Abstract
Real-time reverse transcription PCR (RT-qPCR) relies on a housekeeping or normalizer gene whose expression remains constant throughout the experiment. RT-qPCR is commonly used for characterization of human bone marrow mesenchymal stem cells (hBMSCs). However, to the best of our knowledge, there are no studies validating the expression stability of the genes used as normalizers during hBMSCs differentiation. This work aimed to study the stability of the housekeeping genes β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal protein L13A (RPL13A) during the osteogenic differentiation of hBMSCs. Their stability was evaluated via RT-qPCR in 14 and 20 day differentiation assays to the osteogenic lineage. Different normalization strategies were evaluated to quantify the osteogenic markers collagen type I, bone sialoprotein and osteonectin. Cell differentiation was confirmed via alizarin red staining. The results demonstrated up-regulation of β-actin with maximum fold changes (MFC) of 4.38. GAPDH and RPL13A were not regulated by osteogenic media after 14 days and presented average fold changes lower than 2 in 20 day cultures. RPL13A (MFC < 2) had a greater stability when normalizing as a function of culture time compared with GAPDH (MFC ≤ 2.2), which resulted in expression patterns of the osteogenic markers more consistent with the observed differentiation process. The results suggest that β-actin regulation could be associated with the morphological changes characteristic of hBMSCs osteogenic differentiation, and provide evidence for the superior performance of RPL13A as a normalizer gene in osteogenic differentiation studies of hBMSCs. This work highlights the importance of validating the normalizer genes used for stem cells characterization via RT-qPCR.
Keywords: Mesenchymal stem cells, Osteogenic differentiation, Normalization, Gene expression, Quantitative RT-PCR
Introduction
Since the introduction of real-time (quantitative) reverse transcription PCR (RT-qPCR) and the development of relative quantification models for gene expression assays, many studies in the literature have relied on conventional housekeeping genes, whose expression was long assumed to be stable, as internal references or normalizers (Bustin 2000; Huggett et al. 2005; Szabo et al. 2004). However, an increasing number of reports have demonstrated the instability in the gene expression levels of popular housekeeping genes such as β-actin (ACTB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal RNA (rRNA) among others under different experimental conditions (Bustin 2000; Schmittgen and Zakrajsek 2000; Tricarico et al. 2002; Kok et al. 2005). These reports showed the need to validate the housekeeping gene stability for particular tissues (i.e. cell lineages) and experimental treatments (Bustin 2000; Huggett et al. 2005; Wong and Medrano 2005). Moreover, they suggested that the use of any housekeeping gene without prior validation rendered gene expression assays via RT-qPCR unreliable (Huggett et al. 2005; Szabo et al. 2004; Tricarico et al. 2002; Wong and Medrano 2005). Indeed, instability in the expression levels of housekeeping genes may be misinterpreted as variations in the levels of the genes of interest (i.e. biomarkers) or simply affect their accurate quantification.
Recent developments in the areas of tissue engineering and regenerative medicine have focused on the potential therapeutic use of autologous adult stem cells (Grayson et al. 2004; Kim et al. 2005). In this context, human bone marrow mesenchymal stem cells (hBMSCs) have been widely studied for their relative easy access and their multipotentiality (Bianco et al. 2001; Caterson et al. 2002). Particularly, the osteogenic potential of hBMSCs has been extensively exploited for the biological evaluation of scaffolds or materials with applications in the area of bone tissue engineering (Meinel et al. 2004a, b, 2006). In addition, since 2002 an increasing number of studies using hBMSCs have used RT-qPCR for the characterization of differentiation processes (Frank et al. 2002; Mygind et al. 2007). However, no reports were found validating the stability of housekeeping genes used as normalizer genes in studies of hBMSCs differentiation.
To address this issue, this work aimed to study the stability of two of the most commonly used housekeeping genes in osteogenic differentiation studies of stem cells: ACTB and GAPDH (Vandesompele et al. 2002; Cho et al. 2005; Mbalaviele et al. 2005), in hBMSCs undergoing osteogenic differentiation. The results were compared with the stability of the gene ribosomal protein L13A (RPL13A), which has been shown to have stable expression levels in bone marrow tissues (Vandesompele et al. 2002). Furthermore, various normalization strategies were evaluated for the quantification of the osteogenic markers collagen type I (CI), bone sialoprotein II (BSP) and osteonectin (ON), to assess the effects of housekeeping gene variability on their expression patterns.
Materials and methods
Isolation and expansion of human bone marrow mesenchymal stem cells
Human bone marrow mesenchymal stem cells were extracted from the trabecular bone in the femoral head of patients after total hip arthroplasty, as described elsewhere with some modifications (Schutze et al. 2005; Lee et al. 2003; Sakaguchi et al. 2004; Quiroz et al. 2008). The protocol was approved by the Review Boards of the Hospital Pablo Tobón Uribe and the CES University, and all samples were processed after written informed consent was obtained. Briefly, the trabecular bone was extracted, washed with a phosphate buffered saline solution (PBS) (Gibco, USA) to facilitate the disaggregation of the tissue, and mechanically dissected to obtain fragments of approximately 2 mm3. The obtained solution was recollected and filtered with a 70 μm cell strainer (Falcon, USA) before centrifuging at 400g for 10 min. The pellets were resuspended in non-osteogenic medium (NO) consisting of Dulbecco’s modified Eagle’s Medium (DMEM) (Sigma, USA), supplemented with 10% Fetal Bovine Serum (FBS) (Gibco, USA) and 1% Antibiotics (streptomycin and penicillin) (Gibco, USA); and were cultured in 25 cm2 flasks at 37 °C in a humidified atmosphere containing 5% CO2. At day 4 the cultures were washed with PBS to remove the non-adherent cells and were further expanded until they reached ~80% confluency, when they were harvested and cultured in 75 cm2 flasks. After subculture these cells were designated as passage 1; cells at passage 5 were used to study the stability of the housekeeping genes. Since different mesenchymal stem cell populations are known to have different differentiation potentials, housekeeping stability was studied in a single population to eliminate inter-patient variability.
Osteogenic differentiation
Two differentiation experiments, for 14 and 20 days, were performed, and the differentiation was assessed via Alizarin Red staining and RT-qPCR. To induce the osteogenic differentiation of the established hBMSCs, 7 × 104 cells were seeded in 12 multi-well plates, at an estimated confluency of 80%, and cultured in osteogenic medium (OM), which consisted of NO supplemented with 0.2 mM ascorbic acid (Amresco, USA), 10 mM β-Glycerol Phosphate (Sigma, USA) and 100 nM Dexamethasone (Sigma, USA) (Meinel et al. 2006). Each experiment comprised 12 cultures, 6 under differentiation conditions and 6 controls under normal culture conditions (NO), in order to have triplicate cultures for each evaluation method. Medium was changed every 2–3 days.
Alizarin red staining
Alizarin Red staining was used to verify the state of differentiation in terms of the extracellular matrix mineralization (Colter et al. 2001). Briefly, cells were fixed for 20 min with 70% cold ethanol before 500 μl Alizarin Red S (ARS) (Sigma, USA) at 2% (W/V) pH 4.0 were added for additional 20 min. ARS solution was washed with ultra pure water and the cultures were imaged by phase contrast microscopy (Nikon Eclipse TS100, USA).
RNA extraction and quantification
RNA was extracted from cell monolayers following the instructions of the manufacturer (RNeasy Mini Kit, Qiagen). The isolated RNA was eluted in 40 μl RNase-free water and 4:100 quantification stocks were prepared for RNA quantification, using the Quant-iT RiboGreen Kit (Invitrogen, USA). This method was selected over conventional spectrophotometry to provide optimal accuracy and reproducibility for RNA quantification, considering that RNA input in the RT-qPCR reactions constituted the only source for normalization. Briefly, the reactions were performed in a final volume of 100 μl by incubation in the real-time thermocylcer RotorGene 6000 (Corbett, Australia) for 2 min at 25 °C followed by 10 cycles for fluorescence acquisition for 10 s each at 25 °C. To transform fluorescence data into RNA concentration a calibration curve was constructed, using the standard RNA provided in the kit, diluted from 100 to 500 μg/ml in order to obtain maximum linearity. The quantifications were performed in triplicate.
Quantitative RT-PCR (RT-qPCR)
A two step RT-PCR protocol was used for mRNA quantification. Briefly, 350 ng of RNA were reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen, USA) and following the instructions of the manufacturer. Reverse transcription (RT) controls with no enzyme were prepared in order to discriminate from genomic DNA contaminations. cDNA and RT control samples were evaluated via quantitative PCR following the instructions of the manufacturer (QuantiTect SYBR Green, Qiagen). Reactions were carried out with a final volume of 25 μl and using 300 nM for each primer (Table 1). The reactions were incubated in the real-time thermocycler RotorGene 6000 (Corbett, Australia). The PCR program consisted of an initial hold at 50 °C for 2 min followed by an activation step at 95 °C for 15 min, and 45 cycles at 95° C for 15 s, annealing at 60 °C for 30 s, and 72 °C for 30 s. Then, a melting curve was constructed by heating from 65 °C to 95 °C with temperature steps of 0.4 °C. All reactions included 0.25 U of uracil-DNA-glycosylase (Fermentas, USA) to avoid contamination with PCR products from previous reactions (Kleiboeker 2005). The experiments evaluating the stability of the housekeeping genes were performed in triplicate and the gene expression assays with the osteogenic markers were performed in duplicate (including the normalizer genes).
Table 1.
Primer sequences of the tested housekeeping genes ACTB, GAPDH, RPL13A and of the osteogenic markers collagen type I (CI), osteonectin (ON) and bone sialoprotein II (BSP)
| Gene | Forward primer | Reverse primer | Product (bp) |
|---|---|---|---|
| Housekeeping genes | |||
| ACTBa | CTGGAACGGTGAAGGTGACA | AAGGGACTTCCTGTAACAATGCA | 140 |
| GAPDH | CGACCACTTTGTCAAGCTCA | GAGGGTCTCTCTCTTCCTCT | 150 |
| RPL13A | CTATGACCAATAGGAAGAGCAACC | GCAGAGTATATGACCAGGTGGAA | 121 |
| Osteogenic markers | |||
| CI | TTCGGAGGAGAGTCAGGAAG | CACAAGGAACAGAACAGAACAGTC | 116 |
| ON | TCCACAGTACCGGATTCTCTCT | TCTATGTTAGCACCTTGTCTCCAG | 107 |
| BSP | GCAGTAGTGACTCATCCGAAGAA | GCCTCAGAGTCTTCATCTTCATTC | 121 |
aPrimer sequence described by Vandesompele et al. (2002)
Evaluation of normalizer genes
For the analysis of the expression stability of the genes ACTB, GAPDH and RPL13A, the quantitative RT-PCR data were analyzed using two different approaches: (1) Statistical analysis of the threshold cycle (CT) variance and (2) calculation of average fold changes (AFC) from the mean and average maximum variability (MFC), by transforming threshold cycle values into fold differences with the formula 2−ΔCt and using the mean or maximum/minimum CT from control cultures (calibrators), as previously described (Dheda et al. 2004). Briefly, CT values were calculated as the fraction of PCR cycle where the reaction fluorescence reached a set threshold of 0.02 lying in the exponential region of the amplification curves. The mean, maximum and minimum CT values were then calculated for all the samples corresponding to NO cultures, which were defined as the calibrators for each evaluation time point. The AFC measures the average—conservative—fold change in the expression levels of the normalizer gene by subtracting the CT value for each reaction under osteogenic conditions from the mean CT for the corresponding non-osteogenic cultures, and calculating the fold change as 2−ΔCt. The calculated values are then averaged to yield the AFC for a given gene and time point. The calculation of MFC is analogous to that of AFC values, but using maximum or minimum CT values from the NO cultures to estimate what the maximum—non-conservative—possible variability could be with respect to the calibrator condition. Noteworthy, an ideal normalizer gene has AFC and MFC of 1. Statistical analysis was performed using STATGRAPHICS Software (Statistical Graphics Corp., Version 5), with an unstacked one-way ANOVA at a 95% confidence level.
In order to use the second proposed strategy for evaluating gene stability, as well as quantifying the gene expression levels of the osteogenic markers (as explained below), which both relied on the assumption that for the designed primers the amplification efficiency (E) is nearly 100% (i.e. E ≈ 2), the experimental amplification efficiency was calculated for all the reactions performed in this study using LinRegPCR (Ramakers et al. 2003). A hypothesis test (95% confidence level) with null hypothesis E = 2 demonstrated that the assumption was valid, since all the p-values were above 0.05 (Quiroz et al. 2008).
Expression patterns of osteogenic markers
The quantification of the osteogenic genes was performed with a newly synthesized cDNA for all the evaluated conditions (14/20 days, OM/NO) in order to eliminate inter-assay variability (Hellemans et al. 2007). The analysis of the expression levels of CI, BSP and ON served to study the effects of different normalization strategies on their expression patterns, through normalization by ACTB, GAPDH, RPL13A or a combination of GAPDH and RPL13A. Each gene was evaluated in independent PCR runs including the complete set of samples (i.e. 14/20 days and OM/NO) to allow comparison between all treatments and evaluation times. Data from the osteogenic markers were processed with the qBase software to calculate gene expression levels as fold changes using the 2−ΔCt method, in which the 14 day cultures under non-osteogenic conditions were designated as calibrators (Hellemans et al. 2007).
Results
Cell differentiation and matrix mineralization
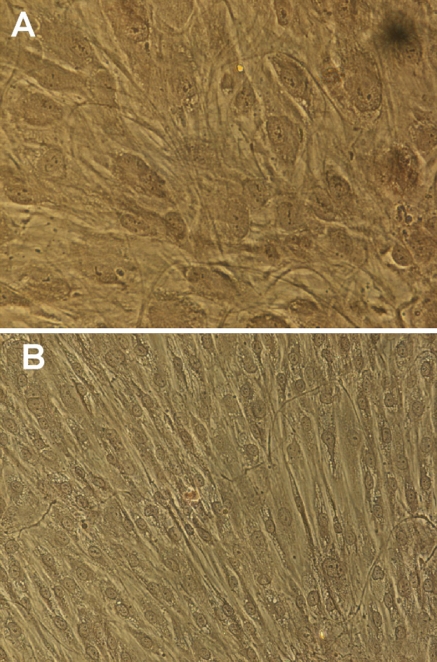
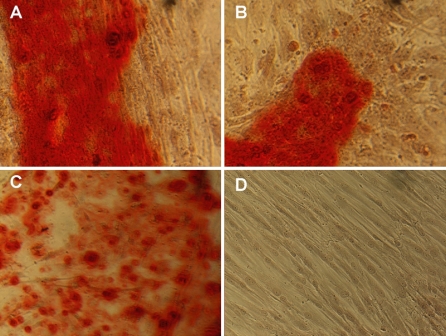
The osteogenic potential of the isolated mesenchymal cells was confirmed qualitatively by evaluating changes in cell morphology and calcium synthesis. Cells under normal culture conditions (NO) maintained an undifferentiated phenotype with a marked fibroblast-like morphology (Fig. 1b). Despite the morphological changes induced by the OM after 5 days of incubation, matrix staining with ARS did not reveal calcium nodules at day 14 (Fig. 1a, b). Cultures evaluated at day 20, however, presented positive ARS staining, indicative of calcium nodule formation and matrix mineralization (Fig. 2a–c).
Fig. 1.
Alizarin Red staining in 14 day cultures. a Cells under osteogenic (×20) or b normal culture conditions (×20). Extracellular matrix mineralization was not observed
Fig. 2.
Alizarin Red staining in 20 day cultures. a–c Cells under osteogenic or d normal culture conditions. Matrix mineralization was only observed (red-stained) in cultures with osteogenic media. a ×40, b–d ×20
Evaluation of normalizer genes
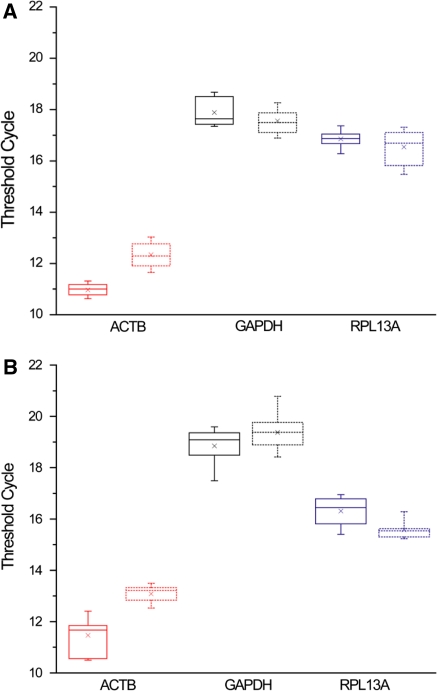
The threshold cycle values obtained for the evaluated housekeeping genes in the 14 and 20 day differentiation experiments are shown in Fig. 3. Statistical analysis of the CT values measured for the 14 day experiment revealed that neither GAPDH (p = 0.2045) nor RPL13A (p = 0.8384) were regulated by the OM, since there were no significant differences between the mean CT for the osteogenic and control cultures. In contrast, ACTB presented statistically significant differences (p < 0.00001). At a more advanced stage of cell differentiation (day 20), CT values of GAPDH remained unaffected by the treatment (p = 0.1393), but RPL13A (p = 0.0059) and ACTB (p < 0.00001) showed significant differences between treated and control cultures.
Fig. 3.
Analysis of the threshold cycle values (CTs). a Results for the 14 and b 20 day differentiation experiments. For each gene the box with solidline corresponds to the osteogenic cultures and the dotted one to the controls, and delimits the 25 and 95 percentile for each data set. The median and mean CT values are indicated by the line and the star within the box, respectively
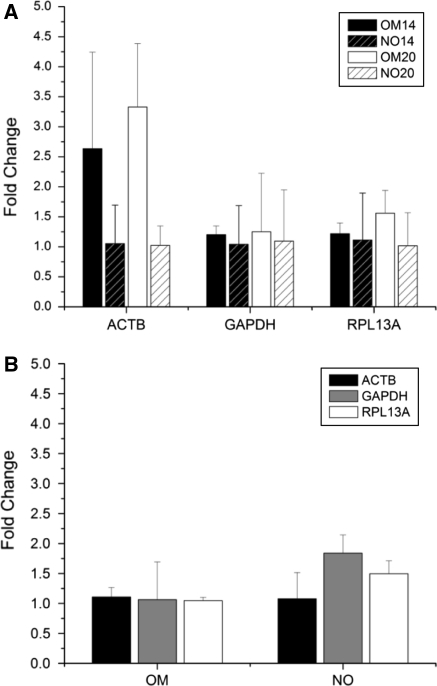
Figure 4 shows the analysis of the normalizer genes stability in terms of the average fold changes from the mean CT (AFC) and the maximum fold change from the maximum/minimum CT (MFC). The data in Fig. 4 do not necessarily represent an up-regulation of the gene expression levels, since for those cases where down regulation occurred, the reciprocal value is displayed in order to facilitate the comparison of the fold changes. Fig. 4b was prepared with data derived from the gene expression assays for the quantification of the osteogenic markers, since it was not valid to establish comparisons as a function of the evaluation time with the data set used in Fig. 4a due to potential inter-run variability.
Fig. 4.
Fold changes in gene expression. a Average fold changes (AFC, columns) calculated for each day and gene evaluated (OM vs. NO). b AFC and maximum fold changes (MFC) as a function of evaluation time (20 vs. 14 days). MFC are presented as errorbars. OMxxd: cultures incubated in osteogenic medium for xx days (i.e. 14 or 20), NOxxd: cultures incubated in non-osteogenic medium for xx days
The results indicated the higher variability of ACTB and the regulation of its expression levels as a function of the incubation time in osteogenic medium, with AFC of 2.64 and 3.33 for the 14 and 20 days experiments, respectively, and MFC < 5. GAPDH and RPL13A showed comparable stability at day 14, since no significant differences were found in their AFC (p = 0.9330) and MFC (p = 0.7865). Both housekeeping genes had an average 20% increase in their expression levels under osteogenic conditions; MFC values for GAPDH and RPL13A were 1.34 and 1.39, respectively. Later in the differentiation process (day 20), a small increase in AFC was observed for GAPDH (AFC = 1.25, 4% increase) and RPL13A (AFC = 1.56, 22% increase). Thus, RPL13A presented an AFC 19.8% higher than GAPDH, but RPL13A had an MFC 12.7% lower (MFC of 1.94 and 2.22, respectively). Fig. 4b shows that all genes had AFC lower than 2, with ACTB and RPL13A as the most stable. For instance, ACTB showed excellent performance in cultures with and without osteogenic medium, with AFC of 1.1 and 1.07, respectively, when comparing the same treatments at the two evaluation times. In contrast, GAPDH presented a higher variability in this assay, particularly in cultures without osteogenic medium (MFC = 2.14).
Expression of osteogenic markers
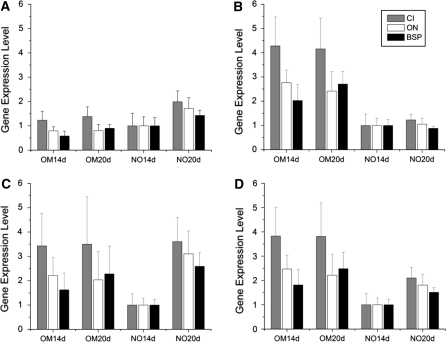
The effects of using different normalization strategies (i.e. selection of normalizer gene) on the expression levels of the tested osteogenic markers (BSP, ON and CI) are shown in Fig. 5. Marked differences in the expression patterns were observed for the different normalization strategies that were applied. For ACTB an up-regulation in the expression levels of the 3 osteogenic markers was observed in NO cultures as a function of incubation time. In contrast, when normalizing with RPL13A all osteogenic genes appeared overexpressed in cultures with OM, without significant changes between the two evaluation times for CI (p = 0.4314) and ON (p = 0.1471) and a 25% increase for BSP (p = 0.0017) at day 20. GAPDH produced a mixed pattern with up-regulation of the 3 markers for osteogenic cultures in the 14 day experiment, and up-regulation in control cultures in the 20 day experiment.
Fig. 5.
Gene expression levels of the osteogenic markers using different combinations of the evaluated normalizer genes. Normalizer used: a ACTB, b RPL13A, c GAPDH and d GAPDH and RPL13A. Errorbars correspond to the standard error of the mean. OMxxd: cultures incubated in osteogenic medium for xx days (i.e. 14 or 20), NOxxd: cultures incubated in non-osteogenic medium for xx days
Discussion
The expression stability of the housekeeping genes used as normalizers determines the sensibility and reliability of mRNA quantifications via RT-qPCR (Szabo et al. 2004; Dheda et al. 2004; Aerts et al. 2004; Glare et al. 2002; Biederman et al. 2004; Johansson et al. 2007; García-Crespo et al. 2005). This work studied the stability of two of the most used housekeeping genes, ACTB and GAPDH, and one previously shown to be stable in bone marrow tissues, RPL13A, as potential candidates to be normalizer genes in hBMSCs differentiation assays to the osteogenic lineage (Vandesompele et al. 2002). The evaluation time points are representative of two different stages of the hBMSCs differentiation process, as demonstrated by the observed differences in the extent of matrix mineralization (Figs. 1, 2). These observations provide support for the selection of the evaluation time points herein studied, as they enable the evaluation of the housekeeping gene stability under two distinct stages of the differentiation process. Similarly, the increasing matrix mineralization can be expected to correlate with an up-regulation in the expression levels of the osteogenic markers CI, ON and BSP (Frank et al. 2002; Mygind et al. 2007).
After an extensive review of the related literature, no previous reports about the validation of normalizer genes for gene expression assays of hBMSCs during osteogenic differentiation were found. The first report, as stated by the authors, studying hBMSCs differentiation to the osteogenic lineage via quantitative RT-PCR corresponds to the work by Frank et al. (2002). In that study, the authors used 18S rRNA as normalizer gene based on preliminary assays comparing its stability against GAPDH. Later reports, as the one by Catelas et al. (2006), also used 18S rRNA as a normalizer gene under similar experimental conditions. Other studies, as recent as the works by Cho et al. (2005), Abdallah et al. (2005) and Friedman et al. (2006), used ACTB for normalizing the gene expression levels of hBMSCs under osteogenic conditions. Other contemporary publications reported the use of GAPDH for normalization purposes with hBMSCs and osteogenic conditions (Mbalaviele et al. 2005; Sumanasinghe et al. 2006; Tsukahara et al. 2006). Furthermore, none of these studies provided supporting information for the selection of the normalizer gene.
Mygind et al. (2007) studied hBMSCs osteogenic differentiation on hydroxyapatite scaffolds, using an immortalized mesenchymal cell line (hMSC-TERT4) (Mygind et al. 2007). They used a normalization strategy (BestKeeper index) based on the geometric mean of the expression levels of three housekeeping genes (GAPDH, Ubiquitin C and RNA Polymerase II) (Mygind et al. 2007; Pfaffl et al. 2004). Unfortunately, the stability data of these normalizer genes was not reported. Noteworthy, the use of several housekeeping genes does not guarantee the reliability of the normalization process, as it depends on the stability of the selected genes, which was also observed in this study (Fig. 5d).
On the other hand, it is important to highlight that the selection of 18S rRNA, as in the pioneering work by Frank et al. (2002), is in opposition with recent normalization studies that have avoided its use as a normalizer gene, even when it has been shown to be more stable than other housekeeping genes evaluated. The main arguments against the use of 18S rRNA are: (1) its high abundance of expression making accurate quantification difficult (i.e. removing fluorescence base line), (2) its relative resistance to degradation compared to mRNA, and (3) the imperfect correlation of expression of rRNA with mRNA, as rRNA transcription is carried out by a different RNA polymerase (Huggett et al. 2005; Aerts et al. 2004; Biederman et al. 2004; Goossens et al. 2005; Zhang et al. 2005).
ACTB is probably the most commonly used housekeeping gene for expression assays via RT-qPCR (Huggett et al. 2005; Schmittgen and Zakrajsek 2000; Glare et al. 2002). After the first reports evaluating housekeeping genes stability numerous authors demonstrated ACTB instability, suggesting its unsuitability for normalization purposes (Huggett et al. 2005; Aerts et al. 2004). However, ACTB continues to be used as a normalizer gene (Oshina et al. 2007). Additional studies have demonstrated that ACTB presents superior stability over a pool of housekeeping genes (including GAPDH) for certain tissues and treatments (Biederman et al. 2004; Johansson et al. 2007). In the present study, ACTB presented the highest variability compared with GAPDH and RPL13A, with AFC > 2 and MFC < 5, and its expression levels were demonstrated to be regulated by osteogenic media (Fig. 4a). It should be noted that the MFC values herein reported for ACTB are considerably low compared with other reports showing fold changes up to 35X (Dheda et al. 2004; Aerts et al. 2004). Interestingly, ACTB regulation by the osteogenic media was found to be intimately related with the morphological changes (i.e. cytoskeleton reorganization) characteristic of the hBMSCs osteogenic differentiation, since ACTB instability appeared only when comparing cultures with cells displaying completely different phenotypes (i.e. undifferentiated vs. differentiated phenotype), as in Fig. 4a, whereas stable expression levels were observed otherwise, even when comparing cultures at different stages of the differentiation process (non-mineralized vs. mineralized cultures) but where the major morphological changes had already occurred (Fig. 4b). This may suggest that ACTB should only be used as normalizer gene in those cases where the evaluation time points and culture conditions are controlled in such a way to ensure that the major morphological changes have already occurred.
The increased expression levels of ACTB induced by osteogenic media resulted in an underestimation of the expression levels of the osteogenic markers in these cultures, suggesting an apparent differentiation process of the hBMSCs cultured without osteogenic supplements (Fig. 5a), which is not in accordance with the observed morphological changes and mineralization process (Figs. 1, 2). Therefore, the expression pattern of the osteogenic markers obtained when using ACTB as the normalizer gene demonstrates the potential bias in the quantification process introduced by normalizer instability.
Dheda et al. (2004) published a study validating housekeeping genes for normalizing mRNA expression in real-time RT-PCR assays in blood samples from patients with tuberculosis. They tested a pool of 13 housekeeping genes (including GAPDH and ACTB) for selecting a normalizer capable of discriminating small changes in the expression levels of low-copy number cytokines associated with this pathology. In this context, they proposed a selection criteria of AFC < 2 to avoid limiting the sensitivity of the assay. As a result, the gene encoding for the human acidic ribosomal protein (HuPO) was selected as an appropriate normalizer, with AFC < 2 and MFC ~5. In addition, GAPDH and ACTB showed the highest variability with AFC > 2 and MFC up to 35 (Dheda et al. 2004).
Based on the analysis of Dheda et al. (2004), the stability showed by GAPDH and RPL13A under the differentiation conditions and mesenchymal stem cell line tested, with AFC < 2 and MFC < 2.5 for all cases, suggests that both genes could be considered as appropriate normalizers to study hBMSCs differentiation. However, a more rigorous selection criteria with MFC < 2 would suggest that only RPL13A could be used as normalizer gene for the evaluated conditions. In addition, both GAPDH (Vandesompele et al. 2002; García-Crespo et al. 2005) and RPL13A (Szabo et al. 2004) have already been validated as appropriate normalizer genes for a considerable number of tissues.
The quantification of the gene expression levels of the ostoegenic markers CI, ON and BSP in the 14 and 20 day differentiations assays, allowed us to discriminate the effects of the small differences in the performance of GAPDH and RPL13A, as well as the effects of ACTB instability. The increased variability of GAPDH in control cultures as a function of evaluation time (Fig. 4b), which in fact corresponds to a reduction in its expression levels (AFC = 0.54 and MFC = 0.30), resulted in an overestimation of the mRNA levels of the three osteogenic markers in the control cultures at day 20 (Fig. 5c, NO20d). However, in cultures with OM (i.e. OM20d and OM14d) normalization with GAPDH produced a similar expression pattern to that of RPL13A for the 3 markers, which is associated with similar stability levels for both genes (Fig. 4b). The combined use of GAPDH and RPL13A as normalizers produced a mixed expression pattern where all differentiation markers were up-regulated in the osteogenic cultures, as expected, although GAPDH variability in the controls introduced a systematic error with an increase of ~42% in the expression levels of the three markers in cultures with NO at day 20 compared with the respective levels at day 14. This apparent increase in the expression levels of the osteogenic markers in cultures with NO was not observed in the data set normalized by RPL13A (Fig 5b). However, the qualitative analysis of the differentiation process did not show signs of differentiation in these cultures. Therefore, these results show the potential problems of normalizing to GAPDH when using calibration conditions in different evaluation time points, particularly in studies without ostoegenic conditions.
Finally, the relative absence of information regarding the selection of normalizer genes to be used in osteogenic differentiation studies, in reports as recent as some of the mentioned, contrasts with the urgent need to validate normalizer genes for specific applications and cell lineages. This warning has already been echoed by many authors, particularly in clinical studies (Schmittgen and Zakrajsek 2000; Kok et al. 2005; Aerts et al. 2004; Biederman et al. 2004; Zhang et al. 2005; Andersen et al. 2004).
The results herein presented demonstrated the unsuitability of β-actin as a normalizer gene for the study of human bone marrow mesenchymal stem cells differentiation to the osteogenic lineage, and suggest the possibility that the regulation of its expression levels may be associated with the morphological changes characteristic of the osteogenic differentiation process. In addition, normalization with GAPDH did not produce results that agreed with the morphological and mineralization observations. Finally, only RPL13A was capable of producing gene expression patterns consistent with the observed differentiation processes, providing evidence for its adequate performance as a normalizer gene in differentiation studies of hBMSCs to the osteogenic lineage. This work exposed the potential pitfalls associated with housekeeping gene instability for the adequate quantification of osteogenic markers during stem cells osteogenic differentiation and therefore, highlights the importance of validating the housekeeping genes used for studying osteogenic differentiation.
Acknowledgments
This project was partially funded by Colciencias (Colombian research foundation). The authors are grateful to the patients that participated in this study and to the healthcare institutions Hospital Pablo Tobón Uribe and Clínica Leon XIII, as well as to Dr. Daniel Estes (University of Michigan, USA) and Professors Juan A. López (University of Antioquia, Colombia) and Mauricio Rojas (University of Antioquia, Colombia) for their critical review of this manuscript.
Abbreviations
- RT-qPCR
Quantitative reverse transcription PCR
- hBMSCs
Human bone marrow mesenchymal stem cells
- ACTB
β-actin
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- RPL13A
Ribosomal protein L13A
- CI
Collagen type I
- ON
Osteonectin
- BSP
Bone sialoprotein
- NO
Non-osteogenic medium
- OM
Osteogenic medium
- ARS
Alizarin red staining
- AFC
Average fold changes from the mean threshold cycle
- MFC
Average maximum fold changes
- CT
Threshold cycle
References
- Abdallah BM, Haack-Sorensen M, Burns JS, Elsnab B, Jakob F, Hokland P, Kassem M. Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene in despite of extensive proliferation. Biochem Biophys Res Commun. 2005;326:527–538. doi: 10.1016/j.bbrc.2004.11.059. [DOI] [PubMed] [Google Scholar]
- Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. BioTechniques. 2004;36:84–91. doi: 10.2144/04361ST04. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-pcr data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets cancer research. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bianco P, Riminuncci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- Biederman J, Yee J, Cortes P. Validation of internal control genes for gene expression analysis in diabetic glomerulosclerosis. Kidney Int. 2004;66:2308–2314. doi: 10.1111/j.1523-1755.2004.66016.x. [DOI] [PubMed] [Google Scholar]
- Bustin S. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assay. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Catelas I, Sese N, Wu BM, Dunn JCY, Helgerson S, Tawil B. Human mesenchymal stem cell proliferation and osteogenic differentiation in fibrin gels in vitro. Tissue Eng. 2006;11:2385–2396. doi: 10.1089/ten.2006.12.2385. [DOI] [PubMed] [Google Scholar]
- Caterson EJ, Nesti LJ, Danielson KG, et al. Human marrow-derived mesenchymal progenitor cells: isolation, culture expansion, and analysis of differentiation. Mol Biotechnol. 2002;20:245–256. doi: 10.1385/MB:20:3:245. [DOI] [PubMed] [Google Scholar]
- Cho HH, Park HT, Kim YJ, Bae YC, Suh KT, Jung JS. Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J Cell Biochem. 2005;96:533–542. doi: 10.1002/jcb.20544. [DOI] [PubMed] [Google Scholar]
- Colter DC, Sekiya I, Prockop DJ. Identification of a subpopulation of rapidly selfrenewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98:7841–7845. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- Frank O, Heim M, Jakob M, Barbero A, Schäfer D, Bendik I, Dick W, Heberer M, Martin I. Real-time quantitative RT-PCR analysis of human bone marrow stromal cells during osteogenic differentiation in vitro. J Cell Biochem. 2002;85:737–746. doi: 10.1002/jcb.10174. [DOI] [PubMed] [Google Scholar]
- Friedman MS, Long MW, Hakenson KD. Osteogenic differentiation of human mesenchymal stem cells is regulated by bone morphogenetic protein-6. J Cell Biochem. 2006;98:538–554. doi: 10.1002/jcb.20719. [DOI] [PubMed] [Google Scholar]
- García-Crespo D, Juste RA, Hurtado A. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet Res. 2005;1:3. doi: 10.1186/1746-6148-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glare EM, Divjak M, Bailey MJ, Walters EH. β-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–770. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens K, Poucke MV, Soom AV, Vandesompele J, Zeveren A, Peelman LC. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol. 2005;5:27. doi: 10.1186/1471-213X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Ma T, Bunnell B. Human mesenchymal stem cells tissue development in 3D PET matrices. Biotechnol Prog. 2004;20:905–912. doi: 10.1021/bp034296z. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Johansson S, Fuchs A, Ökvis A, Karimi M, Harper C, Garrick T, Sheedy D, Hurd Y, Bakalkin G, Ekström TJ. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res. 2007;1132:20–28. doi: 10.1016/j.brainres.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim UJ, Vunjak-Novakovic G, Min BH, Kaplan DL. Influence of macroporous protein scaffolds on bone tissue engineering from bone marrow stem cells. Biomaterials. 2005;26:4442–4452. doi: 10.1016/j.biomaterials.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Kleiboeker SB. Quantitative assessment of the effect of uracil-DNA glycosylase on amplicon DNA degradation and RNA amplification in reverse transcription-PCR. Virol J. 2005;2:29. doi: 10.1186/1743-422X-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- Lee H, Huang G, Chiang H, Chiou LL, Chen MH, Hsieh CH, Jiang CH. Multipotential mesenchymal stem cells from femoral bone marrow near the site of osteonecrosis. Stem Cells. 2003;21:190–199. doi: 10.1634/stemcells.21-2-190. [DOI] [PubMed] [Google Scholar]
- Mbalaviele G, Sheikh S, Stains JP, Salazar VS, Cheng SL, Chen D, Civitelli R. β-Catenin and BMP-2 synergize to promote osteoblast differentiation and new bone formation. J Cell Biochem. 2005;94:403–418. doi: 10.1002/jcb.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinel L, Karageorgiou V, Hofmann S, Fajardo R, Snyder B, Li C, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. Engineering bone-like tissue in vitro using human bone marrow stem cells and silk scaffolds. J Biomed Mater Resv. 2004;71A:25–34. doi: 10.1002/jbm.a.30117. [DOI] [PubMed] [Google Scholar]
- Meinel L, Karageorgiou V, Fajardo R, Snyder B, Shinde-Patil V, Zichner L, Kaplan D, Langer R, Vunjak-Novakovic G. Bone tissue engineering using human mesenchymal stem cells: effects of scaffold material and medium flow. Ann Biomed. 2004;32:112–122. doi: 10.1023/B:ABME.0000007796.48329.b4. [DOI] [PubMed] [Google Scholar]
- Meinel L, Hofmann S, Betz O, Fajardo R, Merkle HP, Langer R, Evans CH, Vunjak-Novakovic G, Kaplan DL. Osteogenesis by human mesenchymal stem cells cultured on silk biomaterials: comparison of adenovirus mediated gene transfer and protein delivery of BMP-2. Biomaterials. 2006;27:4993–5002. doi: 10.1016/j.biomaterials.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Mygind T, Stiehler M, Baatrup A, Li H, Zou X, Flyvbjerg A, Kassem M, Bünger C. Mesenchymal stem cell ingrowth and differentiation on coralline hydroxyapatite scaffolds. Biomaterials. 2007;28:1036–1047. doi: 10.1016/j.biomaterials.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Oshina H, Sotome S, Yoshii T, Torigoe I, Yumi S, Maehara H, Marukawa E, Omura K, Shinomiya K. Effects of continuous dexamethasone treatment on differentiation capabilities of bone marrow-derived mesenchymal cells. Bone. 2007;41:575–583. doi: 10.1016/j.bone.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper—excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Quiroz GF, Posada OM, Gallego D, Higuita N, Sarassa C, Hansford DJ, Agudelo-Florez P, López LE. Isolation of human bone marrow mesenchymal stem cells and evaluation of their osteogenic potential. Revista Ingeniería Biomédica. 2008;3:48–55. [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Sakaguchi Y, Sekiya I, Yagishita K, Ichinose S, Shinomiya K, Muneta T. Suspended cells from trabecular bone by collagenase digestion become virtually identical to mesenchymal stem cells obtained from marrow aspirates. Blood. 2004;104:2728–2735. doi: 10.1182/blood-2003-12-4452. [DOI] [PubMed] [Google Scholar]
- Schmittgen T, Zakrajsek B. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Schutze N, Noth U, Schneidereit J, Hendrich C, Jakob F. Differential expression of CCN-family members in primary human bone marrow-derived mesenchymal stem cells during osteogenic, chondrogenic and adipogenic differentiation. Cell Commun Signal. 2005;3:5. doi: 10.1186/1478-811X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanasinghe RD, Bernacki SH, Loboa EG. Osteogenic differentiation of human mesenchymal stem cells in collagen matrices: effect of uniaxial cyclic tensile strain on bone morphogenetic protein (BMP-2) mRNA expression. Tissue Eng. 2006;12:3459–3465. doi: 10.1089/ten.2006.12.3459. [DOI] [PubMed] [Google Scholar]
- Szabo A, Perou CM, Karaca M, Perreard L, Palais R, Quackenbush JF, Bernard PS. Statistical modeling for selecting housekeeper genes. Genome Biol. 2004;5:R59. doi: 10.1186/gb-2004-5-8-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/S0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Tsukahara S, Ikeda R, Goto S, Yoshida K, Mitsumori R, Sakamoto Y, Tajima A, Yokoyama T, Toh S, Furukawa KI, Inoue I. Tumour necrosis factor α-stimulated gene-6 inhibits osteoblastic differentiation of human mesenchymal stem cells induced by osteogenic differentiation medium and BMP-2. Biochem J. 2006;398:595–603. doi: 10.1042/BJ20060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034.1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. BioTechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ding L, Sandford AJ. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol. 2005;6:4. doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]