Abstract
The immunological synapse (IS) as a concept has evolved from a static view of the junction between T cells and their antigen-presenting cell partners. The entire process of IS formation and extinction is now known to entail a dynamic reorganization of membrane domains and proteins within and adjacent to those domains.
Discussion
The entire process is also intricately tied to the motility machinery—both as that machinery directs “scanning” prior to T-cell receptor engagement and as it is appropriated during the ongoing developments at the IS. While the synapse often remains dynamic in order to encourage surveillance of new antigen-presenting surfaces, cytoskeletal forces also regulate the development of signals, likely including the assembly of ion channels. In both neuronal and immunological synapses, localized Ca2+ signals and accumulation or depletion of ions in microdomains accompany the concentration of signaling molecules in the synapse. Such spatiotemporal signaling in the synapse greatly accelerates kinetics and provides essential checkpoints to validate effective cell–cell communication.
Keywords: T lymphocyte, ion channel, Ca2+ signaling, STIM1, Orai1
Introduction
The immune system relies upon molecular signaling and cellular communication initiated by direct cell to cell contact. Coined by Sherrington from the Greek “syn” (together) and “haptein” (to clasp) to signify neuronal cell to cell junctions that communicate electrically through ion channel activity, the term immunological synapse (IS) has come to represent a wide variety of cell to cell junctions that vary in molecular structure and dynamics. A dozen years ago, Kupfer and colleagues originally described T-cell–B-cell synapses as having a characteristic bull’s-eye pattern with centrally localized T-cell receptors engaging peptide major histocompatibility complex (pMHC) and the co-receptor CD28 engaging CD80/86 in the central supramolecular activating complex (c-SMAC), surrounded by a peripheral zone (p-SMAC) with adhesion molecules lymphocyte function-associated antigen-1 (LFA-1) engaging intercellular adhesion molecule-1 (ICAM-1) [1]. Yet it was quickly realized that the term encompasses a wide variety of molecular assemblies, depending on the cell types [2]. Moreover, synapses can form transiently (kinapses) and promote signaling [3], and even the relatively stable interactions evolve over time, including undergoing continuous surveillance of a chosen antigen-presenting cell (APC).
Our purpose in this mini-review is to point out some organizing principles that promote localized signaling and feedback loops involving ion channels, calcium influx, and the cytoskeleton. Both neuronal and immunological synapses serve to bring molecules into close proximity or direct contact, both laterally within the plasma membrane of pre- and post-synaptic elements and across the synaptic cleft. By focusing signaling into two-dimensional microdomains, synapses raise the effective concentrations of kinases, adapter molecules, and ion channels, thereby promoting local signaling and greatly accelerating the kinetics of reaction networks. Receptor–ligands interactions such as the T-cell receptor (TCR)–pMHC also appear to have fundamentally higher avidities for one another in the two-dimensional synaptic environment, as compared to the same pairs measured in solution phase [4]. Finally, cell motility is an integral part of the formation of these junctions, and thus, a treatment of the process from scanning to calcium signaling must include consideration of the activities of the cytoskeleton.
Synapse Ultrastructure and Microcluster Dynamics
The IS assembles and dissembles over the course of minutes to hours: a much more dynamic contact as compared to neurological synapses. However, over the relatively short period of its existence, it is engaged to produce the same degrees of efficiency and fidelity as compared to the more stable neuronal structure.
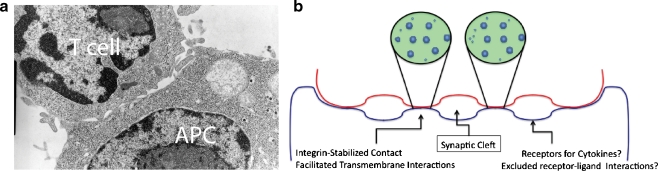
The ultrastructural view of a T cell–dendritic cell (DC), T cell–B cell, or cytotoxic T lymphocyte (CTL) target contacts [5, 6] reveals that the IS is indeed synaptic (see Fig. 1a, b)—namely that the distance between apposing cells varies from approximately 15 nm at their closest to distances on the order of 100 nm. This latter extracellular opening between cells, termed a synaptic cleft, is thought to be the region into which cytolytic granules and cytokines are released, much like neurotransmitters are released into the synaptic space in that structure. The intermembrane distances at the close appositions are then those presumably underlying the T-cell receptor contacts and, at the edges, likely receptor ligand pairs such as LFA-1/ICAM-1 [6–8]. While it has also been proposed that ICAM/LFA-enriched domains must be separated by approximately 40 nm, as this is the intermolecular distance calculated for the LFA-1/ICAM pair from crystal structures [7, 8], ultrastructural analyses failed to find distinct zones matching this outer spacing [6]. This suggests either that ICAM “bends” to accommodate a closer apposition or that these ligands must be densely concentrated at the edge of the contacting region, giving rise to its apparent accumulation there as visualized by fluorescence microscopy. The spacing between membranes in the synaptic clefts is highly variable (averages around 100 nm) but has been observed to account for upwards of 80% of the total length of the membrane in the synapse [6]. The arrangement of these closely and distantly apposed domains is also likely dynamic in nature although this has been difficult to ascertain experimentally.
Fig. 1.
Multifocal and uniform models of IS assembly. a TEM of D10 CD4+ T cell interacting with CH27 APC in the presence of 1 μM antigenic peptide, 15 min after the onset of coupling. Note relative paucity of contact area compared to synaptic regions. b Models for synaptic membrane configurations in cell–cell synapse and T-cell–bilayer synapse models. In this model, each contact region may be thought of as a microcosm of a bilayer contact in which microclusters of TCRs move and coalesce in a membrane patch (expanded views)
Interestingly, the synaptic nature of the contact, particularly the relatively minimal contact region, is likely absent in one of the most highly studied model for the immunological synapse, the supported lipid bilayer. The use of these bilayers, in which high densities of pMHC complexes and ICAM are seeded, has been tremendously important for revealing the fine details of molecular rearrangements in signaling structures, due to its ability to be imaged in the shallow illumination fields of total internal reflection (TIRF) microscopy. However, it is notable that cells form a c-SMAC-like structure in these systems in >90% of cases, whereas T–B-cell contacts do so with lower frequencies varying from 30% as measured by electron microscopy (EM) to 50–80% as observed by confocal microscopy [9]. This existence of “multifocal” synapses by both EM and in microscopy suggests that at least one parameter limiting complete “zippering” of membranes between T cells and APCs must be missing from the supported lipid bilayer models. Good candidates for such would be the highly sialylated glycocalyx of APCs or rigidity and curvature of the opposing antigen-presenting cell membrane.
Despite this pitfall, the benefits of the bilayer system in revealing dynamic signaling assemblies have been extensive. While en face views of IS assembly first characterized “clusters” of approximately 1 μm in minimum diameter that formed during signaling onset [10], TIRF imaging of T cells on bilayers revealed much smaller “microclusters” whose dimensions are on the order of a few hundred nanometers or less in diameter and which coalesce to form larger aggregates, often in the c-SMAC [11, 12]. In lipid bilayers, the microclusters “stream” toward the center of the contact, dependent upon an intact actin cytoskeleton [11, 12]. At the same time as the TCRs are moving inward, ICAM clusters are confined to the exterior of the contact where they remain in the p-SMAC. Such absence of streaming of ICAM may be a function of cluster size as artificially large ICAM clusters can induce ICAM–LFA-1 clusters to also stream toward the center [13]. Given the discrepancies in topology between the systems noted, it is tempting to imagine that each contact at a cell–cell junction may be a small microcosm of what is exemplified in the single larger and flatter contact that is formed in the lipid bilayer system (Fig. 1b).
Insights from In Vivo Imaging: Dynamics of Cellular Interactions and Synapse Formation
Within the past decade, two-photon imaging has permitted real-time visualization of cellular interactions in lymphoid organs and in peripheral tissues as T cells encounter APCs. T cells are more robustly motile in vivo than in vitro and come into contact with DC dendrites in the diffuse cortex of lymph nodes. In the absence of antigen, DCs make frequent but brief contacts with T cells, at a rate of 5–10,000 cells per hour with contact durations of 2–3 min, enabling efficient scanning of T cells of varying antigen specificity by a stochastic mechanism [14]. In the presence of antigen, CD4+ or CD8+ T-cell interactions with DCs evolve in distinct stages [15–17]: (1) prolonged but still intermittent interactions, (2) stable interactions lasting hours, and (3) release and swarming followed by episodes of T-cell proliferation. During stage 1, kinapses are formed with multiple DCs, and Ca2+ signals are evoked in the T cells by TCR engagement of pMHC on the DC surface. During stage 2, stable synapses are formed and interleukin-2 gene expression begins. During stage 3, T cells again make contacts with multiple DCs and a fresh wave of DC can modulate the outcome by increasing cytokine production [18].
An entirely different set of interactions takes place between helper CD4+ T cells and B cells [19]. B cells initially are localized to the follicle region of the lymph node, where they migrate randomly at a slower pace (8 μm/min) than T cells (12–15 μm/min) and are segregated from T cells in the surrounding diffuse cortex. Following antigen engagement, B cells up-regulate functional expression of the chemokine receptor CCR7 and migrate by chemotaxis to the follicle edge, following a gradient of CCL19 and CCL21. At the follicle edge, they encounter and begin to interact with activated helper T cells. Immediately following the initial contact, B cells and T cells begin to migrate as stable conjugate pairs, with B cells leading the way while dragging rounded up T cells behind. This second synaptic encounter is primarily monogamous, in contrast to T cell–DC encounters that may involve up to a dozen T cells forming stable contacts with a single DC. Moreover, the stability of T-cell–B-cell interaction allows migration as a conjugate pair. Although partner exchange can occur, such events are relatively infrequent. Migration may allow the B cell to direct the colonization of germinal centers while ensuring that a captive T cell provides help from the rear. Moreover, it is tempting to speculate that pairwise migration ensures widespread distribution of T-cell cytokines secreted from the rear of the cell.
Activated effector memory T (TEM) cells lose the ability to home into lymphoid organs and instead migrate to sites of inflammation. Imaging in spinal cord during autoimmune-mediated demyelination [20] and during delayed-type hypersensitivity [21] has shown that T cells initially arrest at the site of antigen presentation, enlarge as they become re-activated in the tissue environment, and then resume their motility. A similar choreography of thymocytes has been observed during positive selection in the thymus [22–24].
Recently, one of the T-cell subsynaptic molecules, linker for activation of T cells (LAT; tagged with eGFP), was imaged as T cells encountered DCs or B cells presenting antigen [25]. Under steady-state conditions, LAT was found distributed primarily in the trailing uropod of motile T cells. Upon contact with antigen-bearing DCs, recently activated T cells tended to form kinapses with DCs, whereas the same T cells formed motile conjugates with antigen-bearing B cells. T–B synapses were found to be more stable than T–DC synapses, and redistribution of LAT to the synapse was more apparent. Similar types of studies have been performed in one of our labs using TCR transgenic mice in which the alpha chain was tagged with GFP. Interestingly, these provide direct evidence for TCR internalization, indicative of T-cell signaling, at very transient contacts and in the absence of a stable c-SMAC-like structure (Friedman et al., in submission). These results show the feasibility of imaging synaptic molecules in vivo and reinforce previous imaging results indicating diverse IS characteristics depending upon the cell type.
Motility, Synapse Dynamics, and the Underlying Cytoskeleton
Unlike the neurological synapse, the IS must form, dissolve, form again, and so on through the lifespan of the T cell. It was first observed that during initial signaling, Ca2+ influx resulted in cortical relaxation of T cells and “rounding” [26–28]. In addition, there is then feedback between cell morphology changes as mitochondria pivot toward the synapse to support Ca2+ influx and buffering [29]. The initial Ca2+ rise, as a result of TCR signaling and discussed in further detail below, may function through a number of mechanisms including subsequent phosphorylation of ERM proteins that results in the detachment of the actin cytoskeleton from membrane proteins [30] (and reviewed in [31]) and/or phosphorylation of myosin heavy chains, resulting in loss of membrane tension [32]. The net result is that most IS are “motile” in the sense that that there is continuous movement of the contact, particularly at the edge. To understand this requires a brief review of the motility mechanisms of T cells prior to engagement.
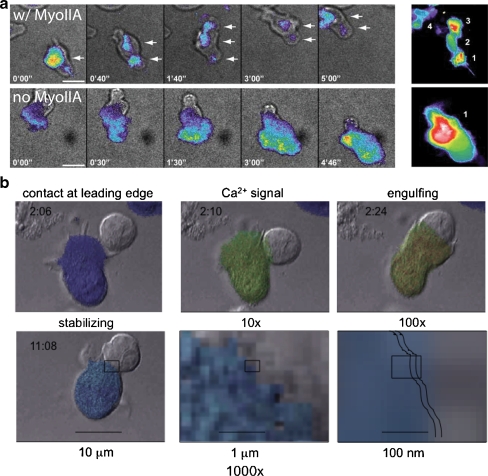
As noted above, the basal state for T-cell motility is one of stochastic scanning in which T cells dynamically move through the T-cell zone [33]. Similar scanning is also observed within the thymus during T-cell development [34]. During these behaviors, the T-cell cytoskeleton is appropriated for amoeboid motion in various ways. Most notably, lymphocytes appear to have integrin-dependent and independent modes of motion, with the latter predominating for lymphocyte motion in vivo [35, 36]. In vitro, two different modes can be directly observed (Fig. 2a), the integrin-independent mode being faster, myosin-dependent, and negatively regulated by the presence of integrin ligands. In contrast, the slower mode uses integrins as part of motility, is slower, and involves greater surface contacts. It is hypothesized that this latter mode may be used in the presence of more inflammatory milieu (chemokines and integrins) and enhanced by TCR-induced inactivation of myosin tension.
Fig. 2.
Motility modes for T cells and control of membrane–membrane contacts and control by calcium signaling. a Two modes of cell–substrate interaction during motility as observed in Jacobelli et al. 2009. Individual timepoints of contacts of actin–GFP bearing cells as visualized in TIRF, to visualize the contact surface, are shown. The right-hand panel is a time-sum of all the positions that the cell made with the surface during the entire time sequence. In the top (“w/ MyoIIA”): In a “walking” mode that gives rise to faster motility rates, T cells make multiple, sequential, and discrete contacts with the substrate. Bottom (“no MyoIIA”): In a “sliding” mode more akin to how mesenchymal cells move, cells make a continuous and large contact with the substrate, gliding along during propagation. b Formation of an immunological synapse. T-cell (fura-2 dye-loaded) and B-cell images from Negulescu et al. [26]. Colors indicate T-cell cytosolic Ca2+ (dark blue ∼50 nM to red >1 μM); times shown in each frame are minute:second. Contact is initiated by the leading edge of the T cell. The leading edge probes the APC for 20 s to several minutes before the contact area expands. Ca2+ signaling begins 10 s later. The Ca2+ signal can be oscillatory, transient, or sustained, as Ca2+ enters through CRAC channel Orai1 pore-forming subunits. The signal is sustained by K+ channel (Kv1.3 or KCa3.1) activity. Ca2+ must remain elevated for synapse stabilization, inhibition of motility, and NF-AT gene expression responses. Scale bars and boxes in lower panels indicate “zooming” into the synaptic region after initial stabilization of the synapse. Jagged lines indicate the approximate width of the synaptic cleft
Thus, T cells can always live in two modes—one which is optimized for navigation and one which is optimized for surface contact. Upon antigen engagement in vivo, T cells probably obligately convert to the latter but then appear to take one of two possible paths. On the one hand, they may simply slow their motility and more actively scan the milieu, repeatedly engaging the same APCs (serial scanning, akin to simply adopting the mesenchymal mode, described above). Alternatively, their dynamic motility may fully arrest as T cells round up and form the synapse with an APC [26–28], as illustrated in Fig. 2b. In multiple studies, cells often start with the former approach and then only later proceed to the more stable T/DC interactions [15–17], and it is now tempting to hypothesize that the serial scanning mode is produced by a change in motility mode, from the fast integrin-independent to the more adherent mode triggered by increased adhesion and decreases in cortical tension. It is notable though that increased stability of the T–APC synapse dynamics are quite generally observed: including in T/B interactions [19, 25], in TEM cells during DTH [21], and in autoimmune models [20].
A possible explanation for these variations in the motile state of the IS is suggested by the observation that the actin cytoskeleton is undergoing continuous polymerization and inward “streaming” in to the IS, even when the contact is relatively stable as mediated by a uniform lipid bilayer [37]. Thus, the cytoskeleton is not at all at rest, even when cells are “arrested”. Such streaming is likely to be mediated by actin polymerization mediated by TCR activation of WAVE or WASP complexes that subsequently trigger the Arp2/3 complex which nucleates actin assembly. At sites where strong stimuli are engaged, it can then be supposed that TCR microclusters themselves “slip” and trigger local integrin activation which prevents polymerization from inducing net forward movement. One attractive hypothesis then is simply that the IS always contains such motility-encouraging elements but that, in a uniform surface such as a lipid bilayer or a highly activated APC loaded with high densities of pMHC, the forces generated by this streaming are balanced such that the cell does not move. In contrast, when a single edge of an IS finds above average levels of pMHC or integrin, an imbalance of traction (relative to the other sites on the IS where traction is lower) results in net movement into that area.
Tying It All Together: Local Ca2+ Signaling Sustained by Ion Channels at the Synapse
The above discussion focuses highly upon the cytoskeletal relaxation that may accompany the transition to signaling, largely in motile synapses. But, what synapse features give rise to microcluster assemblies in the IS? The simplest answer is that stable and motile actin arrays underlie microclusters. Actin has been shown to modulate the ability of specific proteins to interact within the membrane [38] and further has been implicated in generating “island”-like domains in which multiple proteins can co-aggregate [39]. As noted, the actin cytoskeleton undergoes retrograde flow toward the center of individual contacts [37], suggesting that specific strands of actin may generate a flow to which microclusters can couple in order to reach the center of contact zones. The actin formed in the IS is also likely to be responsible for integrin-mediated adhesions which keep the IS from dissolving until signaling is ceased and these bonds are broken.
An as-yet unappreciated aspect of the cytoskeleton, however, is its role in delivering specific contents to the IS, in order to assist, reinforce, or attenuate signaling. To this extent, both the actin and tubulin cytoskeleton may act as rails upon which specific intracellular proteins may be delivered to nascent microclusters. Such has already been observed for the proximal kinase lck, for which a large intracellular pool has been observed relocalizing to the IS, albeit late in the response [40]. What other players need to be moved to the IS?
T lymphocytes express ion channels with a diverse repertoire of biophysical, molecular, and cellular functions [41]. Ca2+ signaling in T cells is linked to cell function in several ways including the rapid inhibition of motility (Fig. 2b) and delayed activation of gene expression via the cyclosporin-dependent calcineurin pathway. The default condition of resting T cells is robust motility with actively probing pseudopodia; active motility is inhibited rapidly by only a three- to sixfold increase in cytosolic Ca2+ [26]. To trigger gene expression responses, the Ca2+ signal must reach a cytosolic concentration tenfold higher than resting and be sustained for at least 2 h [42, 43]. For this purpose, three ion channels in the T-cell plasma membrane (PM) sustain Ca2+ signaling. Most importantly, Ca2+ release-activated Ca2+ (CRAC) channels conduct Ca2+ into the cell following TCR engagement. Although vital for T-cell activation and described biophysically as an inwardly rectifying and Ca2+-selective current more than 20 years ago [44], the CRAC channel eluded molecular identification and mechanistic understanding until recently. RNAi screening for molecules required for store-operated Ca2+ influx led first to the identification of stromal interaction molecule (STIM) proteins that functionally couple the endoplasmic reticulum (ER) to the plasma membrane and subsequently to Orai proteins that form the pore of the Ca2+ channel itself, as reviewed [45]. Upon depletion of Ca2+ from the ER lumen, STIM1 physically translocates from the bulk ER to activate Orai proteins in the plasma membrane. In primary resting T cells, STIM1 couples primarily to Orai1, but other isoforms are up-regulated following activation [46]. Two types of K+ channel provide an electrical counterbalance to the entry of Ca2+. The rise in cytosolic Ca2+ rapidly opens a K+ channel composed of KCa3.1 tetramers with CaM bound to the C-terminus. Ca2+ binds to CaM and rapid conformational changes then open the channels within milliseconds. The resulting K+ efflux helps to keep the membrane potential negative, compensating electrically for Ca2+ influx to prevent PM depolarization. A voltage-gated K+ channel composed of Kv1.3 tetramers also keeps the membrane potential negative and sustains Ca2+ signaling by virtue of its intrinsic voltage dependence. Any tendency toward membrane depolarization opens this K+ channel, and the resulting K+ efflux restores the membrane potential toward rest. A key point discussed previously [41] is that the balance of KCa3.1 and Kv1.3 channels depends upon the state of activation. In recently activated cells, KCa3.1 is up-regulated and predominates in the role of sustaining Ca2+ influx. Chronically activated T cells, however, become TEM cells with up-regulated levels of Kv1.3 and fewer KCa3.1 channels. This shift in channel expression results in TEM cells being more sensitive to channel blockers, enabling TEM cell-mediated autoimmune and inflammatory reactions to be targeted selectively in animal models [41].
Less is known about TRPM7, a ubiquitously expressed channel kinase. TRPM7 is expressed in T lymphocytes and is also regulated by intracellular Mg2+ ions by a surface charge screening mechanism [47]. Although it is Ca2+-permeable, there is no evidence that it participates in generating the Ca2+ signal following TCR engagement. Remarkably for an ion channel, TRPM7 bears a functional α-kinase domain at its C-terminus that autophosphorylates and in turn may alter key cytoskeletal proteins such as MyosinIIA [48]. TRPM7 knockouts are embryonic—lethal and conditional deletion in thymocytes, driven by CD4-CRE, leads to impaired thymopoeisis and block of T-cell development.
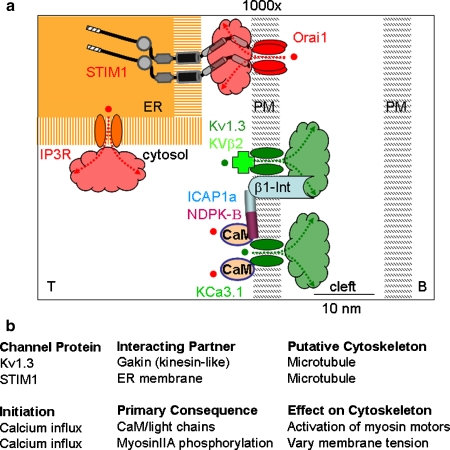
CRAC channels, formed from Orai1 and triggered by STIM1, and the two lymphocyte K+ channels, Kv1.3 and KCa3.1, form a functional network that sustains the Ca2+ signal in T cells. Moreover, within the past 5 years, all three ion channels have been shown to redistribute to the zone of contact at the immunological synapse (Fig. 3a): Orai1/STIM1 [46], KCa3.1 [49], and Kv1.3 [50–55]. The co-localization of ion channels to the synapse may strongly influence signaling by several mechanisms. (1) Most importantly, Ca2+ influx localized to the synapse results in a higher localized concentration in microdomains within the subsynaptic region adjacent to the T-cell plasma membrane. This could alter the activity of Ca2+-dependent enzymes and cytoskeletal elements. (2) Localized influx could produce significant depletion of Ca2+ from the synaptic cleft between the T cell and the APC, providing negative feedback to keep cytosolic Ca2+ from going too high. (3) Localized efflux of K+ into the synaptic cleft may increase extracellular K+ concentration, in turn promoting membrane potential depolarization of T cell, APC, or both. (4) Beta subunits (Kvβ2 and hDig/SAP97) of Kv1.3 may link these channels to β1-integrin, ZIP proteins linking to PKCζ, to GAKIN linking to other cytoskeletal elements, and via Lck to CD4 and the TCR. Kvβ2 also has enzyme activity as an aldo-keto reductase and binds NADP with high affinity. Furthermore, Kv1.3 trafficking to the IS is defective in T cells from patients with systemic lupus erythematosus and may result in excessive Ca2+ signaling [56]. (5) KCa3.1/CaM is coupled to a kinase (nucleoside diphosphate kinase, NDPK-B) and to a phosphatase (histidine phosphatase, PHPT-1), both of which modulate KCa3.1 channel function. NDPK-B interacts with the integrin-associated protein 1 alpha (ICAP-1α), a molecule that may link KCa3.1 to β1-integrin.
Fig. 3.
Calcium channels and the cytoskeleton at the immunological synapse. a Zoomed-in schematic diagram of region shown by box in the lower right panel of Fig. 2a, showing T-cell molecules concentrated at the synapse, separated by a narrow synaptic cleft (∼15–20 nm) from the B cell. Scales are approximate. A STIM1 dimer within the endoplasmic reticulum (ER) is shown activating a tetramer of Orai1 in the plasma membrane by direct contact. Protein domains of STIM1 include (from left to right): signal peptide, EF hand (without bound Ca2+), and SAM domains in the ER lumen; a transmembrane segment, and bipartite coiled-coil in cytosol. Depletion of the ER Ca2+ store by release of Ca2+ ions (red) through the IP3 receptor (IP 3 R) initiates signaling via STIM1 to the PM. Ca2+ first unbinds from the EF hand; STIM1 then forms oligomers that translocate to ER–PM junctions; Orai1 tetramers then open; Ca2+ ions enter the cell and begin to diffuse. Ca2+ opens KCa3.1/CaM; membrane depolarization opens Kv1.3. K+ ions (green), leaves the cytosol, and enters the cleft. Kv1.3 and KCa3.1 are linked through associated subunits to enzymes, to β1-integrin (β1-Int), and to cytoskeletal elements. Clouds indicate diffusion of Ca2+ and K+ ions and accumulation in nanodomains near Orai1 (red) and Kv1.3 and KCa3.1 channels (green). b Putative interplay between channels and the cytoskeleton. Channels themselves are likely assembled as a function of dynamic reorganization of cytoskeletal elements. Conversely, calcium transients and gradients almost certainly integrate at multiple levels to continue to stabilize or destabilize motility modes
Overview and Future Questions
Ion channels have joined the array of molecules that localize at the synapse and have the potential—through local ionic signaling and physical links to receptors, enzymes, and cytoskeletal elements—to effect modulation of a network of other elements. Given the level of potential complexity, there is still much to be learned about the local signaling dynamics in the IS. Some of the key questions are: What are the mechanisms that enable ion channels to redistribute into the synapse? Figure 3b illustrates some of the key interactions that are currently known but where the exact cytoskeletal mechanisms remain undiscovered. Of particular note is the movement of the STIM1 subunit, which resides on the ER membrane, into close proximity with the Orai1 subunit, localized on the plasma membrane—might this ER translocation utilize a cytoskeletal rail system for its assembly? Conversely, what are the feedback loops between Ca2+ signaling and synapse stabilization, notably how does localized calcium gradients affect the local balances of forces that influence cell movement? For example, in motile fibroblasts, “flickers” of calcium are observed at the leading edge, likely in part regulated by TRPM7, perhaps functioning as a stretch-sensitive calcium-permeable channel [57]. Does “balanced” calcium flux at the IS maintain a stable (non-motile) IS and, in contrast, does variegated microcluster formation lead to a destabilization and result in probing motility? Similar questions surround the cessation of signaling at the IS: Does Ca2+ depletion or K+ accumulation in the synaptic cleft initiate or terminate signaling in DCs or B cells as the APC? Finally, how may ion channel modulators (blockers or openers) be harnessed for therapeutic action in autoimmunity and transplantation? As a promising example, Kv1.3 blockade in vivo by a peptide inhibitor has shown efficacy in experimental autoimmune encephalitis and in delayed type hypersensitivity responses, and it completely immobilizes TEM cells in the periphery without altering the motility of naive T cells in the lymph node [21]. It is becoming clear that answering these questions will require an integrated view extending from calcium channel function to localized motility and the action of the T-cell cytoskeleton.
Acknowledgments
This study was supported by National Institutes of Health grants GM-41514 and NS-14609 (MDC) and by AI52116 and the Leukemia and Lymphoma Foundation (MFK).
Conflict of interests
None declared.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Matthew F. Krummel, Email: matthew.krummel@ucsf.edu
Michael D. Cahalan, Email: mcahalan@uci.edu
References
- 1.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–6. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 2.Trautmann A, Valitutti S. The diversity of immunological synapses. Curr Opin Immunol. 2003;15:249–54. doi: 10.1016/S0952-7915(03)00040-2. [DOI] [PubMed] [Google Scholar]
- 3.Gunzer M, Schafer A, Borgmann S, Grabbe S, Zanker KS, et al. Antigen presentation in extracellular matrix: interactions of T cells with dendritic cells are dynamic, short lived, and sequential. Immunity. 2000;13:323–32. doi: 10.1016/S1074-7613(00)00032-7. [DOI] [PubMed] [Google Scholar]
- 4.Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, et al. TCR–peptide–MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–7. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stinchcombe JC, Bossi G, Booth S, Griffiths GM. The immunological synapse of CTL contains a secretory domain and membrane bridges. Immunity. 2001;15:751–61. doi: 10.1016/S1074-7613(01)00234-5. [DOI] [PubMed] [Google Scholar]
- 6.Brossard C, Feuillet V, Schmitt A, Randriamampita C, Romao M, et al. Multifocal structure of the T cell–dendritic cell synapse. Eur J Immunol. 2005;35:1741–53. doi: 10.1002/eji.200425857. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AS, Dustin ML. Making the T cell receptor go the distance: a topological view of T cell activation. Immunity. 1997;6:361–9. doi: 10.1016/S1074-7613(00)80279-4. [DOI] [PubMed] [Google Scholar]
- 8.Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol. 2006;7:803–9. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- 9.Richie LI, Ebert PJ, Wu LC, Krummel MF, Owen JJ, et al. Imaging synapse formation during thymocyte selection: inability of CD3zeta to form a stable central accumulation during negative selection. Immunity. 2002;16:595–606. doi: 10.1016/S1074-7613(02)00299-6. [DOI] [PubMed] [Google Scholar]
- 10.Krummel MF, Sjaastad MD, Wülfing C, Davis MM. Differential assembly of CD3z and CD4 during T cell activation. Science. 2000;289:1349–52. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- 11.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–27. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:117–27. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 13.Hartman NC, Nye JA, Groves JT. Cluster size regulates protein sorting in the immunological synapse. Proc Natl Acad Sci U S A. 2009;106:12729–34. doi: 10.1073/pnas.0902621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc Natl Acad Sci U S A. 2004;101:998–1003. doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bousso P, Robey E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat Immunol. 2003;4:579–85. doi: 10.1038/ni928. [DOI] [PubMed] [Google Scholar]
- 16.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–9. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 17.Miller MJ, Safrina O, Parker I, Cahalan MD. Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J Exp Med. 2004;200:847–56. doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celli S, Garcia Z, Bousso P. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J Exp Med. 2005;202:1271–8. doi: 10.1084/jem.20051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:1047–61. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami N, Nagerl UV, Odoardi F, Bonhoeffer T, Wekerle H, et al. Live imaging of effector cell trafficking and autoantigen recognition within the unfolding autoimmune encephalomyelitis lesion. J Exp Med. 2005;201:1805–14. doi: 10.1084/jem.20050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matheu MP, Beeton C, Garcia A, Chi V, Rangaraju S, et al. Imaging of effector memory T cells during a delayed-type hypersensitivity reaction and suppression by Kv1.3 channel block. Immunity. 2008;29:602–14. doi: 10.1016/j.immuni.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhakta NR, Oh DY, Lewis RS. Calcium oscillations regulate thymocyte motility during positive selection in the three-dimensional thymic environment. Nat Immunol. 2005;6:143–51. doi: 10.1038/ni1161. [DOI] [PubMed] [Google Scholar]
- 23.Witt CM, Raychaudhuri S, Schaefer B, Chakraborty AK, Robey EA. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 2005;3:1062–9. doi: 10.1371/journal.pbio.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrlich LI, Oh DY, Weissman IL, Lewis RS. Differential contribution of chemotaxis and substrate restriction to segregation of immature and mature thymocytes. Immunity. 2009;31:986–98. doi: 10.1016/j.immuni.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azar GA, Lemaitre F, Robey EA, Bousso P. Subcellular dynamics of T cell immunological synapses and kinapses in lymph nodes. Proc Natl Acad Sci U S A. 2010;107:3675–80. doi: 10.1073/pnas.0905901107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negulescu PA, Krasieva TB, Khan A, Kerschbaum HH, Cahalan MD. Polarity of T cell shape, motility, and sensitivity to antigen. Immunity. 1996;4:421–30. doi: 10.1016/S1074-7613(00)80409-4. [DOI] [PubMed] [Google Scholar]
- 27.Delon J, Stoll S, Germain RN. Imaging of T-cell interactions with antigen presenting cells in culture and in intact lymphoid tissue. Immunol Rev. 2002;189:51–63. doi: 10.1034/j.1600-065X.2002.18906.x. [DOI] [PubMed] [Google Scholar]
- 28.Donnadieu E, Bismuth G, Trautmann A. Antigen recognition by helper T cells elicits a sequence of distinct changes of their shape and intracellular calcium. Curr Biol. 1994;4:584–95. doi: 10.1016/S0960-9822(00)00130-5. [DOI] [PubMed] [Google Scholar]
- 29.Quintana A, Kummerow C, Junker C, Becherer U, Hoth M. Morphological changes of T cells following formation of the immunological synapse modulate intracellular calcium signals. Cell Calcium. 2009;45:109–22. doi: 10.1016/j.ceca.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Faure S, Salazar-Fontana LI, Semichon M, Tybulewicz VL, Bismuth G, et al. ERM proteins regulate cytoskeleton relaxation promoting T cell–APC conjugation. Nat Immunol. 2004;5:272–279. doi: 10.1038/ni1039. [DOI] [PubMed] [Google Scholar]
- 31.Burkhardt JK, Carrizosa E, Shaffer MH. The actin cytoskeleton in T cell activation. Annu Rev Immunol. 2008;26:233–59. doi: 10.1146/annurev.immunol.26.021607.090347. [DOI] [PubMed] [Google Scholar]
- 32.Jacobelli J, Chmura SA, Buxton DB, Davis MM, Krummel MF. A single class II myosin modulates T cell motility and stopping but not synapse assembly. Nat Immunol. 2004;5:531–8. doi: 10.1038/ni1065. [DOI] [PubMed] [Google Scholar]
- 33.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–73. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 34.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte–stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–80. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 35.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–5. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 36.Woolf E, Grigorova I, Sagiv A, Grabovsky V, Feigelson SW, et al. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nat Immunol. 2007;8:1076–85. doi: 10.1038/ni1499. [DOI] [PubMed] [Google Scholar]
- 37.Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc Natl Acad Sci U S A. 2007;104:20296–301. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein–protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–50. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–6. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrlich LI, Ebert PJ, Krummel MF, Weiss A, Davis MM. Dynamics of p56lck translocation to the T cell immunological synapse following agonist and antagonist stimulation. Immunity. 2002;17:809–22. doi: 10.1016/S1074-7613(02)00481-8. [DOI] [PubMed] [Google Scholar]
- 41.Cahalan MD, Chandy KG. The functional network of ion channels in T lymphocytes. Immunol Rev. 2009;231:59–87. doi: 10.1111/j.1600-065X.2009.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fanger CM, Hoth M, Crabtree GR, Lewis RS. Characterization of T cell mutants with defects in capacitative calcium entry: genetic evidence for the physiological roles of CRAC channels. J Cell Biol. 1995;131:655–67. doi: 10.1083/jcb.131.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negulescu PA, Shastri N, Cahalan MD. Intracellular calcium dependence of gene expression in single T lymphocytes. Proc Natl Acad Sci U S A. 1994;91:2873–7. doi: 10.1073/pnas.91.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis RS, Cahalan MD. Mitogen-induced oscillations of cytosolic Ca2+ and transmembrane Ca2+ current in human leukemic T cells. Cell Regul. 1989;1:99–112. doi: 10.1091/mbc.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahalan MD. STIMulating store-operated Ca(2+) entry. Nat Cell Biol. 2009;11:669–77. doi: 10.1038/ncb0609-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lioudyno MI, Kozak JA, Penna A, Safrina O, Zhang SL, et al. Orai1 and STIM1 move to the immunological synapse and are up-regulated during T cell activation. Proc Natl Acad Sci U S A. 2008;105:2011–6. doi: 10.1073/pnas.0706122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozak JA, Matsushita M, Nairn AC, Cahalan MD. Charge screening by internal pH and polyvalent cations as a mechanism for activation, inhibition, and rundown of TRPM7/MIC channels. J Gen Physiol. 2005;126:499–514. doi: 10.1085/jgp.200509324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, et al. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. EMBO J. 2006;25:290–301. doi: 10.1038/sj.emboj.7600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicolaou SA, Neumeier L, Peng Y, Devor DC, Conforti L. The Ca(2+)-activated K(+) channel KCa3.1 compartmentalizes in the immunological synapse of human T lymphocytes. Am J Physiol Cell Physiol. 2007;292:C1431–9. doi: 10.1152/ajpcell.00376.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicolaou SA, Neumeier L, Steckly A, Kucher V, Takimoto K, et al. Localization of Kv1.3 channels in the immunological synapse modulates the calcium response to antigen stimulation in T lymphocytes. J Immunol. 2009;183:6296–302. doi: 10.4049/jimmunol.0900613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicolaou SA, Szigligeti P, Neumeier L, Lee SM, Duncan HJ, et al. Altered dynamics of Kv1.3 channel compartmentalization in the immunological synapse in systemic lupus erythematosus. J Immunol. 2007;179:346–56. doi: 10.4049/jimmunol.179.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panyi G, Bagdany M, Bodnar A, Vamosi G, Szentesi G, et al. Colocalization and nonrandom distribution of Kv1.3 potassium channels and CD3 molecules in the plasma membrane of human T lymphocytes. Proc Natl Acad Sci U S A. 2003;100:2592–7. doi: 10.1073/pnas.0438057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panyi G, Vamosi G, Bacso Z, Bagdany M, Bodnar A, et al. Kv1.3 potassium channels are localized in the immunological synapse formed between cytotoxic and target cells. Proc Natl Acad Sci U S A. 2004;101:1285–90. doi: 10.1073/pnas.0307421100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toth A, Szilagyi O, Krasznai Z, Panyi G, Hajdu P. Functional consequences of Kv1.3 ion channel rearrangement into the immunological synapse. Immunol Lett. 2009;125:15–21. doi: 10.1016/j.imlet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Beeton C, Wulff H, Standifer NE, Azam P, Mullen KM, et al. Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc Natl Acad Sci U S A. 2006;103:17414–19. doi: 10.1073/pnas.0605136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicolaou SA, Neumeier L, Takimoto K, Lee SM, Duncan HJ, et al. Differential calcium signaling and Kv1.3 trafficking to the immunological synapse in systemic lupus erythematosus. Cell Calcium. 2010;47:19–28. doi: 10.1016/j.ceca.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei C, Wang X, Chen M, Ouyang K, Song LS, et al. Calcium flickers steer cell migration. Nature. 2009;457:901–5. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobelli J, Bennett FC, Pandurangi P, Tooley AJ, Krummel MF. Myosin-IIA and ICAM-1 regulate the interchange between two distinct modes of T cell migration. J Immunol. 2009;182:2041–50. doi: 10.4049/jimmunol.0803267. [DOI] [PubMed] [Google Scholar]