Abstract
The canonical Wnt pathway is one of the oldest and most functionally diverse of animal intercellular signaling pathways. Though much is known about loss-of-function phenotypes for Wnt pathway components in several model organisms, the question of how this pathway achieved its current repertoire of functions has not been addressed. Our phylogenetic analyses of 11 multigene families from five species belonging to distinct phyla, as well as additional analyses employing the 12 Drosophila genomes, suggest frequent gene duplications affecting ligands and receptors as well as co-evolution of new ligand–receptor pairs likely facilitated the expansion of this pathway’s capabilities. Further, several examples of recent gene loss are visible in Drosophila when compared to family members in other phyla. By comparison the TGFβ signaling pathway is characterized by ancient gene duplications of ligands, receptors, and signal transducers with recent duplication events restricted to the vertebrate lineage. Overall, the data suggest that two distinct molecular evolutionary mechanisms can create a functionally diverse developmental signaling pathway. These are the recent dynamic generation of new genes and ligand–receptor interactions as seen in the Wnt pathway and the conservative adaptation of ancient pre-existing genes to new roles as seen in the TGFβ pathway. From a practical perspective, the former mechanism limits the investigator’s ability to transfer knowledge of specific pathway functions across species while the latter facilitates knowledge transfer.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-010-9337-z) contains supplementary material, which is available to authorized users.
Keywords: Wnt pathway, TGFβ pathway, Co-evolution, Gene loss, Phylogenetics
Introduction
Secreted Wnt ligands perform essential roles during development in all animal species. Among their responsibilities are the regulation of cell polarity and migration, organismal axis formation, cell fate specification, epithelial–mesenchymal interactions, and growth (reviewed in van Amerongen and Nusse 2009). In target cells, Wnt ligands can stimulate three distinct signal transduction systems: canonical, planar cell polarity, and Wnt/Ca2+. The canonical pathway is highly conserved and cross-species functionality has been observed between D. melanogaster and vertebrates (Klingensmith et al. 1996; Rothbächer et al. 1995). Here, we address the question: How did the Wnt pathway achieve its current repertoire of developmental functions?
The canonical Wnt pathway employs a double-negative method of information transfer (Fig. 1a; Angers and Moon 2009). When a Wingless/Wnt ligand (D. melanogaster and vertebrate names, respectively, and abbreviated as Wnt throughout) binds to Arrow/LRP (Arr) and Frizzled (Fz) transmembrane receptors, the Dishevelled (Dsh) signal transducer is recruited to the cytoplasmic side of the membrane. At the membrane, via an unknown mechanism Dsh is phosphorylated which leads it to inhibit the antagonistic activity of a cytoplasmic complex composed of Zw3/GSK3-β a constitutively active serine–threonine kinase, dAxin, and dAPC. In the absence of Wnt signals, Zw3/GSK3-β phosphorylates the transcription factor Armadillo/β-catenin (Arm) targeting it for destruction via the ubiquitin–proteasome pathway. Once the cytoplasmic complex is inhibited by Dsh, Zw3 moves to the membrane to phosphorylate Arr amplifying the Wnt signal and Arm translocates to the nucleus where it affects target gene expression by interaction with transcription factors such as Pygopus (Pygo), Legless/Bcl9 (Lgs), and Pangolin (TCF).
Fig. 1.
The canonical Wnt and TGFβ pathways in D. melanogaster. a Wnt pathway. In the absence of a Wnt signal, the destruction complex composed of Axin, APC, and Zw3 phosphorylates Arm (pArm) to tag it for degradation via the ubiquitin–proteasome pathway. In order to activate the pathway, a secreted Wnt ligand binds to unrelated Fz and Arr receptors. Wnt binding leads to the phosphorylation of the Dsh signal transducer. Dsh then inhibits the antagonistic activity of the destruction complex (a double negative mechanism of action), and Arm accumulates in the nucleus. There Arm joins transcription–activation complexes composed of one or more of the transcription factors Pygo, Lgs, and TCF. Arrows indicate positive, and T-bars negative effects on information transfer. b TGFβ pathway: The Dpp ligand binds to Punt a Type II transmembrane receptor serine–threonine kinase, which then recruits the related Tkv Type I receptor and phosphorylates it. Tkv then phosphorylates the R-Smad Mad (pMad). pMad then translocates to the nucleus as a heteromeric complex with the Co-Smad Medea. This multi-Smad complex then regulates the expression of target genes in cooperation with tissue-specific activators and repressors (a linear positive mechanism of action)
In contrast, the equally ancient and diverse Transforming Growth Factorβ (TGFβ)-signaling pathway employs a unidirectional method of information transfer (Fig. 1b; Derynck and Miyazono 2008; Kahlem and Newfeld 2009). In D. melanogaster, the ligand Dpp is identified and bound by the Type II receptor Punt, a constitutively active serine–threonine kinase. Punt then recruits the closely related Type I receptor Thickveins (Tkv) into a receptor complex. Punt then phosphorylates Tkv (also a serine–threonine kinase) that in turn phosphorylates Mad, a Receptor-associated Smad (R-Smad). Once phosphorylated, Mad translocates to the nucleus as a heteromeric complex with its relative Medea (a Co-Smad). This multi-Smad complex then regulates the transcription of target genes in cooperation with tissue-specific transcription factors. The Smad family also contains I-Smads that antagonize TGFβ signaling.
Over the years several investigators have addressed questions regarding the increasing number and diversification of the Wnt multigene family during evolution. Two early studies suggest that one important event was a whole genome duplication that occurred in the ancestor of all jawed vertebrates. These authors proposed that this duplication provided the raw material for the numerous Wnt family members currently found in vertebrates, explaining the paucity of Wnt proteins seen in invertebrates (Sidow 1992; Jockusch and Ober 2000). The discovery that the cnidarian Nematostella vectensis and the echinoderm Strongylocentrotus purpuratus have nearly as much Wnt diversity as mammals (11 subfamilies in N. vectensis and 10 in S. purpuratus versus 12 in M. musculus) falsified that hypothesis. Instead, a mechanism of repeated gene deletion in specific invertebrate groups such as Drosophila provides a better explanation for their lack of Wnts (Kusserow et al. 2005; Croce et al. 2006). Most recently, a study comparing the insects T. castaneum and D. melanogaster revealed that two Wnt family members have been lost in D. melanogaster just since the separation of these species, a finding that also supports the repeated deletion view (Bolognesi et al. 2008).
Here, we build upon these reports by expanding the analysis to understanding the diversity of the multigene families that comprise Wnt signaling pathways. We studied eleven families in the Wnt signal transduction cascade, and our phylogenetic analyses revealed that recent gene gain and loss affecting Wnt ligands and receptors as well as species-specific ligand–receptor co-evolution likely facilitated the expansion of Wnt pathway roles. By comparison the TGFβ signaling pathway likely achieved its current capabilities via the repeated adaptation to new functions of ancient gene duplications of ligands, receptors, and signal transducers. Overall, the data suggests that these two pathways employed distinct molecular evolutionary mechanisms to achieve their present form: the dynamic generation of new ligands and ligand–receptor interactions as seen in Wnt signaling or the conservative adaptation of pre-existing pathways as seen in TGFβ signaling.
Materials and Methods
Sequences
We examined eleven multigene families in the Wnt signaling pathway: Wnt ligands, Fz receptors, Arr receptors, Dsh signal transducers, destruction complex components (Axin, APC, and Zw3), and transcription activation complex components (Pygo, Lgs, TCF, and Arm). We obtained data from fully sequenced organisms belonging to five distinct phyla. The longest full-length isoform of each protein in N. vectensis (Nv), S. purpuratus (Sp), C. elegans (Ce), D. melanogaster (Dm), and M. musculus (Mm) was retrieved from NCBI (http://www.ncbi.nlm.nih.gov/) as of January 2010. Family members were identified via a variety of methods: Blastp, literature search, genome sequence annotation, and the Wnt home page (http://www.stanford.edu/~rnusse/wntwindow.html). See Table S1 for accession numbers.
In a separate analysis, we examined the same 11 multigene families in the twelve sequenced Drosophila genomes: D. melanogaster (dmel), D. simulans (dsim), D. sechellia (dsec), D. yakuba (dyak), D. erecta (dere), D. ananassae (dana), D. pseudoobscura (dpse), D. persimilis (dper), D. willistoni (dwil), D. mojavensis (dmoj), D. virilis (dvir), and D. grimshawi (dgri). The longest full-length isoform of each protein was identified and retrieved in January 2010 as described above. See Table S2 for accession numbers.
Phylogenetics
For the 22 multigene families (11 families in each of the two analyses) full-length protein sequences were aligned with MAFFT using L-INS-i and BLOSUM62 (Katoh et al. 2005). Three different types of trees were made for each family: Maximum Likelihood (ML), Neighbor-Joining (NJ), and Maximum Parsimony (MP). ML trees were created with PhyML v3.0 (Guindon and Gascuel 2003). BIONJ was used to create the initial trees. NNI improvement was used, and topology and branch lengths were optimized. Branch support was determined using SH-like aLRT (Anisimova and Gascuel 2006). The LG amino acid substitution model (Le and Gascuel 2008) was used with the proportion of invariable sites and the discrete gamma shape parameter with four rate categories estimated. NJ trees with complete deletions were created with MEGA4 using the JTT substitution matrix and 2000 bootstrap replicates (Kumar et al. 2008). MP trees were created utilizing all sites in MEGA4 using CNI level 3 and 1000 bootstrap replicates. A bootstrap (and by analogy an aLRT value) of 70 or above is considered statistically significant (Sitnikova 1996). In all trees, branches with statistical values below 50 were collapsed to emphasize significant branches. As a result in all trees, branch lengths are not to scale and no scale bar is shown.
For the five phyla analysis, ML trees are shown while NJ and MP trees are shown in Figs. S1–S4. For the 12 Drosophila genomes analysis, all trees are shown in Figs. S5–S15. For the Lgs family in the five phyla analysis, the ML and NJ trees have no statistics nor were MP trees constructed because at least four family members are required. For the Wnt, Arr/LRP, and Arm families in the 12 Drosophila genomes analysis, NJ trees are not shown because when employing the complete deletion option (the one utilized in the five phyla analysis), this algorithm did not generate trees with any branches above the bootstrap 50 cutoff.
Results
In order to provide explanatory power, we analyzed five animal species with fully sequenced genomes belonging to distinct phyla. Three of the species are coelomates, metazoans with three germ layers, and a digestive tract with two openings: the arthropod D. melanogaster (a protostome in which the blastopore becomes the mouth), the echinoderm S. purpuratus, and the chordate M. musculus (both deuterostomes in which the blastopore becomes the anus). One species is the nematode C. elegans (a pseudocoelomate) that has three germ layers but a digestive tract with just one opening. Together, these species belong to the superphyla Bilateria. The last species is the cnidarian N. vectensis, an acoelomate with no true digestive tract and only two germ layers. Chordates and echinoderms diverged ~712 million years ago (mya), deuterostomes and protostomes diverged ~826 mya, coelomates and pseudocoelomates diverged ~993 mya, and bilateria and cnidaria diverged ~1036 mya (Hedges et al. 2006; http://www.timetree.org/). In order to provide confidence, we generated three types of trees and compared them. ML trees are shown with NJ and MP trees found in the supplementary data.
Wnt Ligands
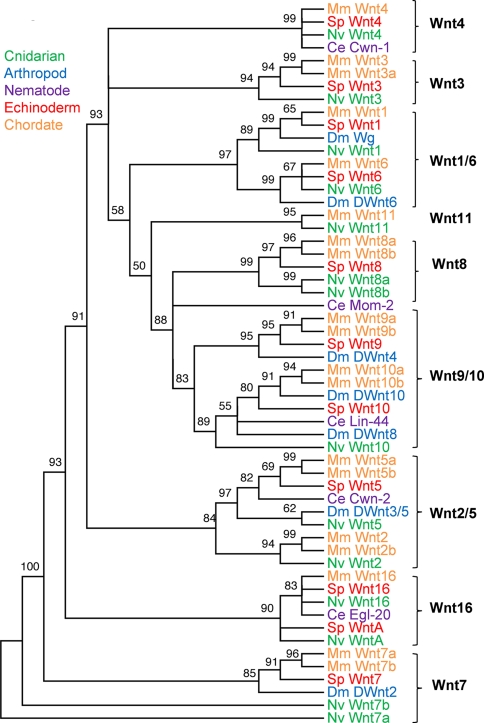
The 56 Wnt family members from N. vectensis (fourteen), C. elegans, (five), D. melanogaster (seven), S. purpuratus (eleven), and M. musculus (nineteen) cluster into 13 distinct subfamilies with strong statistical support (Fig. 2). Here, subfamily names derive from the M. musculus member for 12 of these groups with the 13th consisting solely of Ce Mom-2. Five subfamilies contain a single M. musculus protein while seven subfamilies contain two tightly linked M. musculus proteins reflecting seven recent duplication events. Of the S. purpuratus sequences, 10 belong to distinct subfamilies with one pair (Sp Wnt16 and Sp WntA) in the same subfamily. Of the D. melanogaster proteins, five belong to distinct subfamilies with Dwnt8 and Dwnt10 in the same subfamily. Of the C. elegans proteins, all five belong to distinct subfamilies. The N. vectensis proteins belong to 11 subfamilies (only the Wnt8 subfamily has no cnidarian member) with two of them (Wnt7 and Wnt8 subfamilies) containing tightly linked proteins reflecting lineage-specific duplication events.
Fig. 2.
Maximum likelihood tree of Wnt ligands. Fifty-six Wnt family members from N. vectensis (Nv, green), C. elegans (Ce, purple), D. melanogaster (Dm, blue), S. purpuratus (Sp, red), and M. musculus (Mm, orange) are shown. Twelve small, statistically supported subfamilies are indicated by the name of a M. musculus protein and a bracket (note that Wnt1/6 indicates a statistically supported cluster containing the Wnt1 and Wnt6 subfamilies). The length of the alignment was 1407 amino acids. Statistical confidence was measured via aLRT with values equal to or above 70 considered significant. Branches with aLRT values below 50 are collapsed. The tree is unrooted, and two sequences (Nv Wnt7a and Nv Wnt7b) do not have aLRT values
Two of the subfamilies contain members from all five species (Wnt5 and Wnt10). Three subfamilies contain four species without C. elegans (Wnt1, Wnt6, and Wnt7). Two subfamilies contain four species without D. melanogaster (Wnt4 and Wnt16). Two subfamilies contain three species without C. elegans and D. melanogaster (Wnt3 and Wnt8). The Wnt9 subfamily contains three species without N. vectensis and C. elegans. Two subfamilies contain only N. vectensis and M. musculus sequences (Wnt2 and Wnt11) and Ce Mom-2 is alone in its subfamily. Six of these subfamilies cluster together into three groups with statistical confidence: Wnt1/Wnt6, Wnt9/Wnt10, and Wnt2/Wnt5. In addition, Wnt9/10 clusters with Wnt8 and Ce Mom-2 to form the largest supported subgroup. With at least two members from each species, the Wnt8/9/10 cluster contains 17 members, the largest highly conserved group of sequences.
In the NJ and MP trees, the 12 M. musculus subfamilies were present but each contained fewer members. In these trees, branch resolution was poor with 17 (NJ) or 22 (MP) sequences, rather than just one as in the ML tree, now outside any subfamily (Fig. S1). An examination of the Wnt family in twelve Drosophila species revealed that the gene tree for each family member matched the species tree (Tamura et al. 2004) with the exception that D. simulans Wnt6 and D. virilis Wnt3/5 were not identifiable in their genome sequence (Fig. S5). The absence of D. simulans Wnt6 led to the placement of D. simulans Wg between the Wg and Wnt6 subfamilies that form a tight cluster rather than explicitly with the Wg subfamily.
Fz and Arrow Receptors
The 26 Fz family members from N. vectensis (four), C. elegans (four), D. melanogaster (four), S. purpuratus (four), and M. musculus (ten) are organized into six subfamilies (Fig. 3a). Five have a M. musculus member, and Ce Fz-2 is alone in the sixth subfamily. Of the multimember subfamilies, all except Fz4 contain multiple tightly linked M. musculus sequences indicative of five recent duplication events. S. purpuratus sequences are present in four subfamilies. D. melanogaster proteins are present in three subfamilies with one (Fz4) containing two tightly linked family members indicative of a lineage-specific duplication. C. elegans proteins belong to three subfamilies with one (Fz3/6) containing two tightly linked family members indicative of a lineage-specific duplication. N. vectensis proteins belong to four subfamilies. In contrast to the Wnt family, the Fz family contains evidence of lineage-specific duplications in both D. melanogaster and C. elegans. All of the subfamilies except Fz1/2/7 cluster together into a larger group but the presence of N. vectensis sequences in the Fz1/2/7 subfamily suggests this grouping is an ancestral one.
Fig. 3.
Maximum likelihood trees of Fz and Arrow receptors. Sequences are colored and presented as in Fig. 2. a Twenty-six Fz family members. Six small subfamilies are present. Five subfamilies contain a M. musculus member (brackets), and Ce Fz-2 is alone in the sixth subfamily. The length of the alignment was 1232 amino acids. The tree is unrooted, and two sequences (Mm Fz6 and MmFz3) do not have aLRT values. Note that Ce Fz-1 is also known as Mig-1 and Dm Smoothened does not bind Wg so it was excluded. b Eighteen Arrow family members. Five subfamilies are present. Four subfamilies contain a M. musculus member (brackets), and Dm Yolkless is alone in the fifth subfamily. The alignment was 6113 amino acids. The tree is unrooted and two sequences (Mm LRP5 and Mm LRP6) do not have aLRT values. Note that Mm LRP3 was excluded because the complete sequence was not available
In the NJ tree, only four M. musculus subfamilies were present because Mm Fz4 was placed in the Fz9/10 subfamily. Also, two D. melanogaster and two additional C. elegans proteins are now outside any subfamily (Fig. S2). In the MP tree, only the Fz5/8 subfamily is present with all other proteins unresolved except the recently duplicated M. musculus sequences. An examination of the Fz family in 12 Drosophila species revealed that the gene tree for each family member matched the species tree (Fig. S6).
The 18 Arrow family members from N. vectensis (one), C. elegans (two), D. melanogaster (five), S. purpuratus (one), and M. musculus (ten) are organized into five subfamilies (Fig. 3b). The full complement of M. musculus sequences is presented for this poorly characterized family, but to date only Mm LRP5 and Mm LRP6 are implicated in Wnt signaling. Four subfamilies have at least one M. musculus sequence, and Dm Yolkless is the sole member of the fifth subfamily. The LRP1/2/8/11 subfamily has an unusual arrangement of M. musculus sequences—none of the four are tightly linked to each other. Mm LRP11 is linked to Dm CG33087 (note that D. melanogaster genes with CG prefixes are predictions) and Mm LRP8 is unlinked to any other member of the subfamily. A typical arrangement is found in the LRP9/10/12 and LRP5/6 subfamilies with the M. musculus genes tightly linked (note Mm LRP9/10 is a single gene that has been given two different names in the literature). The only S. purpuratus sequence belongs to the LRP5/6 subfamily suggesting that it may play a role in Wnt signaling in that species. Two of the D. melanogaster proteins belong to the LRP1/2/8/11 subfamilies though they are not tightly linked. Dm Arrow that participates in Wg signaling belongs to the LRP5/6 subfamily with the M. musculus Wnt signal transducers while Dm Yolkless has no close relatives. The two C. elegans proteins belong to the LRP1/2/8/11 subfamilies though they are not tightly linked. The sole N. vectensis sequence is only loosely contained within the LRP9/10/12 subfamily.
A larger group containing LRP4/5/6 proteins is separated from the other sequences that are themselves clustered with statistical significance. The presence of N. vectensis and C. elegans proteins suggests that LRP1/2/8/9/10/11/12 is the older of the two subfamilies. In the NJ tree, no significant branches are seen (Fig. S3). In the MP tree, the LRP4 cluster is loosely attached to the Wnt signaling LRP5/6 cluster, and Dm Yolkless remains isolated but no other subfamilies are visible. An examination of the Arr family in 12 Drosophila species revealed that the gene tree for each family member matched the species tree with minor exceptions (Fig. S7). There appears to be a duplication of D. pseudoobscura CG8909 while CG34352 was missing from D. simulans and D. sechellia, and Yolkless was missing from D. simulans.
Dsh Signal Transducers
The nine Dsh family members from N. vectensis (one), C. elegans (three), D. melanogaster (one), S. purpuratus (one), and M. musculus (three) are organized into a single family (Fig. 4a). In this family, the three M. musculus proteins form a cluster as do the three C. elegans proteins indicating two duplications in each lineage. In addition, the N. vectensis sequence groups most closely with the M. musculus sequences. The clustered duplications but not the association of N. vectensis and M. musculus are seen in the NJ and MP trees (Fig. S3A). The Dsh family has experienced far less duplication than the Wnt, Fz, or Arr families. An examination of Dsh in 12 Drosophila species revealed that the gene tree matched the species tree except that Dsh is missing from D. simulans (Fig. S8).
Fig. 4.
Maximum likelihood trees of Dsh and the destruction complex. Sequences are colored and presented as in Fig. 2. a Nine Dsh family members. A single family of all sequences is present. The tree is unrooted and two sequences (Ce Dsh-1 and Ce Dsh-2) do not have aLRT values. The alignment was 978 amino acids. b Four Axin family members. A single family of all sequences is present. The alignment was 1136 amino acids. The tree is unrooted, and two sequences (Mm Axin1 and Mm Axin2) do not have aLRT values. c Six APC family members. A single family of all sequences is present. The alignment was 3852 amino acids. The tree is unrooted, and thus two sequences (Dm APC1 and Dm APC2) do not have aLRT values. Note that APC is missing from the N. vectensis genome. d Seven Zw3 family members. The alignment was 1091 amino acids. The grouping of Ce Gsk-2 with Mm GSKα is not significant. Note that Dm Zw3 is also known as Shaggy. The tree is unrooted, and two sequences (Dm Zw3 and Dm Gskt) do not have aLRT values
Axin, APC, and Zw3 Destruction Complex
Although each of these proteins has a roughly contemporaneous role in Wnt signaling (as part of the cytoplasmic complex targeting Arm for degradation) the Axin, APC, and Zw3 families are not related by sequence. The six Axin family members, one from each species with a single duplication in M. musculus, are organized into a single family (Fig. 4b). The Axin family has experienced less duplication than Dsh. This topology is seen in the NJ and MP trees (Fig. S3B). An examination of Axin in 12 Drosophila species revealed that the gene tree matched the species tree (Fig. S9).
The six APC family members, duplicated in M. musculus and D. melanogaster but absent from N. vectensis, are organized into a single family (Fig. 4c). As seen for the Axin and Dsh families, the duplications are tightly linked indicating lineage-specific events. This topology is seen in the NJ and MP trees (Fig. S3C). An examination of APC in 12 Drosophila species revealed that the gene tree matched the species tree (Fig. S10).
The seven Zw3 family members, duplicated in M. musculus and D. melanogaster, are organized into a single family (Fig. 4d). Note that the grouping of MmGSK3-α with Ce Gsk-3 is not significant and each of the other family members should be considered statistically equidistant from the GSK3-β cluster. In contrast to Dsh, Axin, and APC, the Zw3 duplications are not tightly linked suggesting an origin after the divergence of the cnidarian and nematode lineages from the arthropod–echinoderm–vertebrate lineage with a loss (or sequence gap) in echinoderms. This topology is seen in the MP but not the NJ tree where Mm GSK3-α and Mm GSK3-β cluster together suggesting a recent duplication (Fig. S3D). An examination of 12 Drosophila species revealed that the gene tree matched the species tree for Dm Gskt but that Dm Zw3 was missing from D. sechellia, D. yakuba, D. persimilis, and D. willistoni (Fig. S11).
Pygo, Lgs, TCF, and Arm Transcription Factors
Although each has a roughly analogous role in Wnt signaling (as part of a transcription complex with Arm), the Pygo, Lgs, and TCF families are not related by sequence. Further, these proteins are not required together (as are the proteins in the destruction complex) to fulfill their roles in Wnt signaling.
The five Pygo family members from D. melanogaster (one), S. purpuratus (two), and M. musculus (two) are not resolved except the M. musculus proteins are tightly linked (Fig. 5a). No members in N. vectensis or C. elegans were identified suggesting an origin in the arthropod–echinoderm–vertebrate lineage. This topology is seen in the NJ and MP trees (Fig. S4A). An examination of 12 Drosophila species revealed that the gene tree matched the species tree (Fig. S12).
Fig. 5.
Maximum likelihood trees of the transcription activation complex. Sequences are colored and presented as in Fig. 2. a Five Pygo family members. Two subfamilies are present, but only the M. musculus subfamily is significant. The alignment was 1159 amino acids. The tree is unrooted, and thus one sequence (Sp Pygo2) does not have an aLRT value. Note that a potential Pygo sequence in N. vectensis, XP_001629729.1 was not included as it is incomplete. b Three Lgs family members. The alignment was 1822 amino acids. No statistics were possible due to small family size. c Nine TCF family members. The alignment was 910 amino acids. A single poorly resolved family of all sequences in present. The tree is unrooted, and two sequences (Mm TCF3 and Mm TCF4) do not have aLRT values. Note that Ce Son-1 is also known as Hmg-1.2 and that Mm TCF1, 3, and 4 are also known as Mm TCF7, 7L, and 7L2, respectively. d Six Arm family members. The alignment was 1024 amino acids. A single poorly resolved family of all sequences is present. The tree is unrooted and two sequences (Ce Wrm-1 and Ce Bar-1) do not have aLRT values. Note that Ce Hmp-2 and Mm Plakoglobin are not involved in Wnt signaling and were excluded from the analysis
The three Lgs family members from D. melanogaster (one) and M. musculus (two) cannot be analyzed statistically, but the most parsimonious explanation of the tree is that the two M. musculus proteins resulted from a recent duplication. Note that only the two Mm Bcl-9 sequences are members of the Lgs family because mammalian Bcl proteins are functionally related, associated with chromosome aberrations found in B-cell chronic lymphocytic leukemia, but they are not similar sequences (Ohno et al. 2005). No family members in N. vectensis, C. elegans, or S. purpuratus were identified suggesting an origin in the arthropod–echinoderm–vertebrate lineage with a loss (or sequence gap) in echinoderms. This topology is seen in the NJ tree (Fig. S4B). An examination of Lgs in 12 Drosophila species revealed a duplication event in D. simulans, but otherwise the gene tree matched the species tree (Fig. S13).
The nine TCF family members from N. vectensis (one), C. elegans (two), D. melanogaster (one), S. purpuratus (one), and M. musculus (four) are organized into a single family (Fig. 5c). The C. elegans proteins are tightly linked together as are the M. musculus proteins (though the relationship between the M. musculus proteins is not clear) indicating lineage-specific duplications. This topology is seen in the NJ and MP trees (Fig. S4C). An examination of 12 Drosophila species revealed that the gene tree matched the species tree (Fig. S14).
The six Arm family members form two subfamilies with the pair of tightly linked C. elegans proteins distinct from the others. This topology is seen in the NJ and MP trees (Fig. S4D). An examination of 12 Drosophila species revealed that the gene tree matched the species tree except that Arm is missing from D. persimilis (Fig. S15).
Discussion
Sponges are the simplest multicellular animals (a few cell types derived from a single germ layer), and the split with all other animals occurred roughly 1.5 billion years ago (Hedges et al. 2006). Wnt ligands, Fz receptors, and Dsh signal transducers are present in a single sponge species, and they are all expressed during larval stages. This suggests that the pathway existed in the common ancestor of all animals. From unknown roles in the earliest metazoans, the pathway has grown in complexity and diversified to perform a myriad of functions.
Wnt Family Diversification
The analysis showed that each of the other species is absent from at least one of the twelve Wnt subfamilies found in M. musculus. Based on the presence/absence of species, we infer that 10 subfamilies are ancestral (present in N. vectensis) with one subfamily (Wnt 9) originating after the split of cnidarians and nematodes from the arthropod–echinoderm–vertebrate lineage. Overall, six lineage-specific losses occurred in C. elegans, six in D. melanogaster, and two in S. purpuratus while seven lineage-specific duplications occurred in M. musculus, two in N. vectensis with one in D. melanogaster (Dm DWnt10–Dm DWnt8), and one in S. purpuratus (Sp Wnt16 -Sp WntA). Thus, a total of 25 gain or loss events are evident in the Wnt multigene family.
The losses in D. melanogaster are evident in the five phyla analyses via subfamilies that contain the other four species (Wnt4 and Wnt16), subfamilies that contain three species without C. elegans and D. melanogaster (Wnt3 and Wnt8), and subfamilies that contain only N. vectensis and M. musculus sequences (Wnt2 and Wnt11). The absence of these six Wnt sequences in D. melanogaster was previously noted in a study of the Wnt family in the beetle T. castaneum (Bolognesi et al. 2008). Interestingly, those authors found that T. castaneum has three of the Wnts missing in D. melanogaster (Wnt8, Wnt11, andWnt16) indicating that the other three Wnts were lost in the Drosophila lineage after the separation of holometabolous and hemimetabolous insects (250 mya; Hedges et al. 2006). Our analysis of the Wnt family in twelve Drosophila genomes revealed that all species contain the same Wnt complement. Thus, the loss of the three Wnts in the Drosophila lineage must have occurred before the Sophophora/Drosophila subgeneric divergence 65 mya (Tamura et al. 2004). Taken together, these two studies suggest that three Wnts were lost from Drosophila in a span of 185 million years.
This example of Wnt gene loss in the Drosophila lineage can be viewed in light of published observations that the D. melanogaster genome undergoes the rapid loss of unconstrained sequences (Petrov 2002; Petrov et al. 1996). The Wnt results suggest that even protein-coding genes belonging to large multigene families can be lost in this lineage. One can imagine that a redundant protein could arise as the result of the convergence of two independent events. Utilizing Wnts as an example, first a cis-acting regulatory mutation (or a mutation in a trans-acting factor) generates a new expression pattern for one Wnt that mimics or at least overlaps the pattern of a second Wnt. Second, due to pre-existing sequence similarity, the Wnt with the new expression pattern is capable of performing the job of the other Wnt. Thus, the Wnt without the regulatory mutation now performs a redundant function and is amenable to being eliminated without incurring any selective disadvantage.
A prediction of this hypothesis is that individual D. melanogaster Wnts will have more complex regulation and more distinct functions than Wnts in a species with a larger complement of ligands. Suggestive evidence for this prediction is presented by Bolognesi et al. (2008) who showed that T. castaneum Wnt11 (absent from D. melanogaster) is expressed in the embryo at the border of the dorsal ectoderm in a location analogous to the expression of D. melanogaster, DWnt8. From this, we infer that DWnt8 may be fulfilling the role of T. castaneum Wnt11 in ectoderm differentiation in addition to its other roles in Drosophila development.
Examination of published data on Wnt–Fz biochemical and genetic interactions (summarized in Table S3) from a phylogenetic perspective suggest a number of new hypotheses regarding the function of currently poorly characterized Wnt family members. First, biochemical and genetic studies have shown that Dm DWnt3/5 binds to the tyrosine kinase receptor Derailed. The strong clustering of Dm Dwnt3/5 in the Wnt2/5 combined subfamily implies that subfamily members in other species may also interact with tyrosine kinase receptors. Second, biochemical studies have shown that Dwnt8 only interacts with the Dm Fz4 receptor (Wu and Nusse 2002). Dm DWnt8 belongs to the only Wnt subfamily with two D. melanogaster members (with DWnt10), and the Fz4 receptor also occurs in the only subfamily with two D. melanogaster members (with Dm Fz3). The specificity of the DWnt8 and Dm Fz4 interaction suggests that this ligand–receptor pair is co-evolving. The presence of N. vectensis sequences within subfamilies containing recently duplicated D. melanogaster Wnt and Fz sequences does not detract from our contention that the lineage-specific D. melanogaster sequences are co-evolving. Third, the recent origins of ligand–receptor pairs in M. musculus also suggest they may be co-evolving. For example, Mm Wnt3a and Mm Fz8 biochemically interact (Zhu et al. 2008) and both show evidence of recent origins (Mm Wnt3/Wnt3a and Mm Fz5/Fz8 are tightly linked).
Wnt Pathway Diversification
The Fz analysis showed that four subfamilies are ancestral (present in N. vectensis). While each of the species except M. musculus has four family members, their distribution reveals that three lineage-specific losses occurred in C. elegans, two in D. melanogaster, and two in S. purpuratus while five lineage-specific duplications occurred in M. musculus, two in C. elegans, and one in D. melanogaster. Thus, a total of 15 gain or loss events are evident in the Fz multigene family.
The presence of one N. vectensis sequence in the Arr analysis revealed that this family experienced only gains —C. elegans (one), D. melanogaster (four), and M. musculus (eight). A total of 13 duplication events are evident in the Arr multigene family. In addition, as the Arr family was not included, in our phylogenetic analysis of lysine conservation in the Wnt pathway (Konikoff et al. 2008), we determined that there are 10 conserved lysines in the Wnt signaling Mm LRP5, Mm LRP6, and Dm Arrow sequences. These fall within β-propeller domains 1–3 of the extracellular portion of the protein. As these conserved lysines are not accessible to intracellular enzymatic manipulation, it appears that ubiquitination and sumoylation may not be important regulatory mechanisms for Arr family members.
The four trees of the Dsh and destruction complex proteins reveal seven duplications: four in M. musculus, two in D. melanogaster, and one in C. elegans. The four trees of the transcription activation complex proteins reveal eight duplications: five in M. musculus, two in C. elegans, and one in S. purpuratus. Thus, a total of 15 duplication events are evident in these nine “downstream families” that function in signal transduction beyond the receptors.
Given the value of a comparison between T. castaneum and D. melanogaster Wnts (Bolognesi et al. 2008), we then examined the T. castaneum genome for the remaining components of the Wnt pathway (summarized in Table S4). We found substantial similarity. For example, a subfamily of receptor (either Fz or Arr) that is not found in D. melanogaster was likely also absent from T. castaneum. In addition, Dsh and all of the destruction and transcription complex proteins were present in T. castaneum except for Pygo.
Wnt Versus TGFβ Pathway Diversification
A comparison of the phylogenetics of these two signaling pathways reveal strong differences in ligand, receptor, and signal transducer families that appear to have a common theme. The Wnt family’s broad but shallow topology with 12 small subfamilies stands in contrast to the deeply conserved tree for TGFβ family members (Kahlem and Newfeld 2009; Newfeld et al. 1999). Unlike the Wnt tree, most of the duplications that link TGFβ proteins are ancient and result in five readily identifiable groups. In addition, the loss of Wnts in C. elegans and D. melanogaster is without parallel in the TGFβ family, though the loss of accessory proteins in TGFβ signaling in D. melanogaster has been reported (Van der Zee et al. 2008).
In contrast to reported and proposed ligand–receptor interactions in the Wnt pathway, in the TGFβ family there is no evidence for tyrosine kinase receptor binding or species-specific co-evolution of ligands and receptors. Instead, TGFβ family ligand and receptor co-evolution is ancient and predates the common ancestor of C. elegans, D. melanogaster, and M. musculus. This is shown by the fact that BMP family members in these species interact only with BMP Type I and Type II receptors in their respective species but not TGFβ/Activin subfamily receptors (Derynck and Miyazono 2008). This specialization holds even across species—the BMP Type I receptor Saxophone from D. melanogaster together with the BMP-4 Type II receptor Daf-4 from C. elegans bind to human BMP2 with high affinity (Brummel et al. 1994). This deep conservation extends to Smads beginning with C. elegans that can be functionally apportioned into the TGFβ/Activin and BMP pathways while Wnt ligands, receptors, and signal transducers cannot be assigned to functional units.
Viewing the receptors from a larger perspective, the lack of a structural or evolutionary relationship between the Arr and Fz families is distinct from that of TGFβ Type I and Type II receptors. The Wnt receptor families are structurally dissimilar (seven pass for Fz and single pass for Arr transmembrane proteins), and neither has any identifiable enzymatic activity. Alternatively, all of the TFGβ receptors are structurally similar (single-pass transmembrane proteins with serine–threonine kinase activity), and the level of amino acid similarity between them indicates descent from a common ancestor. Thus, another contrast between the pathways is that TGFβ receptor diversification occurs within an ancient receptor paradigm while the diversification of Wnt receptors resulted from the convergence of two unrelated proteins.
The Dsh family’s lack of diversification contrasts in two ways with the diversification of the Smad family of TGFβ signal transducers. First, Smads have experienced many more duplications in each species than the Dsh family. This is true even if one only focuses on R-Smads, those functionally analogous to receptor-activated Dsh signal transducers (Newfeld and Wisotzkey 2006). Second, while Dsh proteins are solely positive components of the Wnt pathway, numerous ancient duplications allowed Smads to assume positive, negative, and cooperative roles in signal transduction.
The long-term stability of destruction complex families is similar to the I-Smad subfamily in TGFβ signaling (Newfeld and Wisotzkey 2006). In the transcription complex, the Arm and TCF families closely resemble the transcriptional activator R-Smad and Co-Smad subfamilies in TGFβ signaling. Lastly, the Pygo and Lgs families are the only ones in either the Wnt or TGFβ pathways that we have studied without a member in C. elegans.
Overall, the phylogenetic analysis of the Wnt pathway in five phyla revealed that dynamic recent gene duplications affecting Wnt ligands and Fz receptors likely facilitated the expansion of this pathway’s capabilities. The data also suggested that co-evolution of ligand–receptor pairs in D. melanogaster and M. musculus contributed to the pathway’s current composition. This two-part molecular evolutionary mechanism (recent duplication and subsequent co-evolution of new ligand–receptor pairs) and the evidence of loss of Wnts in C. elegans and D. melanogaster stand in contrast to the mechanism employed by the TGFβ signaling pathway to achieve its current array of roles. TGFβ signaling appears to have achieved its diverse functional capabilities via two sets of ancient events—duplications of ligands, receptors, and Smads followed by the diversification of Smad signal transducer functions. In the TGFβ pathway, recent duplication events are restricted to vertebrates, but these new family members remain within the ancient framework of functionally linked ligand, receptor, and Smad interactions (Newfeld et al. 1999).
In summary, the data extend our knowledge of evolutionary developmental biology in two ways. First, that natural selection can generate a complex and functionally diverse signaling pathway via two (at least) distinct molecular mechanisms. This discovery has practical implications as the dynamic mechanism visible in the Wnt pathway limits the investigator’s ability to transfer knowledge of specific pathway functions across species while the conservative mechanism evident in the TGFβ pathway facilitates knowledge transfer. Second, that gene loss and regulatory mutations likely play sizeable roles in shaping the size and content of the genomes of many organisms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Sudhir Kumar for his valuable comments. The laboratory of SJN is supported by grants from NIH (HG002516), ENFIN—a European Commission Network of Excellence in Systems Biology and the Intertribal Council of Arizona.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Angers S, Moon R. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Beermann A, Farzana L, Wittkopp N, Lutz R, Balavoine G, Brown S, Schröder R. Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev Genes Evol. 2008;218:193–202. doi: 10.1007/s00427-007-0170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Twombly V, Marqués G, Wrana J, Newfeld S, Attisano L, Massagué J, O’Connor M, Gelbart W. Characterization and relationship of Dpp receptors encoded by the saxophone and thick veins genes in Drosophila. Cell. 1994;78:251–261. doi: 10.1016/0092-8674(94)90295-X. [DOI] [PubMed] [Google Scholar]
- Croce J, Wu S, Byrum C, Xu R, Duloquin L, Wikramanayake A, Gache C, McClay D. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin S. purpuratus. Dev Biol. 2006;300:121–131. doi: 10.1016/j.ydbio.2006.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Miyazono K. The TGFβ family. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hedges S, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Jockusch E, Ober K. Phylogenetic analysis of the Wnt gene family and discovery of an arthropod Wnt-10 orthologue. J Exp Zool. 2000;288:105–119. doi: 10.1002/1097-010X(20000815)288:2<105::AID-JEZ3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Kahlem P, Newfeld S. Informatics approaches to understanding TGFβ pathway regulation. Development. 2009;136:3729–3740. doi: 10.1242/dev.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingensmith J, Yang Y, Axelrod J, Beier D, Perrimon N, Sussman D. Conservation of dishevelled structure and function between flies and mice: isolation and characterization of Dvl2. Mech Dev. 1996;58:15–26. doi: 10.1016/S0925-4773(96)00549-7. [DOI] [PubMed] [Google Scholar]
- Konikoff C, Wisotzkey R, Newfeld S. Lysine conservation and context in TGFβ and Wnt signaling suggest new targets and general themes for posttranslational modification. J Mol Evol. 2008;67:323–333. doi: 10.1007/s00239-008-9159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt H, Technau U, von Haeseler A, Hobmayer B, Martindale M, Holstein T. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Le S, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Newfeld S, Wisotzkey R. Molecular evolution of Smad proteins. In: Heldin C, ten Dijke P, editors. Smad signal transduction. Dordrecht, Netherlands: Springer; 2006. pp. 15–35. [Google Scholar]
- Newfeld S, Wisotzkey R, Kumar S. Molecular evolution of a developmental pathway: phylogenetic analyses of TGFβ family ligands, receptors and Smad signal transducers. Genetics. 1999;152:783–795. doi: 10.1093/genetics/152.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Nishikori M, Maesako Y, Haga H. Reappraisal of BCL3 as a molecular marker of anaplastic large cell lymphoma. Int J Hematol. 2005;82:397–405. doi: 10.1532/IJH97.05045. [DOI] [PubMed] [Google Scholar]
- Petrov D. DNA loss and evolution of genome size in Drosophila. Genetica. 2002;115:81–91. doi: 10.1023/A:1016076215168. [DOI] [PubMed] [Google Scholar]
- Petrov D, Lozovskaya E, Hartl D. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- Rothbächer U, Laurent M, Blitz I, Watabe T, Marsh J, Cho K. Functional conservation of the Wnt signaling pathway revealed by ectopic expression of Drosophila dishevelled in Xenopus. Dev Biol. 1995;170:717–721. doi: 10.1006/dbio.1995.1249. [DOI] [PubMed] [Google Scholar]
- Sidow A. Diversification of the Wnt gene family on the ancestral lineage of vertebrates. Proc Natl Acad Sci USA. 1992;89:5098–5102. doi: 10.1073/pnas.89.11.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnikova T. Bootstrap test for phylogenetic trees. Mol Biol Evol. 1996;13:605–611. doi: 10.1093/oxfordjournals.molbev.a025620. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of Drosophila evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- Van der Zee M, da Fonseca RN, Roth S. TGFβ signaling in Tribolium: vertebrate-like components in a beetle. Dev Genes Evol. 2008;218:203–213. doi: 10.1007/s00427-007-0179-7. [DOI] [PubMed] [Google Scholar]
- Wu C, Nusse R. Ligand receptor interactions in the Wnt signaling pathway in Drosophila. J Biol Chem. 2002;277:41762–41769. doi: 10.1074/jbc.M207850200. [DOI] [PubMed] [Google Scholar]
- Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, Naito A, Nishi J, Ueno H, Umezawa A, Minamino T, Nagai T, Kikuchi A, Asashima M, Komuro I. IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature. 2008;454:345–349. doi: 10.1038/nature07027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.