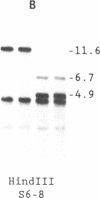
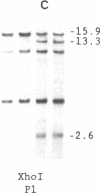
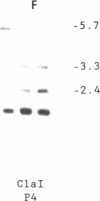
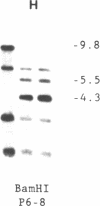
Abstract
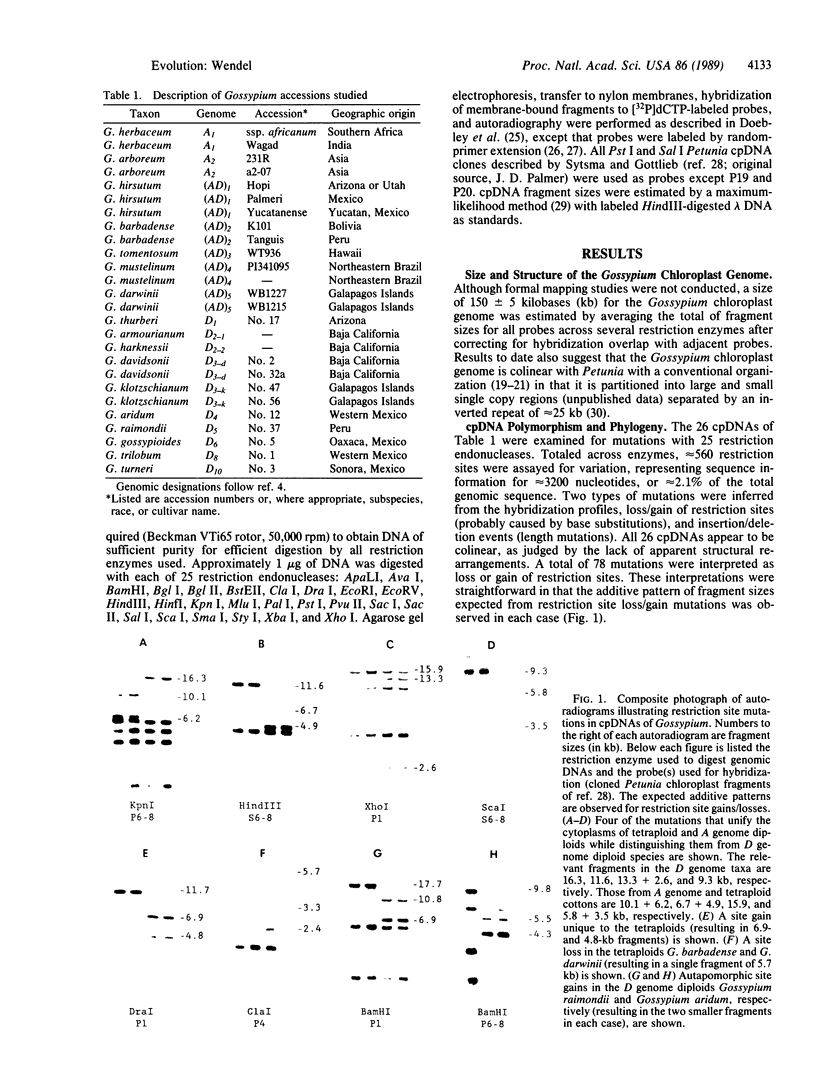
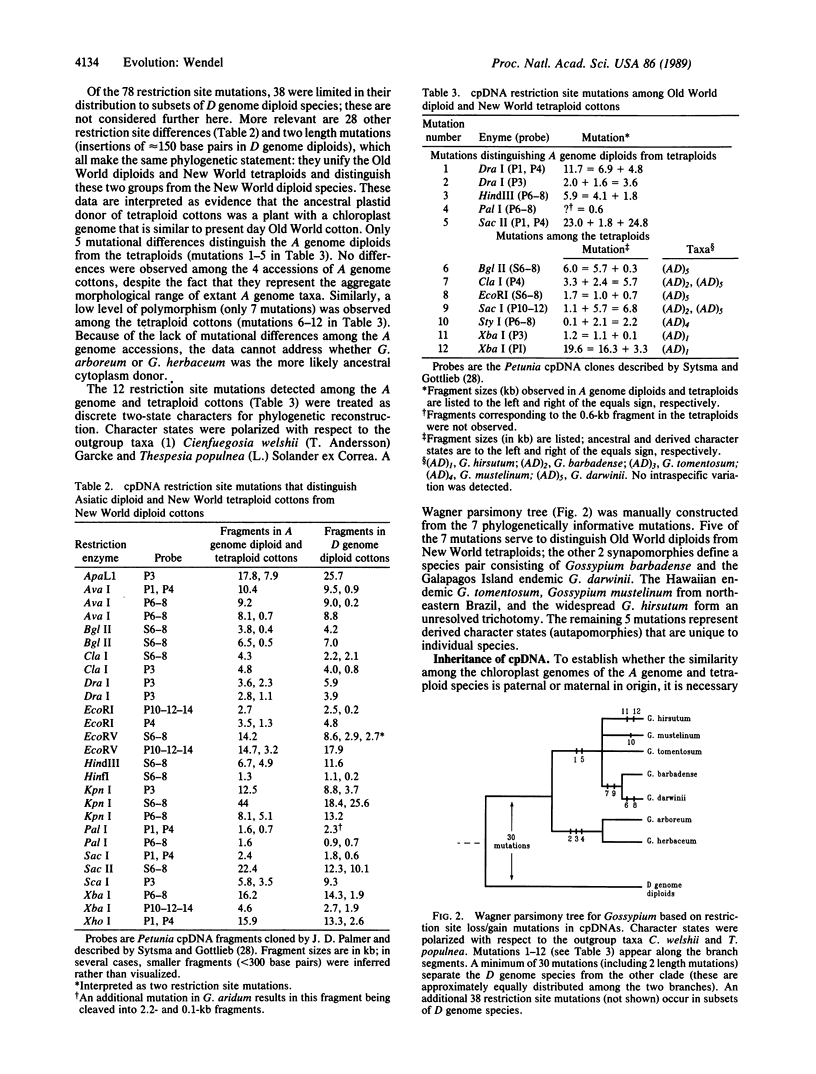
New World tetraploid cottons (Gossypium spp.) originated through hybridization of ancestral diploid species that presently have allopatric ranges in Asia-Africa (the A genome) and the New World tropics and subtropics (the D genome). Despite intensive study, the identity of the parental diploids and the antiquity of polyploidization remain unresolved. In this study, variation in the maternally inherited chloroplast genome was assessed among species representing both of the parental genomes and the tetraploids. Approximately 560 restriction sites were assayed in each accession, representing sequence information for about 3200 nucleotides. The resulting maternal phylogeny has no convergent restriction site mutations and demonstrates that the cytoplasm donor for all tetraploid species was an A genome diploid with a chloroplast genome that is similar to Gossypium arboreum and Gossypium herbaceum. No mutational differences were detected between these two species, and few mutations distinguish the chloroplast genomes of A genome diploids from those of tetraploid taxa. In contrast to expectations based on extensive taxonomic, geographic, and genetic diversity, a surprisingly low level of sequence divergence has accumulated subsequent to polyploidization. Chloroplast genomes of tetraploid species are distinguished from each other by between one and six apparent point mutations. The data suggest that tetraploid cotton originated relatively recently, perhaps within the last 1-2 million years, with subsequent rapid evolution and diversification throughout the New World tropics.
Keywords: chloroplast DNA, molecular evolution, Gossypium, phylogeny, allopolyploidy
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clegg M. T., Rawson J. R., Thomas K. Chloroplast DNA variation in pearl millet and related species. Genetics. 1984 Mar;106(3):449–461. doi: 10.1093/genetics/106.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Renfroe W., Blanton A. Restriction site variation in the zea chloroplast genome. Genetics. 1987 Sep;117(1):139–147. doi: 10.1093/genetics/117.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Menzel M Y, Brown M S. The Significance of Multivalent Formation in Three-Species Gossypium Hybrids. Genetics. 1954 Jul;39(4):546–557. doi: 10.1093/genetics/39.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Tajima F. Maximum likelihood estimation of the number of nucleotide substitutions from restriction sites data. Genetics. 1983 Sep;105(1):207–217. doi: 10.1093/genetics/105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Palmer J. D., Jorgensen R. A., Thompson W. F. Chloroplast DNA variation and evolution in pisum: patterns of change and phylogenetic analysis. Genetics. 1985 Jan;109(1):195–213. doi: 10.1093/genetics/109.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Zamir D. Chloroplast DNA evolution and phylogenetic relationships in Lycopersicon. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5006–5010. doi: 10.1073/pnas.79.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer H. E., Sederoff R. R. Improved estimation of DNA fragment lengths from Agarose gels. Anal Biochem. 1981 Jul 15;115(1):113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]
- Sears B. B. Elimination of plastids during spermatogenesis and fertilization in the plant kingdom. Plasmid. 1980 Nov;4(3):233–255. doi: 10.1016/0147-619x(80)90063-3. [DOI] [PubMed] [Google Scholar]
- Sytsma K. J., Gottlieb L. D. Chloroplast DNA evidence for the origin of the genus Heterogaura from a species of Clarkia (Onagraceae). Proc Natl Acad Sci U S A. 1986 Aug;83(15):5554–5557. doi: 10.1073/pnas.83.15.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. B., Furnier G. R., Saghai-Maroof M. A., Williams S. M., Dancik B. P., Allard R. W. Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc Natl Acad Sci U S A. 1987 Apr;84(7):2097–2100. doi: 10.1073/pnas.84.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Clegg M. T., Brown A. H. The Nature of Nucleotide Sequence Divergence between Barley and Maize Chloroplast DNA. Genetics. 1984 Apr;106(4):735–749. doi: 10.1093/genetics/106.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]