-R16Cδ | 100 ms DARR | long range | 2.5 – 5.4 |
| R16Cα-A28Cβ | 100 ms DARR | long range: β2 – β3 | 2.5 – 5.4 |
| I21Cα-L26Cα | 100 ms DARR | long range: β2 – β3 | 2.5 – 5.4 |
| R6Cδ-C30Cβ | 200 ms DARR | long range: β1 – β3 | 2.5 – 6.3 |
| R16Cβ-F29Cα | 200 ms DARR | long range: β2 – β3 | 2.5 – 6.3 |
| R6Cγ-A28Cα | 200 μs CHHC | long range: β1 – β3 | 2.5 – 8.0 |
| C5Cβ-F29Cβ | 300 μs CHHC | long range: β1 – β3 | 2.5 – 8.0 |
| Y4N-G13N | 3 s 15N PDSD | long range | 3.0 – 6.0 |
| C5N-T19N | 3 s 15N PDSD | long range: β1 – β2 | 3.0 – 6.0 |
| P8N-Y22N | 3 s 15N PDSD | long range | 3.0 – 6.0 |
| P8N-W27N | 3 s 15N PDSD | long range | 3.0 – 6.0 |
| P8N-A28N | 3 s 15N PDSD | long range | 3.0 – 6.0 |
| C10N-G13N | 3 s 15N PDSD | medium range: i – (i+3) | 3.0 – 6.0 |
| Y17N-C30N | 3 s 15N PDSD | long range: β2 – β3 | 3.0 – 6.0 |
| G18N-W27N | 3 s 15N PDSD | long range: β2 – β3 | 3.0 – 6.0 |
| Y22N-R25N | 3 s 15N PDSD | medium range: β2 – β3 | 3.0 – 6.0 |
| G24N-L26N | 3 s 15N PDSD | medium range: i – (i+2) | 3.0 – 6.0 |
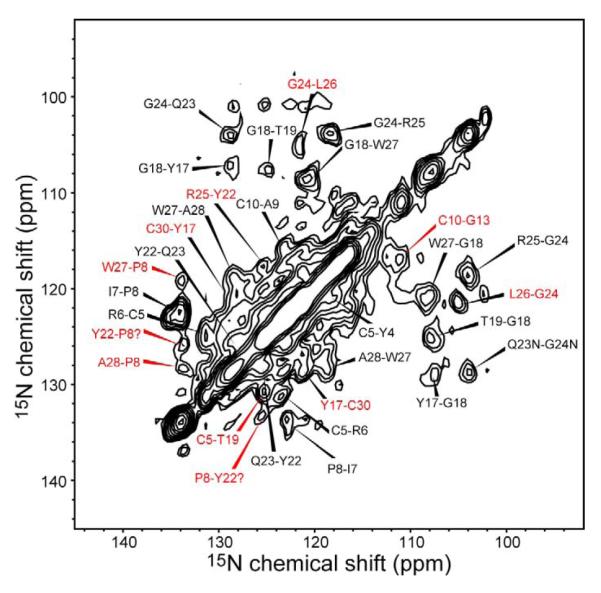
We also measured a 2D 15N-15N correlation spectrum to obtain additional inter-residue distance restraints (Figure 9). The Gly region was the best resolved and provided many useful sequential and long-range cross peaks. P8 also gave well-resolved cross peaks due to its unique imine 15N chemical shift. The 110 – 130 ppm region is more congested, thus we only assigned the intensity maxima and gave them tentative assignments in the same fashion as the CC constraints. The non-sequential NN contacts are included Table 2.
Figure 9.
15N-15N 2D PDSD spectrum with a mixing time of 3 s. Black and red assignments indicate sequential and non-sequential correlations, respectively.
Since the above distance extraction strategy used the HNP-3 structure as partial input, potential differences of HNP-1 from HNP-3 can only be discerned based on missing correlations for HNP-1, and mis-assignment cannot be ruled out. To extract distance restraints in a de novo fashion, without input from the HNP-3 structure, we carried out further 3D 13C-13C-13C correlation experiments, which removed the problem of resonance overlap. As we report in a separate publication, the 3D CCC experiment yielded 270 unique inter-residue distances, among which 129 were sequential, 45 were medium range, and 96 were long-range constraints. These distances verified and refined the current structure, as we show below.
Three-dimensional structure of HNP-1
Using the distance and angular restraints obtained from the above SSNMR experiments, we calculated the HNP-1 structure using the XPLOR-NIH program. We assigned the distance restraints to specific ranges based on the mixing times for the first appearance of cross peaks that correspond to well-defined secondary structures (Table 3). Sequential Cα-Cα distances are fixed by the peptide plane geometry to be 3.8 Å and were thus placed in the range of 3.5-4.1 Å. Sequential N-N distances depend on the ψ torsion angle and fall within the range of 2.5 – 3.6 Å. All inter-residue peaks first observed in the 100 ms 13C-13C DARR spectrum were assigned to the range 2.5 – 5.4 Å, those first appearing in the 200 ms DARR spectrum assigned to 2.5 – 6.3 Å, and those inter-residue peaks appearing in the 2D CHHC spectra assigned to 2.5 – 7.5 Å. In addition, the few inter-residue correlations observed in the 40 ms 13C-13C DARR spectrum were assigned to the range 2.5 – 4.8 Å. All non-sequential 15N-15N contacts were placed in the 3.0 – 6.0 Å range.
Table 3.
Structure calculation summary of HNP-1.
| Restraints | |
|---|---|
| Total inter-residue CC and NN restraints | 60 |
| Sequential (∣i-j∣ = 1) | 36 |
| Medium range (1< ∣i-j∣ <4) | 4 |
| Long range (∣i-j∣ > 4) | 20 |
| β1 – β3 | 6 |
| β2 – β3 | 6 |
| β1 – β2 | 1 |
| CC Restraints in the class 2.5 – 4.8 Å | 4 |
| CC Restraints in the class 2.5 – 5.4 Å | 18 |
| CC Restraints in the class 2.5 – 6.3 Å | 5 |
| CC Restraints in the class 2.5 – 8.0 Å | 7 |
| NN restraints in the class 2.5 – 4.0 Å | 10 |
| NN restraints in the class 3.0 – 6.0 Å | 10 |
| Total (φ, ψ) torsional angle restraints | 56 |
|
| |
| rms deviation of 11 lowest-energy structures from the crystal structure | |
|
| |
| Backbone atoms: | 2.7 Å |
| Heavy atoms: | 3.8 Å |
|
| |
| Eglobal(kcal mol−1) | 264 |
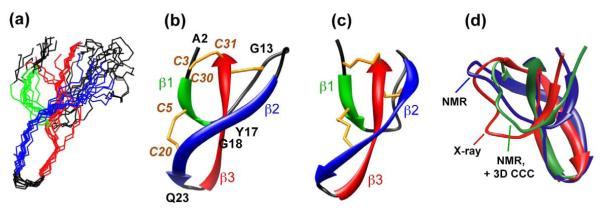
In total, we obtained 60 inter-residue distance restraints, 56 (φ, ψ) torsion angles, and three disulfide bond restraints, and subjected them to structure calculation by XPLOR-NIH. The 10 lowest-energy structures out of 200 calculated structures (Figure 10a) cluster closely. Compared to the crystal structure, the average NMR structure has a backbone atomic root-mean-square deviation (RMSD) of 2.7 Å and a heavy-atom RMSD of 3.8 Å. Figure 10 compares the NMR structure ensemble with the X-ray structure of HNP-3. The NMR structure shows three well-defined β-strands connected by a long loop between the β1 and β2 strands and a tight turn between the β2 and β3 strands. Both the HNP-1 NMR structure and the HNP-3 crystal structure have the same topology: the β3 strand is inserted between the β1 and β2 strands. Beyond these similarities, we found two main differences: the β1-β2 loop has a very different orientation to the rest of the protein in the two structures, and the β2 strand has a different twist and orientation. The origins of these differences will be discussed below.
Figure 10.
SSNMR structures of HNP-1 compared with the X-ray crystal structure of HNP-3. (a) Eleven minimum energy SSNMR structures of HNP-1. (b) Average structure of (a). (c) Crystal structure of HNP-3. The disulfide bonds are shown in orange. (d) Comparison of the β1-β2 loop conformation between the crystal structure (red), the current solid-state NMR structure (blue), and the NMR structure with the additional distance restraints from the 3D CCC experiment (green).
To ascertain whether large-amplitude motion exists in the HNP-1 backbone, we measured the 1H-13C one-bond dipolar couplings using a 2D LG-CP experiment (Figure 11). The spectrum shows that most backbone Cα sites have rigid-limit couplings of ~12.5 kHz after scaling by the homonuclear decoupling scaling factor. The only exception is the signal at 56 ppm, which shows a splitting of 9.8 kHz. This value corresponds to a smaller C-H order parameter of 0.78, consistent with moderate-amplitude dynamics of some of the Cα sites, including I7, Y17, and F29.
Discussion
HNP-1 conformational homogeneity and SSNMR structural quality
The microcrystalline HNP-1 samples are well ordered on the nanometer and micrometer scale. The 13C linewidths of 0.4 – 0.8 ppm and 15N linewidths 1.0-1.5 ppm are overall comparable to those observed in other microcrystalline proteins 49 and are better than some amyloid proteins 31. The 15N linewidths are particularly sensitive to the conformational heterogeneity. Compared to one of the well-studied globular proteins, ubiquitin 49, whose 15N linewidths were reported as 0.3 – 0.5 ppm, the microcrystalline HNP-1 still has residual conformational heterogeneity. However, since the eventual goal is to study HNP-1 in the lipid membrane, whose thermal disorder generally promotes conformational heterogeneity in small proteins, it is not essential to produce HNP-1 with the highest conformational homogeneity.
Variable-temperature 1D 13C and 15N spectra showed quantitatively similar intensities and linewidths in the range of 293 – 253 K (Figure S5), indicating that the HNP-1 backbone is largely immobilized at room temperature and only small-amplitude segmental motions exist. This observation is supported by the 2D LG-CP spectrum, which exhibits near rigid-limit Cα - Hα dipolar couplings for most sites. The persistence of narrow linewidths at mild low temperatures is consistent with the behavior of other microcrystalline proteins but contrasts with that of membrane peptides, which manifest gel-phase-induced line broadening in the same temperature range.
The quality of the HNP-1 structure can be compared with the SSNMR structures of other recently studied globular proteins, such as the α-spectrin SH3 domain 25, ubiquitin 26, and GB1 50. In terms of agreement with the crystal structure, the SH3 domain and HNP-1 are comparable: the SH3 domain NMR structure has a backbone RMSD of 2.6 Å from the crystal structure for the β-strand segments, while the HNP-1 NMR structure has an all-segment backbone RMSD of 2.7 Å from the crystal structure of HNP-3. However, this similarity belies the difference that a larger number of restraints were obtained (292) for SH3, which corresponded to ~5 restraints per residue, and all constraints were independently extracted from NMR. In comparison, 116 restraints were obtained for HNP-1, thus about 4 restraints were available per residue. The HNP-1 structure constraints also contain a smaller number of medium and long-range distances (24) compared to SH3 (170 non-sequential restraints).
For ubiquitin, structure determination using a combination of uniformly 13C-labeled protein and 2-13C selectively labeled protein resulted in 336 inter-residue contacts for the 76-residue protein 49, among which 149 were sequential, 74 were medium range and 113 were long range. Combined with 122 torsion angle restraints, the high-resolution solid-state NMR structure had a ▯1 Å RMSD for secondary structure elements compared to the crystal structure. For the 56-residue GB1, a very high resolution NMR structure was obtained from 888 unique distance restraints and dihedral angles, resulting in a backbone RMSD of 1.32 Å from one of the crystal structures 50.
Conformation of the inter-strand β1-β2 loop and comparison with other human α-defensins
Compared to the structures of other human α-defensins 4, the HNP-1 NMR structure has the same overall fold, with three antiparallel β-strands arranged into a β-sheet. A tight β-hairpin connects β2 and β3 strands while a loop connects the β1 and β2 strands, so that the N- and C-termini lie close together. This structural fold separates the charged and hydrophobic regions spatially, thus may facilitate the insertion of the protein into the phospholipid bilayer with the hydrophobic region buried in the membrane interior while the charged region interacting with the polar surface of the bilayer and with water 3.
Within the context of this general similarity, the SSNMR structure of HNP-1 shows two main differences from the crystal structures of various HNPs. First, the β2 strand in the NMR structure has less orientation change between the N- and C-terminal half compared to the crystal structure (Figure 10b, c), and the strand twist is also less significant. We attribute these differences largely to insufficient distance restraints between the β2 strand and the rest of the protein (9 distances for 7 residues). When the much larger number of inter-residue distances restraints from the 3D CCC experiment were used for structure calculation, the β2 strand was found to have a more similar twist and orientation to those of the HNP-3 structure. Thus, there appears to be no real conformational difference in the β2 strand of HNP-1 and HNP-3.
The second, more important, difference is the conformation of the loop between residues I7 and R15, which controls the relative orientation of the β1 and β2 strands. In the HNP-3 structure, the loop is bent at P8-A9 and protrudes significantly from the rest of the protein, while in the HNP-1 structure, the P8-A9 segment is extended and the loop is tugged closer to the β2 strand (Figure 10d). Several lines of evidence suggest that these are real conformational differences between HNP-1 and HNP-3, rather than artifacts due to insufficient constraints of the NMR structure. The structural inputs for the loop region include the chemical-shift based torsion angles (Table S1) and eleven inter-residue distances (Table 2). The P8 and C10 torsion angles were the main factor that determined the presence or absence of a turn at this position. Although we have not directly measured the P8 ψ torsion angle due to sensitivity limitations, the fact that the C10 φ angle is confirmed by HNCH experiments to have a regular β-strand value suggests that there is a significant possibility that the P8-C10 segment in HNP-1 does not adopt a turn conformation as in HNP-3. Second, eleven inter-residue distances established close contacts between the N-terminal region of the loop (residues I7-P8) and the β3 strand (residues L26-W27), between residue C10 and residues G13-E14 within the loop, and between the end of the loop (residues E14-R15) and the β3 strand (residues F29-C30) (Table 2). Thus, this segment has a reasonable number of distance restraints. When the larger number of independently assigned restraints from 3D CCC experiments were inputted for structure calculation, the loop conformational difference largely remained (Figure 10d), supporting the hypothesis that the conformational difference is real.
It is noteworthy that a previous 1H solution NMR structure of HNP-1 also concluded a different loop orientation 51. Similar to the current result, the solution structure showed the HNP-1 loop to be well defined by itself but that its relation to the rest of the protein was different from that of HNP-3. Moreover, when the HNP-1 solution structure was compared with the structures of the related rabbit defensins NP-2 and NP-5, it was revealed that the loop conformation also differs among these three proteins. Thus, the solution NMR study suggested that the β1-β2 loop conformation may be highly sequence-specific and may play a systematic role in regulating the activities of different α-defensins.
What might be the origin of the loop conformation difference between HNP-1 and HNP-3? It cannot result from fast motion, since the 2D LG-CP spectrum (Figure 11) shows nearly rigid-limit C-H dipolar couplings and 13C variable-temperature spectra (Figure S5) also indicate little temperature dependence. Thus, this conformational difference must be attributed to intrinsic structural differences caused either by the single-residue change at the N-terminus or by external environmental factors such as the ionic content, hydration level, and intermolecular packing. Since the loop conformation difference was also seen in the solution NMR structure 51 where there was no crystal packing effects or hydration problems, the single-residue difference at the N-terminus is likely the main reason for the loop conformational variability. The N-terminus residue is Ala in HNP-1, Asp in HNP-3, and absent in HNP-2, which thus starts its sequence with Cys. Thus, HNP-3 is more polar than HNP-1 or HNP-2 at the N-terminus. Correspondingly, the antimicrobial activities of HNP-1 and HNP-2 are similar but the HNP-3 activity is distinct 2. A recent study of HNP 1-3 against a panel of six bacteria of both Gram-positive and Gram-negative origins showed that the average LD50 of HNP-2 is 1.10 ±0.25 times that of HNP-1 8, indicating very similar potencies, while the average LD50 of HNP-3 is 2.20 ± 1.10 that of HNP-1, indicating that HNP-3 is about 2-fold less active than HNP-1 8. Molecular dynamics simulations comparing the HNP 1-3 structures suggested that Asp2 in HNP-3 may facilitate electrostatic interactions between the two monomers of the dimer, thus making the basket-shaped dimer more compact for HNP-3 than for HNP-1 and 2 52. This difference may in turn cause distinct interactions of HNPs with the lipid membrane.
Whatever the exact molecular reason for the loop conformation difference, given the tight structural constraints imposed by the disulfide bonds and the dominant β-strand motifs for these proteins, one would expect that it is precisely the non-strand residues, without backbone hydrogen bonds, that should modulate the interactions of these proteins with the lipid membrane and tune their antimicrobial activities. Indeed, preliminary spectra of membrane-bound HNP-1 containing site-specific isotopic labels showed that A12 in the loop undergoes an interesting chemical shift change between a more helical conformation and the conformation seen in the microcrystalline state (Figure S6). This data suggests that the β1-β2 loop conformation is sensitive to the environment, in addition to the sequence. Further comparative structural studies are necessary to fully understand the structure-activity relationship of the HNPs and elucidate the functionally relevant conformation of the β1-β2 loop. Solid-state NMR experiments that determine the conformation, orientation and depth of insertion of HNP-1 in lipid bilayers will be crucial for this purpose.
Materials and Methods
HNP-1 expression and microcrystalline sample preparation
Recombinant HNP-1, whose amino acid sequence is shown in Figure 1, was obtained as a cleavage product from its precursor protein, proHNP1, which was expressed as a GST-fusion protein in E. coli and folded. Uniformly 13C, 15N-enriched media Spectra 9 (Cambridge Isotope Laboratories) was used to label the protein. Briefly, the fusion protein GST-proHNP1 was expressed in E. coli BL21 with IPTG induction. The insoluble inclusion bodies were denatured by 8 M urea and then folded in 2 M urea, 3 mM cysteine and 0.3 mM cystine. The folded fusion protein was dialyzed in a pH 7 buffer containing 20 mM Tris-HCl, 150 mM NaCl and 1.5 mM CaCl2. The fusion protein was then cleaved by thrombin, producing proHNP1, which was purified by reversed-phase HPLC. ProHNP1 was further cleaved by cyanogen bromide to yield the correctly folded HNP-1. The crude HNP-1 was purified by reversed-phase HPLC and analyzed by electrospray ionization mass spectrometry to confirm the mass (3634 Da). The yield of HNP-1 was ~3 mg per liter culture. Antimicrobial assays confirmed the activity of the protein. For example, 100% killing of S. aureus is reached at 64 μg/ml HNP-1.
Microcrystalline HNP-1 was precipitated from a polyethylene glycol 400 (PEG-400) solution containing 30 mM cacodylate and 60 mM Li2SO4 at pH 6.5. The PEG-400 solution was added slowly to a protein stock solution to reach a final PEG-400 concentration of 30 wt %. Two protein stock solutions, at concentrations of 30 mg/ml and 36 mg/ml, were used. The protein precipitants were centrifuged in a sealed pipette tip, the supernatant removed, the pipette tip cut open, and the precipitant was centrifuged into a 4 mm MAS rotor. Two HNP-1 samples, one in a 4 mm MAS rotor containing ~4 mg protein and the other in a 2.5 mm MAS rotor (~2.7 mg protein) were used in this study.
Solid-state NMR spectroscopy
Most SSNMR experiments were carried out on a Bruker AVANCE-600 (14.1 Tesla) spectrometer (Karlsruhe, Germany) using triple-resonance MAS probes. Most spectra were measured at 253 – 268 K under MAS frequencies of 8 – 15 kHz. 2D CM5RR spectra were acquired on a 900 MHz NMR spectrometer at the Harvard/MIT Center for Magnetic Resonance. The 900 MHz spectra were measured on a 2.5 mm MAS probe under 20 kHz MAS.
Typical radio-frequency (rf) pulse lengths were 3.5 μs for 13C, 6.0 μs for 15N, and 2.5-4.0 μs for 1H. 1H decoupling fields were typically 70 kHz on the 4 mm rotor sample and 100 kHz on the 2.5 mm rotor sample. 13C chemical shifts were referenced externally to the α-Gly 13CO signal at 176.49 ppm on the TMS scale, and 15N chemical shifts were referenced to the 15N signal of N-acetylvaline at 122.0 ppm on the liquid ammonia scale. The TMS scale differs from the DSS scale by 1.7 ppm, thus all 13C chemical shift values reported here should be increased by 1.7 ppm before comparing with DSS-referenced solution NMR chemical shifts. For torsion angle prediction by the TALOS software, we converted the TMS-based chemical shifts to DSS-referenced values.
2D 13C-13C dipolar-assisted rotational resonance (DARR) experiments 53 were acquired under 8 kHz MAS on the 4 mm rotor sample and 10-15 kHz on the 2.5 mm rotor sample. Four spectra were acquired with mixing times of 20 ms, 40 ms, 100 ms, and 200 ms. For the 4 mm rotor sample, the maximum evolution time for the indirect dimension was 5.6 ms, corresponding to 300 t1 points. The number of scans per t1 slice was 96 and the recycle delay was 2 s, giving an experimental time of ~16 hour per 2D spectrum. For the 2.5 mm rotor sample, the maximum 13C evolution time was increased to 7.2 – 7.4 ms due to the better homogeneity and spectral resolution of the sample. 380 t1 points were measured for the 20 ms and 40 ms experiments and 580 t1 points were measured for the 100 ms and 200 ms mixing times. The experimental time was 18 – 26 hours per 2D spectrum.
Sequence-specific 13C and 15N resonance assignment was carried out by 3D NCOCX and NCACX experiments 34; 35; 54 under 8 kHz MAS at 268 K. Magnetization transfer between 15N and 13CO or 15N and 13Cα was achieved using band-selective SPECIFIC-CP 55. In the intra-residue NCACX experiment, the Cα magnetization was selected by a 17-kHz 15N spin-lock field and a 25-kHz 13C spin lock field on resonance with Cα. In the inter-residue NCOCX experiment, the spin-lock fields were 27 kHz for 15N and 30 kHz for 13C on resonance with Cα. In the latter case, since the CO offset was 18 kHz, the effective field of the CO resonances was ~35 kHz, thus satisfying the sideband matching condition with 15N. After SPECIFIC-CP, 13C-13C magnetization transfer occurred during a DARR mixing time, which was 40 ms for NCACX and 60 ms for NCOCX.
For the NCOCX experiment, the 15N spectral width was 4 kHz and the maximum t1 evolution time was 4.75 ms, corresponding to 38 t1 points. The 13CO spectral width was 2 kHz and the number of t2 points was 22, giving a maximum t2 of 5.56 ms. The experimental time was about 2.5 days. For the NCACX experiment, the 15N spectral width was the same as that of NCOCX, while the 13Cα (ω2) spectral width was 4.5 kHz and the acquisition time was 5.56 ms, corresponding to 50 t2 points. The experimental time was 2 days.
The 2D 15N-15N correlation experiment was conducted using 1H-driven spin diffusion (PDSD) under 6 kHz MAS at 253 K. The mixing time was 3 s to obtain inter-residue 15N-15N correlation peaks. The indirect dimension has a spectral width of 16 kHz and an acquisition time of 6.88 ms, corresponding to 220 t1 points. The number of scan per t1 slice was 144 and the recycle delay was 1.5 s, giving an experimental time of 40 hours.
The N-H doubled HNCH experiment to determine φ torsion angles 36; 47 were carried out on the 2.5 mm rotor sample under 8 kHz MAS at 263 K. 1H homonuclear decoupling during the N-H and C-H evolution periods were achieved using the FSLG sequence 56. The Cα peaks of the chemical shift dimension were assigned based on the 2D and 3D correlation spectra. Although resonance overlap was unavoidable, most residues have consistent φ angles between the TALOS and the crystal structure results, thus allowing the residues of interest to be measured. For sensitivity reasons, only the intensities at 0 and half a rotor period of dipolar evolution times were measured. Simulations (Figure S4) indicate that the intensity at half a rotor period is the most sensitive to φ angle variations.
Structure calculation
The 13Cα, 13Cβ, 13CO and 15N chemical shifts were inputted into the TALOS program 46 to obtain (φ, ψ) torsion angles. Structure calculations were performed using the simulated annealing protocol of the XPLOR-NIH program 57; 58. Input constraints included 60 torsion angles, 9 distances associated with the disulfide bonds, 46 distances between sequential residues, and 24 non-sequential inter-residue distances. An ensemble of 200 structures were first calculated by performing molecular dynamics at 3500 K for 40 ps, followed by slow cooling from 3500 K to 25 K in 12.5 K steps. At each temperature 0.4 ps of dynamics was performed using a soft square NOE potential. Each structure was then refined with the same protocol but with 10 ps of initial annealing using a hard square potential with the kNOE force constant held at 30 kcal. After refinement, the 10 lowest-energy structures were chosen to represent the final structure of HNP-1. All structures were visualized in Chimera (UCSF).
Accession numbers
The HNP-1 SSNMR structure coordinates have been deposited in the Protein Data Bank (PDB code: 2KHT). The chemical shifts and distance restraints have been deposited in the Biological Magnetic Resonance Bank (accession number: rcsb101138).
Supplementary Material
Acknowledgement
The authors thank Chih-Chia Su for assistance with microcrystal visualization, Professor Chad Rienstra for sharing the design of microcrystal transfer tools, Professor Robert Griffin, Dr. Jozef Lewandowski, Dr. Gaël De Paëpe and Dr. Tony Bielecki for assistance with the 900 MHz NMR experiments, and Dr. Andrew Severin and Dr. Shenhui Li for help with the XPLOR-NIH structure calculations.
This work is supported by the NIH grant GM66976 to M.H. and P41-EB-002026 for the 900 MHz NMR time at MIT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 2.Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, Lehrer RI. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 4.Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–60. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabay JE, Scott RW, Campanelli D, Griffith J, Wilde C, Marra MN, Seeger M, Nathan CF. Antibiotic proteins of human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5610–5614. doi: 10.1073/pnas.86.14.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DE, Bevins CL. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 7.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 8.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob. Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Ericksen B, Tucker K, Lubkowski J, Lu W. Synthesis and characterization of human alpha-defensins 4-6. J. Pept. Res. 2004;64:118–125. doi: 10.1111/j.1399-3011.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer RI, Lichtenstein AK, Ganz T. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 11.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 12.Hong M. Structure, topology, and dynamics of membrane peptides and proteins from solid-state NMR spectroscopy. J. Phys. Chem. B. 2007;111:10340–10351. doi: 10.1021/jp073652j. [DOI] [PubMed] [Google Scholar]
- 13.Doherty T, Waring AJ, Hong M. Dynamic structure of disulfide-removed linear analogs of tachyplesin-I in the lipid bilayer from solid-state NMR. Biochemistry. 2008;47:1105–1116. doi: 10.1021/bi701390t. [DOI] [PubMed] [Google Scholar]
- 14.Doherty T, Waring AJ, Hong M. Membrane-bound conformation and topology of the antimicrobial peptide tachyplesin-I by solid-state NMR. Biochemistry. 2006;45:13323–13330. doi: 10.1021/bi061424u. [DOI] [PubMed] [Google Scholar]
- 15.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a b-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Membrane-bound dimer structure of a b-hairpin antimicrobial peptide from rotational-echo double-resonance solid-state NMR. Biochemistry. 2006;45:8341–8349. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- 17.Tang M, Waring AJ, Hong M. Mechanism of Arg insertion into lipid membranes and pore formation by a cationic peptide. J. Am. Chem. Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 18.Tang M, Waring AJ, Lehrer RI, Hong M. Effects of Guanidinium-Phosphate Hydrogen Bonding on the Membrane-Bound Structure and Activity of an Arginine-Rich Membrane Peptide from Solid-State NMR. Angew. Chem. Int. Ed. Engl. 2008;47:3202–3205. doi: 10.1002/anie.200705993. [DOI] [PubMed] [Google Scholar]
- 19.Tang M, Waring AJ, Lehrer RI, Hong M. Orientation of a b-hairpin Antimicrobial Peptide in Lipid Bilayers from 2D Dipolar Chemical-Shift Correlation NMR. Biophys. J. 2006;90:3616–3624. doi: 10.1529/biophysj.105.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buffy JJ, Hong T, Yamaguchi S, Waring A, Lehrer RI, Hong M. Solid-State NMR Investigation of the Depth of Insertion of Protegin-1 in Lipid Bilayers Using Paramagnetic Mn2+ Biophys. J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buffy JJ, McCormick MJ, Wi S, Waring A, Lehrer RI, Hong M. Solid-State NMR Investigation of the Selective Perturbation of Lipid Bilayers by the Cyclic Antimicrobial Peptide RTD-1. Biochemistry. 2004;43:9800–9812. doi: 10.1021/bi036243w. [DOI] [PubMed] [Google Scholar]
- 22.Kagan BL, Selsted ME, Ganz T, Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl. Acad. Sci. U. S. A. 1990;87:210–214. doi: 10.1073/pnas.87.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohner K, Latal A, Lehrer RI, Ganz T. Differential scanning microcalorimetry indicates that human defensin, HNP-2, interacts specifically with biomembrane mimetic systems. Biochemistry. 1997;36:1525–1531. doi: 10.1021/bi961300p. [DOI] [PubMed] [Google Scholar]
- 25.Castellani FR,B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 26.Igumenova T, Wand A, McDermott A. Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126:5323–31. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
- 27.Franks W, Zhou D, Wylie B, Money B, Graesser D, Frericks H, Sahota G, Rienstra C. Magic-angle spinning solid-state NMR spectroscopy of the beta1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J. Am. Chem. Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 28.Marulanda D, Tasayco M, Cataldi M, Arriaran V, Polenova T. Resonance assignments and secondary structure analysis of E. coli thioredoxin by magic angle spinning solid-state NMR spectroscopy. J. Phys. Chem. B. 2005;109:18135–45. doi: 10.1021/jp052774d. [DOI] [PubMed] [Google Scholar]
- 29.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 2002;99:16742–7. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 31.Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cady SD, Hong M. Amantadine-Induced Conformational and Dynamical Changes of the Influenza M2 Transmembrane Proton Channel. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M. Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature. 2006;440:959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 34.Hong M. Resonance Assignment of 13C/15N Labeled Proteins by Two- and Three-Dimensional Magic-Angle-Spinning NMR. J. Biomol. NMR. 1999;15:1–14. doi: 10.1023/a:1008334204412. [DOI] [PubMed] [Google Scholar]
- 35.Rienstra CM, Hohwy M, Hong M, Griffin RG. 2D and 3D 15N-13C-13C NMR chemical shift correlation spectroscopy of solids: assignment of MAS spectra of peptides. J. Am. Chem. Soc. 2000;122:10979–10990. [Google Scholar]
- 36.Hong M, Gross JD, Griffin RG. Site-resolved determination of peptide torsion angle phi from the relative orientations of backbone N-H and C-H bonds by solid-state NMR. J. Phys. Chem. B. 1997;101:5869–5874. [Google Scholar]
- 37.Costa PR, Gross JD, Hong M, Griffin RG. Solid-State NMR Measurement of psi in Peptides: a NCCN 2Q-Heteronuclear Local Field Experiment. Chem. Phys. Lett. 1997;280:95–103. [Google Scholar]
- 38.Feng X, Eden M, Brinkmann A, Luthman H, Eriksson L, Graslund A, Antzutkin ON, Levitt MH. Direct determination of a peptide torsion angle psi by double-quantum solid-state NMR. J. Am. Chem. Soc. 1997;119:12006–12007. [Google Scholar]
- 39.Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. Measurement of 13C-15N distances in uniformly 13C labeled biomolecules: J-decoupled REDOR. J. Am. Chem. Soc. 1999;121:10237–10238. [Google Scholar]
- 40.Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. Through the looking glass: Mechanistic insights from enantiomeric human defensins. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.018085. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hohwy M, Jakobsen HJ, Eden M, Levitt MH, Nielsen NC. Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: a compensated C7 pulse sequence. J. Chem. Phys. 1998;108:2686–2694. [Google Scholar]
- 42.De Paëpe G, Bayro MJ, Lewandowski J, Griffin RG. Broadband homonuclear correlation spectroscopy at high magnetic fields and MAS frequencies. J. Am. Chem. Soc. 2006;128:1776–1777. doi: 10.1021/ja0550430. [DOI] [PubMed] [Google Scholar]
- 43.Kornhaber GJ, Snyder D, Moseley HN, Montelione GT. Identification of zinc-ligated cysteine residues based on 13Calpha and 13Cbeta chemical shift data. J. Biomol. NMR. 2006;34:259–269. doi: 10.1007/s10858-006-0027-5. [DOI] [PubMed] [Google Scholar]
- 44.Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 45.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 46.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 47.Hong M, Gross JD, Rienstra CM, Griffin RG, Kumashiro KK, Schmidt-Rohr K. Coupling Amplification in 2D MAS NMR and Its Application to Torsion Angle Determination in Peptides. J. Magn. Reson. 1997;129:85–92. doi: 10.1006/jmre.1997.1242. [DOI] [PubMed] [Google Scholar]
- 48.Huster D, Yamaguchi S, Hong M. Efficient beta-sheet identification in proteins by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2000;122:11320–11327. [Google Scholar]
- 49.Zech S, Wand A, McDermott A. Protein structure determination by high-resolution solid-state NMR spectroscopy: application to microcrystalline ubiquitin. J Am Chem Soc. 2005;127:8618–26. doi: 10.1021/ja0503128. [DOI] [PubMed] [Google Scholar]
- 50.Franks WT, Wylie BJ, Schmidt HL, Nieuwkoop AJ, Mayrhofer RM, Shah GJ, Graesser DT, Rienstra CM. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pardi A, Zhang XL, Selsted ME, Skalicky JJ, Yip PF. NMR studies of defensin antimicrobial peptides. 2. Three-dimensional structures of rabbit NP-2 and human HNP-1. Biochemistry. 1992;31:11357–11364. doi: 10.1021/bi00161a013. [DOI] [PubMed] [Google Scholar]
- 52.Lourenzoni MR, Namba AM, Caseli L, Degrève L, Zaniquelli ME. Study of the interaction of human defensins with cell membrane models: relationships between structure and biological activity. J. Phys. Chem. B. 2007;111:11318–11329. doi: 10.1021/jp067127g. [DOI] [PubMed] [Google Scholar]
- 53.Takegoshi K, Nakamura S, Terao T. 13C - 1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 54.Straus S, Bremi T, Ernst R. Experiments and strategies for the assignment of fully 13C/15N-labelled polypeptides by solid state NMR. J. Biomol. NMR. 1998;12:39–50. doi: 10.1023/a:1008280716360. [DOI] [PubMed] [Google Scholar]
- 55.Baldus M, Geurts DG, Hediger S, Meier BH. Efficient N-15-C-13 polarization transfer by adiabatic-passage Hartmann-Hahn cross polarization. J. M. R. 1996;118:140–144. [Google Scholar]
- 56.Bielecki A, Kolbert AC, Levitt MH. Frequency-switched pulse sequences: homonuclear decoupling and dilute spin NMR in solids. Chem. Phys. Lett. 1989;155:341–346. [Google Scholar]
- 57.Schwieters C, Kuszewski J, Tjandra N, Clore G. The Xplor-NIH NMR Molecular Structure Determination Package. J. Magn. Res. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 58.Schwieters C, Kuszewski J, Clore G. Using Xplor-NIH for NMR molecular structure determination. Progr. NMR Spectroscopy. 2006;48:47–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.