Abstract
Despite the unique brain imaging capabilities and advantages of functional near-infrared spectroscopy (fNIRS), including portability and comprehensive hemodynamic measurement, widespread acceptance in the neuroimaging community has been hampered by low spatial resolution and image localization errors. While recent technical developments such as high-density diffuse optical tomography (HD-DOT) have, in principle, been shown to have superior in silico image quality, the majority of optical imaging studies are still conducted with sparse fNIRS arrays, perhaps partially because the performance increases of HD-DOT appear incremental. Without a quantitative comparative analysis between HD-DOT and fNIRS, using both simulation and in vivo neuroimaging, the implications of the new HD-DOT technology have been difficult to judge. We present a quantitative comparison of HD-DOT and two commonly used fNIRS geometries using (1) standard metrics of image quality, (2) simulated brain mapping tasks, and (3) in vivo visual cortex mapping results in adult humans. The results show that better resolution and lower positional errors are achieved with HD-DOT and that these improvements provide a substantial advancement in neuroimaging capability. In particular, we demonstrate that HD-DOT enables detailed phase-encoded retinotopic mapping, while sparse arrays are limited to imaging individual block-design visual stimuli.
Keywords: medical imaging, biomedical optics, image quality, imaging systems, diffusion, diffuse optical tomography, optical neuroimaging
Introduction
Near-infrared spectroscopy (fNIRS) holds the promise to extend functional neuroimaging methods into new settings, such as the assessment of brain function in clinical patients unable to be transported for functional magnetic resonance imaging (fMRI). However, the successful transition of optical techniques from intriguing concept to useful neuroscience tool has been hampered by difficulties in acquiring measurements through the scalp and skull. The standard fNIRS method of acquiring arrays of sparsely distributed measurements has limited spatial resolution and irregular spatial sensitivity, resulting in subsequent mislocalization of cortical hemodynamics. Initial simulation studies of an emerging technique known as high-density diffuse optical tomography (HD-DOT) have shown potential improvements for neuroimaging in both resolution and localization errors.1, 2, 3, 4 Human brain mapping studies have shown that HD-DOT studies have been able to generate detailed activation maps.5, 6, 7, 8, 9 However, the link between theoretical comparisons and in vivo results remains circumstantial. In this paper, we provide a thorough evaluation of the abilities of sparse fNIRS and HD-DOT from simulation through in vivo cortical mapping. This analysis consists of three parts: (1) a comparison of resolution and localization errors in dense and sparse diffuse optical imaging using point-spread functions; (2) simulations of more realistic activation patterns similar to those seen in vivo; and (3) in vivo cortical mapping using both the HD-DOT system and a sparse subset of the same imaging array.
fNIRS systems use diffusely scattered near-infrared light to measure changes in tissue absorption, scattering, and fluorescence. Within the context of brain imaging, changes in absorption are primarily due to variance in blood volume and oxygenation. Task-induced changes in the concentrations of oxy-, deoxy-, and total hemoglobin can then be used to access neural activation in much the same manner as the blood oxygenation level dependent (BOLD) contrast for fMRI.10, 11 The majority of such fNIRS studies are conducted using distinct measurements of sources and detectors arranged in a sparse grid. Since sources and detectors generally need to be displaced by approximately 3 cm in order for the sensitivity function to have significant brain sensitivity, this separation becomes a primary factor limiting spatial resolution.
HD-DOT systems use a grid of overlapping source–detector measurements to allow multiple measurements within each voxel of the imaged volume.12, 13, 14 This technique provides better spatial sampling and a more robust approach to image reconstruction, which in turn provides better localization and resolution of the imaged activations.1, 2 The goal of the present work is to provide multiple analyses that quantitatively evaluate the imaging improvements possible with HD-DOT. First, we examine standard simulation metrics in order to understand the ideal performance of multiple fNIRS systems in current use. It is not possible to reproduce such point activations in vivo due to the large spatial extent of brain activations. However, as shown in our earlier publication,6 the visual cortex provides an ideal control system for such a comparison due to its highly organized and detailed structure. For these reasons, the visual cortex has served to validate past improvements in optical neuroimaging analysis.9, 15, 16, 17, 18, 19 Thus, second, we examine the performance of multiple imaging strategies during a simulated retinotopic mapping paradigm. This task synthesizes information from multiple stimulus presentation frames to create a complete map of cortical organization. Here, subtle imaging defects can become apparent. Then, third, these results are compared with our adult human data mapping the structure of the visual cortex. Both simulations and human data show that sparse optode arrangements are unable to provide high enough image quality to perform detailed neuromapping studies, such as retinotopy, that move beyond individual activations to resolving the pattern’s cortical organization. In contrast, we show that HD-DOT has superior performance and is able to reproduce expected in silico performance during in vivo applications.
Methods
Optode Array Arrangements
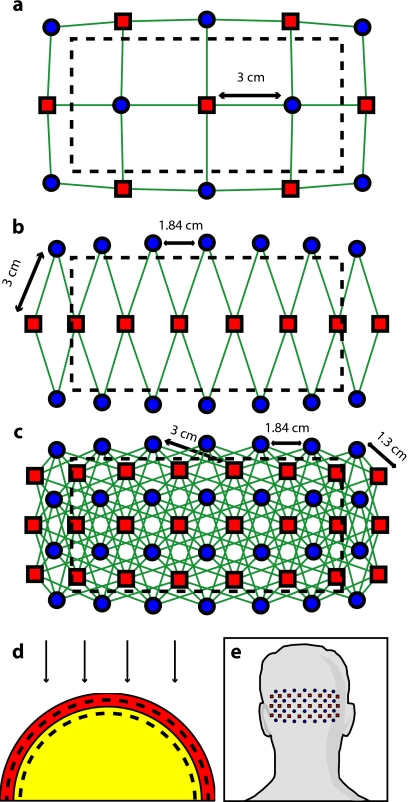
In order to test the performance of various fNIRS optode grids for in vivo brain imaging, we created three arrangements of sources and detectors. All of the grids were first created in two dimensions with the standard interoptode spacing: 3 cm between sources and detectors in the sparse grids, and 1.3 cm for first-nearest neighbors and 3 cm for second-nearest neighbors in the high-density grid. The grids were then conformed to an 8-cm-radius hemisphere preserving distance and angle between each optode and the center point of the grid, which was located at the apex of the hemisphere. The first array was a standard square sparse array of interleaved sources and detectors [Fig. 1a; 7 sources and 8 detectors for 22 total measurements], which is the most commonly used fNIRS geometry.20, 21 The second array was a second sparse array using a line of sources flanked by two rows of detectors [Fig. 1b; 8 sources and 14 detectors for 28 total measurements], which is another commonly used arrangement with the goal of increasing resolution along one axis.22, 23 (We will refer to this arrangement as the triangular sparse array.) The third array was a high-density grid developed for the visual cortex5 [Fig. 1c; 24 sources and 28 detectors for 212 first- and second-nearest neighbor measurements].
Figure 1.
Diffuse optical imaging optode arrays. Sources are red squares, detectors are blue circles, and measurements are green lines. (a) Schematic of the square sparse imaging array. The two-dimensional (2-D) grid with 3-cm spacing was conformed to an 8-cm-radius sphere. Shown is a projection of the resulting three-dimensional (3-D) optode locations. (b) Schematic of the triangular sparse imaging array. (c) Schematic of the high-density imaging array. First-nearest neighbors have 1.3-cm spacing and second-nearest neighbors have 3-cm spacing. (d) Cross section through the 3-D head model. The inner brain region (yellow) has a radius of 7 cm, and the outer skin/skull region is 1 cm thick. All images shown in later figures are posterior coronal projections (direction of view noted by arrows) of a 1-cm-thick shell around the cortical surface (dashed lines). (e) Schematic of the high-density imaging array placed over the occipital cortex of a human subject. (Color online only.)
Forward Model
A two-layer, hemispherical head model (80-mm radius) was used for forward light modeling. The model consisted of an inner brain region (70-mm radius; optical properties at 750 nm: μa=0.15 cm−1, ; at 850 nm: μa=0.17 cm−1, ) surrounded by an outer skin∕skull region (10-mm thickness; optical properties at 750 nm: μa=0.19 cm−1, ; at 850 nm: μa=0.19 cm−1,). These optical properties were derived by linear interpolation from the values used by Strangman et al.24 A tetrahedral mesh (162,040 nodes) was created in NetGen. Finite-element forward light modeling was performed using NIRFAST25 to generate Green’s functions for the sensitivities of each source and detector. These Green’s function were converted from the tetrahedral geometry to a voxelized space (voxels were cubic with a side length of 2 mm) and cropped to the region with high sensitivity (140-mm width, 80-mm height, 48-mm depth, for a total of 67,200 voxels). Sensitivity matrices, A, for each array were constructed using the adjoint formulation and normalized consistent with the Rytov approximation: y=Ax, where y is a vector of differential light measurements [yi=−log(Ii∕⟨Ii⟩); where i runs over all measurements], and x is a vector of the discrete representation of absorption perturbations in the volume.
Inverse Formulation and Reconstruction
The finite-element model was directly inverted for image reconstruction.26, 27 The inverse problem minimizes the objective function: . A solution, x=A#ymeas, was obtained using a Moore-Penrose generalized inverse with, A#=AT(AAT+α2I)−1ymeas. The optimal value of α=10−2s (where s is the maximal singular value of A) was found to provide a good balance between resolution and contrast-to-noise by evaluating in vivo activation data.6 The same values were used for reconstructing all three optode arrays.
Visualization
From the resulting three-dimensional (3-D) inverted A-matrix, we selected a 10-mm-thick cortical shell centered over the surface of the cortex (radius 65 mm to 75 mm). Voxels within this shell were averaged in depth to create a two-dimensional (2-D) posterior cortical projection [Fig. 1d] of dimensions 140 mm by 80 mm. All reconstructions were performed using this shell. (Note that this is different from a cortically constrained reconstruction,28 since the cortical region is chosen after rather than before inversion.) For visualization, we display a smaller, centered field of view of 102-mm width and 50-mm height in order to focus on the regions under the arrays with the highest sensitivities and lower image artifacts. This region is shown in Figs. 1a, 1b, 1c by the black dashed line.
Simulation and Point-Spread Function Analysis
Point-spread functions (PSFs) were created by simulating a perturbation at a single point. The simulated absorption perturbation, xsim, is a vector of zeros except for one target pixel with a value of one. We then generated simulated measurements ysim=Axsim and reconstructed an image xrecon=A#ysim. With this particular choice of xsim, this is equivalent to examining a row from the matrix A#A (known as the resolution matrix).
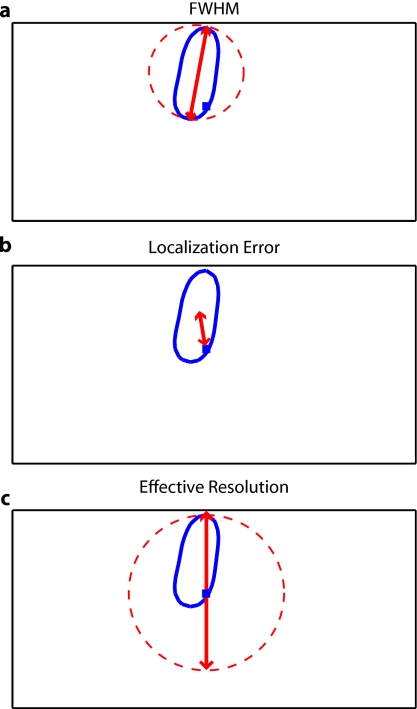
Since the sparse arrays often reconstruct asymmetric and elliptical responses, using the square root of the area above half maximum to construct a full width at half maximum (FWHM) tends to be a poor measure of a response’s characteristic length. Thus, we defined FWHM as the maximum separation between all pairs of points in the activation, which is a lower bound on the diameter of a circle needed to completely enclose the activation [Fig. 2a]. Another standard image metric is the localization error, defined as the distance between the centroid of the response and the known location of the target [Fig. 2b]. In neuroscience applications where the location of the activation is unknown, mislocalization and an overly large activation are both equivalent in that they cause brain hemodynamics to be measured where none actually occurred. Thus, we combined these two image metrics into a single “effective resolution” defined as the diameter of a circle centered at the known perturbation position needed to enclose the entire activation [Fig. 2c], which should more closely characterize expected in vivo performance. For simplicity, all images shown were made using the 850-nm A-matrices.
Figure 2.
Definitions of imaging metrics for point-spread function analysis shown using a simulated image reconstruction from the triangular sparse array. The target perturbation is the blue square, and the contour at half-maximum of the reconstruction is the blue line. (a) Full width at half maximum (FWHM) is defined as the maximum separation between any two points above half maximum in the reconstruction (red arrows). This is a lower bound on the diameter of a circle needed to enclose the reconstruction (red circle). (b) Localization error is the separation of the known target position and the centroid of the reconstruction (red arrows). (c) Effective resolution is the diameter of a circle centered at the known target position needed to enclose all points above half maximum in the reconstruction (red circle and arrows). (Color online only.)
Note that we do not explicitly include noise in the simulated data. However, we do set the regularization based on in vivo imaging. Thus, the reconstructed images implicitly reflect the smoothing necessary to accommodate real-world measurement noise. Working with noiseless simulated data allows us to focus the image quality evaluations on the systematic modeling and data sampling errors which are the central theme of this paper. The evaluation of image quality in the presence of noise is addressed in full with the in vivo data evaluations.
Simulated Cortical Activations
As a bridge between point-spread functions and in vivo results, we also simulated the type of target brain activations one would expect from a retinotopic mapping study. Because each area of the visual cortex contains a map of the visual field (with adjacency preserved), moving visual stimuli create traveling waves of neuronal activity. We thus created a sequence of target activations xsim(t), where each frame has a target similar to the activations seen in our previous retinotopic mapping study.6 The visual angle mapping sequence consists of target activations 10 mm in radius traveling in an ellipse (20-mm and 15-mm major and minor axes) with a center slightly off center from the center of the field of view (from here on referred to as the elliptical target). The eccentricity mapping sequence consists of two rectangular targets (30-mm width, 14-mm height) moving upward in the field of view (to be referred to as the bar target). Reconstructed responses xrecon(t) were then constructed as earlier. Phase-encoded mapping of these periodic responses was performed as explained in below with the in vivo visual cortex data.
Retinotopic Imaging Paradigm
The data for this analysis was acquired in a previous experiment;6 for this analysis, the data has been reimaged using the A-matrices described earlier. The study was approved by the Human Research Protection Office of the Washington University School of Medicine, and informed consent was obtained from all participants prior to scanning. Subjects were seated in an adjustable chair in a sound-isolated room facing a 19-in LCD screen at a viewing distance of 90 cm. The imaging pad was placed over the occipital cortex, and the optode tips were combed through the subject’s hair. Hook-and-loop strapping around the forehead held the array in place. All stimuli were phase-encoded, black-and-white reversing logarithmic checkerboards (10-Hz contrast reversal) on a 50% gray background.29, 30 Polar angle within the visual field was mapped using counterclockwise and clockwise rotating wedges: minimum radius 1 deg, maximum radius 8 deg, width 60 deg, and a rotation speed of 10 deg∕s for a cycle of 36 s. This rotation frequency allows for each stimulated brain region to return to baseline before subsequent activations.31 Eccentricity within the visual field was mapped with expanding and contracting rings: minimum radius 1 deg, maximum radius 8 deg, width 1.4 deg (three checkerboard squares), and 18 positions with 2 s per position for a total cycle of 36 s. Subjects were instructed to fixate on a central crosshair for all experiments. All stimuli started with 5 s of a 50% gray screen, continued with 10 cycles of the phase-encoded stimulus, and concluded with 15 s of a 50% gray screen.
DOT System
Our high-density DOT instrument uses light-emitting diode (LED) sources at 750 nm and 850 nm (750-03AU and OPE5T85, Roithner Lasertechnik) and avalanche photo diode (APD, Hamamatsu C5460-01) detectors.5 Each detector has a dedicated 24-bit analog-to-digital converter (MOTU HD-192). Sources and detectors were coupled with fiber-optic bundles to a flexible imaging cap held on to the back of the head with hook-and-loop strapping [Fig. 1e]. After being digitized, the APD measurements were written directly to hard disk at 96 kHz. With temporal, frequency, and spatial encoding, the system worked in continuous-wave mode with a frame rate of 10.78 Hz.
Processing of In Vivo Data
Source-detector pair data [Vi(t)] were converted to log-ratio data [yi(t)=−log(Vi(t)∕⟨V⟩)] and bandpassed (0.02 Hz to 0.5 Hz) to remove long-term trends and pulse. A subset of the data corresponding to the second (triangular) sparse grid [Fig. 1b] was imaged using the inverted A-matrix constructed earlier. Since the square sparse array [Fig. 1a] was not a subset of the high-density array used for data collection, it was not used in the in vivo analysis. Channels with high variance (>7.5%) are excluded from further analysis and not included during A-matrix inversion. This data was then inverted using the high-density A-matrix constructed earlier. For both imaged data sets, hemoglobin species concentrations were determined using their extinction coefficients. As earlier, all DOT images are posterior coronal projections of a 1-cm-thick cortical shell with the same 102 mm by 50 mm field of view. For simplicity, all images shown are ΔHbO2.
The phase-encoded stimuli create a traveling wave of neuronal activity along the cortical surface as the stimuli move through the visual field. Relative to the stimulus onset, each cortical position will be periodically activated with a different delay. Since we know the position of the stimulus at each time, we can match each pixel’s measured delay to the area of the visual field to which it corresponds.6, 32 In order to perform this analysis, the series of activations due to each stimuli (as well as the simulated cortical activation time courses corresponding to these stimuli) were down-sampled to 1 Hz (36 time points per stimulus cycle). Every pixel’s time course was Fourier transformed and the phase at the stimulation frequency (0.0278 Hz=1∕36 s) found. This phase then corresponds to the delay between stimulus onset and the pixel’s activation. We additionally need to correct for the finite neurovascular (hemodynamic) response time. Assuming that this lag time remains fixed at each cortical position, we correct for this delay using counterpropagating stimuli.32 Our convention is to define zero phase as the center of the visual field for the ring stimuli and as the lower vertical meridian for the wedge stimuli. Both before and after averaging, the phase maps were smoothed using a 3 pixel by 3 pixel moving box average.
Results
Point-Spread Function Simulations
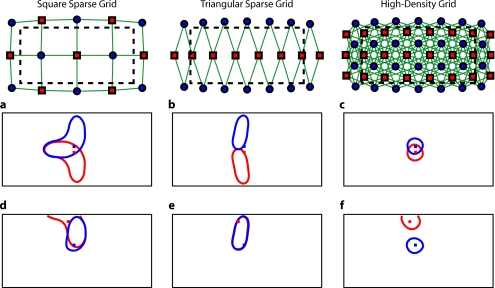
First, we analyzed the point-spread functions (PSFs) for every pixel within our imaging domain (Video 1). Moving the target activation point sequentially through the domain showed that the dense array was able to reconstruct the response with good localization and a slightly blurred but symmetric PSF. Both sparse arrays displaced activations to the nearest area of high sensitivity (i.e., onto the line between the nearest source–detector pair). These errors can result in the reconstructed responses to multiple targets appearing either artificially far apart upon inversion [Figs. 3a, 3b, 3c] or nearly identical despite large true separations [Figs. 3d, 3e, 3f]. Additionally, while the triangular sparse array performs well in horizontal localization, it has essentially only two voxels vertically: it is binary, placing responses either above or below the center source line with no further discrimination. Both sparse arrays have multiple lines of symmetry, points that fall equidistant between two measurements. The location of activations at these points cannot be distinguished and are projected equally into the two measurements, resulting in large reconstructed responses. One particularly notable symmetry is along the source line in the triangular sparse array; a target directly beneath this line is reconstructed as a large vertical activation stretching the entire height of the pad.
Video 1.
Point-spread functions of targets placed at every location in the field of view for the sparse and high-density grids. (upper left) Simulated targets (2 mm×2 mm×10 mm) to be measured and reconstructed by the different arrays. (lower left) The response reconstructed with the high-density array. Note that the response tracks the movement of the target and that there is minimal blurring. (upper right) The response reconstructed with the square sparse array. The response jumps to the high sensitivity underneath the nearest source-detector pair, causing mislocalization and large response sizes. (lower right) The response reconstructed with the triangular sparse array. While the array can localize the target well horizontally, it has only two major positions vertically (MPEG, 19 MB). .
Figure 3.
Representative image performance of sparse and high-density arrays. Targets are shown as red and blue squares. Half-maximum contours of the reconstructed responses are shown with the appropriately colored lines. (a) to (c) Here, two targets have been placed 4 mm apart adjacent to the horizontal midline of the arrays. The high-density array can correctly place the responses with a relatively small PSF (separation 4.4 mm). However, the triangular sparse array artificially displaces the two reconstructions (separation 23.2 mm) and enlarges the PSFs. The square sparse array results in reconstructions with an L-shape, which gives a large activation size, but reasonably close placement of the centroids (separation 11.5 mm). (d) to (f) Here, two targets have been placed far apart. Although the triangular sparse array reconstructs them as superimposed (separation between centroids of only 0.6 mm), the high-density array correctly separates them (17.5 mm). The square sparse array again reconstructs one of the activations in an L-shape, resulting in intermediate separation values (8.2 mm) with a large response size. (Color online only.)
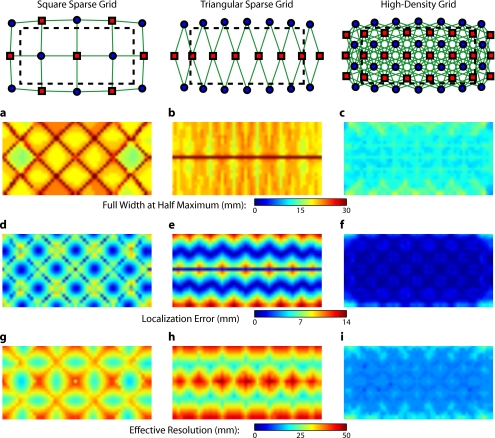
Image errors can be quantified using standard metrics of imaging performance (Tables 1, 2). The size of the reconstructed response is measured with the full effective width at half maximum (FWHM) of each target’s point-spread function [Figs. 4a, 4b, 4c]. Displacement of the response from the target point is calculated using localization error [Figs. 4d, 4e, 4f]. Since both broadening and misplacement of activations are similar in the sense that they both cause reconstructed responses where no activity should be measured, we combined resolution and localization error into a single metric of effective resolution, defined as the diameter of a circle centered at the known target location needed to enclose all activated points above half maximum [Figs. 4g, 4h, 4i]. Judging by all three metrics, the high-density system has two advantages over the sparse arrays. First, the average image quality is higher (an improvement of 1.76× in FWHM, 5.4× in localization error, and 2.27× in effective resolution). Second, this high performance is relatively even over the entire field of view. The performance maps of the sparse arrays alternate between areas of good and poor quality. Since most brain activations have an extent on the order of 1 cm, it is likely that sparse arrays will perform only up to the quality of their worst pixel.
Table 1.
Examination of point-spread functions of sparse and dense fNIRS arrays (mean±standard deviation).
| Array | FWHM (mm) | Localization error (mm) | Effective resolution (mm) |
|---|---|---|---|
| Square sparse | 21.6±4.6 | 5.3±2.0 | 29.9±5.2 |
| Triangular sparse | 20.9±4.3 | 5.4±3.6 | 31.7±7.1 |
| High density | 12.1±1.4 | 1.0±0.9 | 13.5±2.2 |
Table 2.
Performance ranges for point-spread functions of sparse and dense fNIRS arrays (minimum to maximum).
| Array | FWHM (mm) | Localization error (mm) | Effective resolution (mm) |
|---|---|---|---|
| Square sparse | 14.6–42.0 | 0.4–10.7 | 16.5–44.2 |
| Triangular sparse | 16.5–48.2 | 0.0–13.5 | 16.5–48.2 |
| High density | 8.2–17.2 | 0.0–5.8 | 8.9–25.3 |
Figure 4.
Image quality metrics for the point-spread functions of targets placed at every location in sparse and high-density grids. (a) to (c) Full width at half maximum of the imaging arrays. The FWHM was defined as the maximum separation of all pairs of points above half maximum contrast. For the sparse arrays, there is overall poor resolution (high FWHM) with worse resolution along lines of symmetry in the grid geometry. Also, note that the triangular array has the worst resolution directly beneath the source line, where the system cannot constrain the activation vertically. The resolution for the high-density array is high across the entire imaging domain with little variation. (d) to (f) Localization error of the imaging arrays. The localization error was defined as the separation between the known target location and the centroid of the voxels reconstructed above half-maximum contrast. While the sparse arrays have low localization error directly under measurements and along points of symmetry, between measurements, it is high. Localization error for the high-density array is uniformly low across the entire field of view. (g) to (i) Effective resolution of imaging arrays. The effective resolution was defined as the diameter of the circle centered at each target position needed to enclose the response. The sparse arrays have poor effective resolution between the measurements, and only good effective resolution directly between adjacent sources and detectors. Effective resolution for the high-density array is good across the entire imaging domain, with little variation.
Simulated Cortical Activations
In order to provide an intermediary evaluation between in silico point-spread functions and in vivo retinotopy experiments, which necessarily excite extended areas of cortex, we performed simulations of activations designed to approximate neuronal activation patterns seen with a retinotopic mapping experiment.6
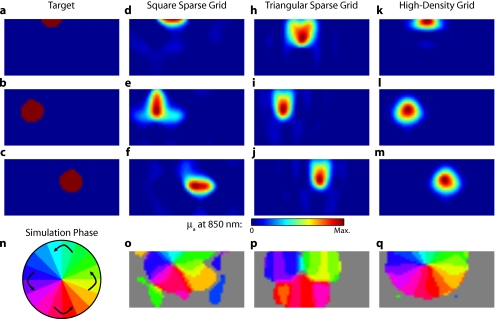
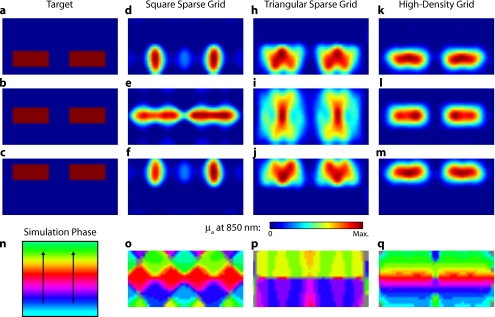
In the first experiment, the target was a 1-cm-radius circle, which moved in an elliptical pattern through the field of view [Figs. 5a, 5b, 5c and upper-left panel of Video 2]. In the second experiment, the target was a bilateral pair of rectangles moving vertically in the field of view [Figs. 6a, 6b, 6c and upper-left panel of Video 3]. A reconstructed time series was generated for both series of targets and for each of the optode arrays. The square sparse grid results in linear reconstructions along the nearest source–detector measurement [Figs. 5d, 5e, 5f, 6d, 6e, 6f and upper-right panel of Videos 2 and 3]. The triangular sparse array localizes the activations well horizontally, but has only binary vertical discrimination—it can only tell whether the target is above or below the source line [Figs. 5h, 5i, 5j, 6h, 6i, 6j and lower-right panel of Videos 2 and 3]. The high-density grid performs well in localizing the targets throughout both series [Figs. 5k, 5l, 5m, 6k, 6l, 6m and lower-left panel of Videos 2 and 3].
Figure 5.
Simulations of activations similar to those from phase-encoded mapping of visual angle. The stimulus is of 1-cm radius and moves in an elliptical pattern, with the center of the ellipse displaced from the center of the field of view. There are a total of 36 activations in the entire rotation series. (a) to (c) Three equally spaced frames from the sequence of targets. (d) to (f) These three activations are reconstructed with the square sparse array. The activations are displaced to the nearest measurement location. (h) to (j) Reconstructions using the triangular sparse array. The activations are located correctly horizontally, but displaced to the same vertical location. (k) to (m) Reconstructions using the high-density array. Activations are correctly placed with the correct size. (n) Legend defining the phase of the target phase-encoded stimulus. (o)The phase of each pixel’s activation at the rotation frequency using the square sparse array. This measure gives the delay between the start of the stimulus and the maximum activation of each pixel. Areas with gray have low signal-to-noise and are discarded. The square sparse array is able to correctly reconstruct the pinwheel of phase from the original stimulus, with some lobes of abnormal phases near the edges and an asymmetric shape. (p) Phase-encoded mapping using the triangular sparse array. Vertical gradients have been incorrectly reconstructed as horizontal, and some phases have been placed in the incorrect quadrant. (q) Phase mapping with the high-density array, which correctly locates all phases in the pinwheel.
Video 2.
Simulations of activations similar to those from phase-encoded mapping of visual angle. The stimulus is of 1 cm radius and moves in an elliptical pattern, with the center of the ellipse displaced from the center of the field of view. There are a total of 36 activations in the entire rotation series. (upper left) The video of 36 targets. (lower left) Reconstructions using the high-density array. Activations are correctly placed with the correct size. (upper right) Activations reconstructed with the square sparse array. The activations are displaced to the nearest measurement location. (lower right) Reconstructions using the triangular sparse array. The activations are located correctly horizontally but jump from above the source line to below it (MPEG, 1 MB). .
Figure 6.
Simulations of activations similar to those from phase-encoded mapping of eccentricity. The stimulus is two 1.4-cm-tall rectangles moving upward in the field of view. There are a total of 18 activations in the entire rotation series. (a) to (c) Three equally spaced frames from the sequence of targets. (d) to (f) These three activations are reconstructed with the square sparse array. The activations are displaced to the nearest measurement location, often resulting in squeezing in the horizontal direction. (h) to (j) Reconstructions using the triangular sparse array. Activations under the source planes are unconstrained vertically due to the pad’s symmetry. (k) to (m) Reconstructions using the high-density array. Activations are correctly placed with the correct size. (n) Legend defining the phase of the target phase-encoded stimulus. (o) The phase of each pixel’s activation at the rotation frequency using the square sparse array. The square sparse array is able to find the general trend of increasing phase vertically, but with many artifacts in shape. (p) Phase-encoded mapping using the triangular sparse array. Due to the inability to vertically localize activations, the array can only define two general regions of phase, and it converts gradients that should be vertical to be horizontal. (q) Phase mapping with the high-density array, which correctly locates all phases in the target.
Video 3.
Simulations of activations similar to those from phase-encoded mapping of eccentricity. The stimulus is two 1.4-cm-tall rectangles moving upward in the field of view. There are a total of 18 activations in the entire rotation series. (upper left) The video of 18 targets. (lower left) Reconstructions using the high-density array. Activations are correctly placed with the correct size. (upper right) Activations reconstructed with the square sparse array. The activations are displaced to the nearest measurement location, often resulting in squeezing in the horizontal direction. (lower right) Reconstructions using the triangular sparse array. Activations under the source planes are unconstrained vertically due to the pad’s symmetry (MPEG, 1 MB). .
By Fourier transforming each pixel’s time traces and finding the phases at the rotation frequency, we can perform in silico phase-encoded retinotopic mapping. By convention, zero phase of the elliptical target is defined as its most vertical position [Fig. 5n] and for the bar target as the bottom of the field of view [Fig. 6n]. Although the square sparse grid mislocalized many of the reconstructed targets, we see that it generates the correct pinwheel phase pattern for the elliptical target [Fig. 5o, although around the edges there are some abnormalities due to the uneven sampling sensitivity] and a distorted but recognizable general phase trend from bottom to top for the bar target [Fig. 6o]. To the contrary, while the triangular sparse array appeared to accurately reconstruct individual target frames, this in fact masked an inability to create a proper phase pattern for either target. With the elliptical target [Fig. 5p], there are three main errors: (1) what should be vertical gradients in phase (e.g., yellow to green and blue to purple along the horizontal midline) are instead reported as horizontal gradients (in the upper field of view); (2) these gradients are reconstructed as bidirectional, resulting in local extrema of phase (yellow between two regions of green and purple between two regions of blue); and (3) in the lower field of view, the phase pattern is actually reversed, with orange appearing on the lower left and pink on the lower right. With the bar target, the triangular sparse array is unable to reconstruct the expected phase pattern [Fig. 6p], again resulting in incorrect horizontal gradients and local extrema. Additionally, there is a discontinuity in phase between the lower and upper halves of the array. The high-density array is able to accurately replicate both expected phase patterns throughout the field of view [Figs. 5q, 5q].
In Vivo Activations
To complete the comparison of sparse and high-density imaging arrays, we analyzed data from our in vivo retinotopic mapping experiment6 using the full high-density system as well as a subset that forms the triangular sparse array. Since data was not collected with an array having the square sparse array as a subset, that array is not included in this analysis. This paradigm included four stimuli: a counterclockwise rotating wedge, a clockwise rotating wedge, an expanding ring, and a contracting ring.
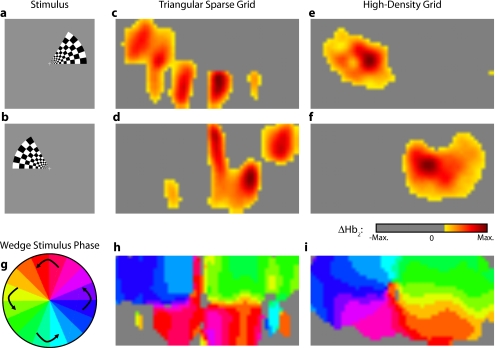
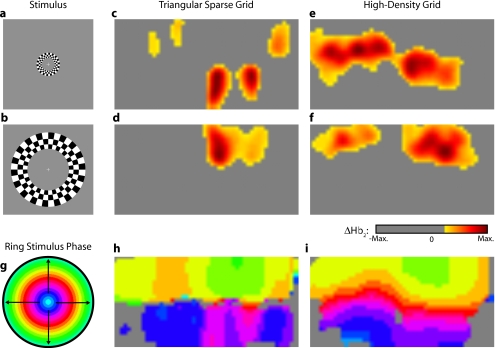
Two frames from the counterclockwise wedge and expanding ring stimuli are shown in Figs. 7a, 7b, 8a, 8b, respectively. (The entire stimuli are shown in the left frame of Videos 4 and 5.) From previous electrophysiology, fMRI, positron emission tomography (PET), and DOT studies, we expect each wedge stimulus to produce an activation in the opposite visual cortex. From our previous DOT studies, we expect the ring stimulus to produce an activation moving upward in the field of view. While the triangular sparse array can somewhat place the wedge activations in the correct quadrant, the activations are extended and have strange shapes [Figs. 7c, 7d and upper-right frame of Video 4], often with components in the wrong quadrants. Similarly, this array often mislocalizes the ring responses or fails to reconstruct entire hemispheres of activity [Figs. 8c, 8d and the upper-right frame of Video 5]. The high-density array accurately places both activation series throughout the field of view with tighter localization [Figs. 7e, 7f, 8e, 8f and lower-right frame of Videos 4 and 5]. We then performed retinotopic mapping using Fourier analysis, with the zero phase defined for the wedge stimulus at the stimulus directly downward [Fig. 7g] and for the ring stimulus at the center of the subject’s vision [Fig. 8g]. The triangular sparse array suffers the same problems in vivo that were predicted in simulation [Figs. 7h, 8h; compare to Figs. 5p, 6p]. The high-density array is able to accurately reconstruct the full range of phases with the correct localization for both stimuli [Figs. 7i, 8i].
Figure 7.
In vivo measurement from a phase-encoded mapping study of visual angle. The stimulus is a counterclockwise rotating, counterphase flickering wedge. There are a total of 36 activations in the entire rotation series. In order to line up stimuli and activations, we used our measured 6-s neurovascular lag. (a) and (b) Two frames from the stimulus. (c) and (d) Activations from these stimuli reconstructed with the triangular sparse array. Note the poor localization and strange activation shapes. (e) and (f) Reconstructions using the high-density array. Activations are correctly placed with reasonable sizes. (g) Legend defining the phase of the target phase-encoded stimulus. Note the 180-deg shift from Fig. 5n due to the transfer from visual field to cortex. (h) The phase of each pixel’s activation at the rotation frequency using the triangular sparse array. Note the ability to reconstruct a pinwheel of phase. The errors are similar to those from the simulation in Fig. 5p. (i) Phase mapping with the high-density array, which correctly locates all phases.
Figure 8.
In vivo measurement from a phase-encoded mapping study of visual eccentricity. The stimulus is an expanding counterphase flickering ring. There are a total of 36 activations in the entire rotation series. In order to line up stimuli and activations, we used our measured 6-s neurovascular lag. (a) and (b) Two frames from the stimulus. (c) and (d) Activations from these stimuli reconstructed with the triangular sparse array. Note the poor localization, especially when the activation passes beneath the source line, and the inability to always locate activations in both hemispheres. (e) and (f) Reconstructions using the high-density array. Activations are correctly placed with reasonable sizes. (g) Legend defining the phase of the target phase-encoded stimulus. (h) The phase of each pixel’s activation at the rotation frequency using the triangular sparse array. Note the ability to distinguish only two regions, and the conversion of vertical gradients into horizontal gradients, similar to Fig. 6p. (i) Phase mapping with the high-density array, which correctly locates all phases.
Video 4.
In vivo measurement from a phase-encoded mapping study of visual angle. (left) The stimulus is a counterclockwise rotating, counterphase flickering wedge. There are a total of 36 activations in the entire rotation series. In order to line up stimuli and activations, we used our measured 6-s neurovascular lag. Here, the stimulus is shown without flicker and at 10× actual presentation speed. (upper right) Activations from these stimuli reconstructed with the triangular sparse array. Note the poor localization (especially in the lower half of the field of view) and strange activation shapes. (lower right) Reconstructions using the high-density array. Activations are correctly placed with reasonable sizes. The videos have been thresholded such that changes in hemoglobin concentration below baseline are not shown (MPEG, 1 MB). .
Video 5.
In vivo measurement from a phase-encoded mapping study of visual eccentricity. (left) The stimulus is an expanding counterphase flickering ring. There are a total of 36 activations in the entire rotation series. In order to line up stimuli and activations, we used our measured 6-s neurovascular lag. Here, the stimulus is shown without flicker and at 10× actual presentation speed. (upper right) Activations from these stimuli reconstructed with the triangular sparse array. Note the poor localization, especially when the activation passes beneath the source line, and the inability to always locate activations in both hemispheres. (lower right) Reconstructions using the high-density array. Activations are correctly placed with reasonable sizes. The movies have been thresholded such that changes in hemoglobin concentration below baseline are not shown (MPEG, 1 MB). .
Discussion
We have conducted a quantitative comparison of the imaging performance of multiple imaging arrays for diffuse optical imaging. The goal of this exercise was to evaluate the potential improvements in image resolution and localization error of HD-DOT over sparse imaging geometries in the context of detailed in vivo neuroimaging tasks. This work builds on previous literature that has evaluated the theoretical performance of neuroimaging DOT systems. Boas et al. analyzed the resolution and localization error of a square and a hexagonal HD-DOT grid.1 In contrast to this paper, both grids were high density, and the subject of the comparison was the use of back-projection versus tomographic imaging techniques. Similarly, Joseph et al. demonstrated qualitative improvement of performing tomography over using single source–detector distances within the context of an HD-DOT array and simple focal activations.2 Since no sparse array was included in the comparison, it is difficult from those results to judge possible improvements over the sparse arrangements that are in widespread current use. Tian et al. examined multiple optode arrangements (of both sparse and dense styles); however, their geometries have a limited field of view, and the arrays do not correspond to those in standard use.4 Furthermore, while they found a useful rule-of-thumb for determining whether a new array geometry is “sparse” or “dense,” their data are not analyzed with standard metrics and are not quantified over the entire field of view. Thus, it is difficult to directly compare their data against either our data or the data from most other publications and systems. Additionally, all three of these studies have two algorithmic limitations: they assume a semi-infinite head geometry, and the reconstructions are depth-constrained to a plane with a known perturbation location. These assumptions limit translation of the simulations to evaluating neuroscience results. For further reference, the performance of DOT has also been analyzed in other applications, including infant neuroimaging by Heiskala et al.,3 slab transmission geometries by Culver et al.,33 and breast imaging by Azizi et al.34 Other papers have also addressed other sources of error in optical imaging, including a mismatch in optical properties3 and the modeling of cerebral spinal fluid.35 While we have dealt with the spatial analysis of diffuse optical imaging, quantification of hemoglobin concentrations is an important clinical parameter subject to different sources of error and deserving of its own evaluation.22, 24
Our first analysis judged three imaging geometries based on their performance on standard metrics of image quality. These simulations yield a FWHM of 12 mm for the high-density array and 21 mm for the two sparse arrays. Boas et al. report their resolution using characteristic areas: 2.0 cm2 for high-density DOT and 4.5 cm2 for a back-projection.1 (This is not the same as using a sparse grid, but it serves as a useful comparison for our results.) Converting our characteristic diameters to areas yields 1.1 cm2 for high density and 3.5 cm2 for sparse. Assuming similar regularization, our simulation might have higher resolution since we used first-nearest-neighbor separations of 1.3 cm and second-nearest-neighbor separations of 3.0 cm, compared to 1.9 cm and 4.25 cm for the Boas et al. study. Additionally, this paper shows the high-density array to have much better localization error than sparse arrays, 1.0 mm versus 5.3 mm. Boas et al. measured localization errors of 2 mm for DOT versus 5 mm for back-projection.
While activation size and mislocalization can be easily separated in simulations since the target is known a priori, this rigor does not translate to in vivo separations. When performing a brain activation study, the goal is to locate the area of the brain that responds preferentially to a given stimuli. This challenge is especially problematic with sparse arrays, which mislocalize many different, widely separated areas to the same area of high sensitivity. From a neuroimaging perspective, it is equivalent if two distinct areas are superimposed due to a broad reconstruction or due to them both being artificially displaced to the same area. Since fNIRS systems currently sample only superficial regions of cortex,36 their brain sensitivity jumps from gyri to gyri. Thus, seemingly small mislocalizations laterally can actually result in large errors in terms of position along the two-dimensional cortical surface. So, to combine knowledge of FWHM and localization error into a single metric of expected in vivo performance, we used the effective resolution (the diameter of a circle centered at the known activation point needed to circumscribe all voxels above half-maximum contrast). The high-density array has an effective resolution of about 1.3 cm, which should prevent identification to the wrong gyrus. However, the sparse arrays both have effective resolutions greater than 3 cm (as expected heuristically based on their source–detector separation).
In order to move beyond previous simulation studies of high-density DOT, we examined the performance of these arrays during a task to map the detailed organization of the visual cortex. This revealed subtle effects that limit wider utility of sparse systems. When examining individual point activations from individual stimuli, it might not appear that the worse resolution of the sparse systems is a problem. For example, in Figs. 5a, 5b, 5c, 5d, 5e, 5f, 5g, 5h, 5i, 5j, 5k, 5l, 5m, the sparse arrays seem to do an acceptable job at reconstructing the target activations. However, due to heterogeneous localization errors and resolution, they are unable to move beyond individual stimuli to assimilating that information to perform retinotopic mapping. The triangular sparse array is unable to properly decode phase in either the elliptical [Fig. 5p] or bar [Fig. 6p] retinotopy simulations. While the square sparse array can do a reasonable job with the elliptical target [Fig. 5o], it suffers many artifacts when attempting to map the bar target [Fig. 6o]. Only the high-density array can reconstruct both simulations properly [Figs. 5q, 6q]. This in silico performance is duplicated in vivo, where a triangular sparse subset of the high-density array is unable to generate visual cortex maps of either visual angle or eccentricity that capture the correct global neural architecture (Figs. 78).
The advantages quantified here, in resolution and localization error, are in addition to the other benefits of high-density arrays, which include lower sensitivity to modeling errors,3 better contrast-to-noise,4 and the ability to regress out a measure of superficial and systemic noise.5, 37 This last benefit can be particularly important to analyzing data from single subjects. Since one of the main benefits of fNIRS technology is its potential to translate to a bedside neuroimaging tool, it is important to develop techniques that can perform effectively in neuromapping paradigms in single subjects. Noninvasive optical techniques have spatial resolution between fMRI and EEG. While improvements such as high-density arrays are unlikely to result in image quality that surpasses the high resolution of fMRI, improving image quality is always a fundamental goal, allowing researchers to access the advantages of optical neuroimaging (e.g., portability and comprehensive hemodynamic measurements) without being hampered by insufficient imaging performance. A reasonable objective within the field of neuroimaging is 1-cm resolution, which would allow the distinguishing of gyri and the ability to perform classical human brain mapping paradigms, such as retinotopy. Our results show that common and traditional sparse array approaches have limited performance when attempting such detailed neuroimaging studies. In contrast, new high-density DOT techniques extend the capabilities of optical methods to meet these challenges.
Acknowledgments
We thank Benjamin Zeff, Gavin Perry, and Martin Olevitch for help with DOT instrumentation and software. This work was supported in part by NIH Grant Nos. R21-HD057512 (J.P.C.), R21-EB007924 (J.P.C.), R01-EB009233 (J.P.C.), and T90-DA022871 (B.R.W.).
References
- Boas D. A., Chen K., Grebert D., and Franceschini M. A., “Improving the diffuse optical imaging spatial resolution of the cerebral hemodynamic response to brain activation in humans,” Opt. Lett. 29(13), 1506–1508 (2004). 10.1364/OL.29.001506 [DOI] [PubMed] [Google Scholar]
- Joseph D. K., Huppert T. J., Franceschini M. A., and Boas D. A., “Diffuse optical tomography system to image brain activation with improved spatial resolution and validation with functional magnetic resonance imaging,” Appl. Opt. 45(31), 8142–8151 (2006). 10.1364/AO.45.008142 [DOI] [PubMed] [Google Scholar]
- Heiskala J., Hiltunen P., and Nissila I., “Significance of background optical properties, time-resolved information and optode arrangement in diffuse optical imaging of term neonates,” Phys. Med. Biol. 54, 535–554 (2009). 10.1088/0031-9155/54/3/005 [DOI] [PubMed] [Google Scholar]
- Tian F., Alexandrakis G., and Liu H., “Optimization of probe geometry for diffuse optical brain imaging based on measurement density and distribution,” Appl. Opt. 48(13), 2496–2504 (2009). 10.1364/AO.48.002496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeff B. W., White B. R., Dehghani H., Schlaggar B. L., and Culver J. P., “Retinotopic mapping of adult human visual cortex with high-density diffuse optical tomography,” Proc. Natl. Acad. Sci. U.S.A. 104(29), 12169–12174 (2007). 10.1073/pnas.0611266104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. R. and Culver J. P., “Phase-encoded retinotopy as an evaluation of diffuse optical neuroimaging,” Neuroimage 49(1), 568–577 (2010). 10.1016/j.neuroimage.2009.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. R., Snyder A. Z., Cohen A. L., Petersen S. E., Raichle M. E., Schlaggar B. L., and Culver J. P., “Resting-state functional connectivity in the human brain revealed with diffuse optical tomography,” Neuroimage 47(1), 148–156 (2009). 10.1016/j.neuroimage.2009.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson A. P., Austin T., Everdell N. L., Schweiger M., Arridge S. R., Meek J. H., Wyatt J. S., Delpy D. T., and Hebden J. C., “Three-dimensional whole-head optical tomography of passive motor evoked responses in the neonate,” Neuroimage 30(2), 521–528 (2006). 10.1016/j.neuroimage.2005.08.059 [DOI] [PubMed] [Google Scholar]
- Wylie G. R., Graber H., Voelbel G. T., Kohl A. D., DeLuca J., Pei Y., Xu Y., and Barbour R. L., “Using co-variation in the Hb signal to detect visual activation: a near infrared spectroscopy study,” Neuroimage 47, 473–481 (2009). 10.1016/j.neuroimage.2009.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strangman G., Culver J. P., Thompson J. H., and Boas D. A., “A quantitative comparison of simultaneous BOLD fMRI and NIRS recordings during functional brain activation,” Neuroimage 17(2), 719–731 (2002). 10.1016/S1053-8119(02)91227-9 [DOI] [PubMed] [Google Scholar]
- Steinbrink J., Villringer A., Kempf F. C. D., Haux D., Boden S., and Obrig H., “Illuminating the BOLD signal: combined fMRI-fNIRS studies,” Magn. Reson. Imaging 24(4), 495–505 (2006). 10.1016/j.mri.2005.12.034 [DOI] [PubMed] [Google Scholar]
- Barbour R. L., Graber H., Aronson R., and Lubowsky J., “Model for 3-D optical imaging of tissue,” in Proc. 10th Int. Geoscience and Remote Sensing Symposium, pp. 1395–1399, IEEE, Piscataway, NJ: (1990).
- Barbour R. L., Graber H. L., Aronson R., and Lubowsky J., “Imaging of subsurface regions of random media by remote sensing,” in Time-Resolved Spectroscopy and Imaging of Tissues, Proc. SPIE 1431, 192–203 (1991). 10.1117/12.44190 [DOI] [Google Scholar]
- Arridge S. R., “Optical tomography in medical imaging,” Inverse Probl. 15(2), R41–R93 (1999). 10.1088/0266-5611/15/2/022 [DOI] [Google Scholar]
- Schroeter M. L., Bucheler M. M., Muller K., Uludag K., Obrig H., Lohmann G., Tittgemeyer M., Villringer A., and von Cramon D. Y., “Towards a standard analysis for functional near-infrared imaging,” Neuroimage 21(1), 283–290 (2004). 10.1016/j.neuroimage.2003.09.054 [DOI] [PubMed] [Google Scholar]
- Plichta M. M., Herrmann M. J., Baehne C. G., Ehlis A.-C., Richter M. M., Pauli P., and Fallgatter A. J., “Event-related functional near-infrared spectroscopy (fNIRS): are the measurements reliable?” Neuroimage 31, 116–124 (2006). 10.1016/j.neuroimage.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Toronov V. Y., Zhang X., and Webb A. G., “A spatial and temporal comparison of hemodynamic signals measured using optical and functional magnetic resonance imaging during activation in the human primary visual cortex,” Neuroimage 34, 1136–1148 (2007). 10.1016/j.neuroimage.2006.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert T. J., Diamond S. G., and Boas D. A., “Direct estimation of evoked hemoglobin changes by multimodality fusion imaging,” J. Biomed. Opt. 13(5), 054031 (2008). 10.1117/1.2976432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert T. J., Hoge R. D., Diamond S. G., Franceschini M. A., and Boas D. A., “A temporal comparison of BOLD, ASL, and NIRS hemodynamic response to motor stimuli in adult humans,” Neuroimage 29, 368–382 (2006). 10.1016/j.neuroimage.2005.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M. J., Herrmann M. J., Dan I., Obrig H., Conrad M., Kuchinke L., Jacobs A. M., and Fallgatter A. J., “Differential activation of frontal and parietal regions during visual word recognition: an optical topography study,” Neuroimage 40(3), 1340–1349 (2008). 10.1016/j.neuroimage.2007.12.037 [DOI] [PubMed] [Google Scholar]
- Taga G., Asakawa K., Maki A., Konishi Y., and Koizumi H., “Brain imaging in awake infants by near-infrared optical topography,” Proc. Natl. Acad. Sci. U.S.A. 100(19), 10722–10727 (2003). 10.1073/pnas.1932552100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boas D. A., Dale A. M., and Franceschini M. A., “Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy,” Neuroimage 23(Suppl. 1), S275–S288 (2004). 10.1016/j.neuroimage.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Huppert T. J., Diamond S. G., Franceschini M. A., and Boas D. A., “HomER: a review of time-series analysis methods for near-infrared spectroscopy of the brain,” Appl. Opt. 48(10), D280–D298 (2009). 10.1364/AO.48.00D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strangman G., Franceschini M. A., and Boas D. A., “Factors affecting the accuracy of near-infrared spectroscopy concentration calculations for focal changes in oxygenation parameters,” Neuroimage 18(4), 865–879 (2003). 10.1016/S1053-8119(03)00021-1 [DOI] [PubMed] [Google Scholar]
- Dehghani H., Pogue B. W., Poplack S. P., and Paulsen K. D., “Multiwavelength three-dimensional near-infrared tomography of the breast: initial simulation, phantom, and clinical results,” Appl. Opt. 42(1), 135–145 (2003). 10.1364/AO.42.000135 [DOI] [PubMed] [Google Scholar]
- Culver J. P., Siegel A. M., Stott J. J., and Boas D. A., “Volumetric diffuse optical tomography of brain activity,” Opt. Lett. 28(21), 2061–2063 (2003). 10.1364/OL.28.002061 [DOI] [PubMed] [Google Scholar]
- Culver J. P., Durduran T., Furuya D., Cheung C., Greenberg J. H., and Yodh A. G., “Diffuse optical tomography of cerebral blood flow, oxygenation and metabolism in rat during focal ischemia,” J. Cereb. Blood Flow Metab. 23, 911–923 (2003). 10.1097/01.WCB.0000076703.71231.BB [DOI] [PubMed] [Google Scholar]
- Boas D. A. and Dale A. M., “Simulation study of magnetic resonance imaging-guided cortically constrained diffuse optical tomography of human brain function,” Appl. Opt. 44(10), 1957–1968 (2005). 10.1364/AO.44.001957 [DOI] [PubMed] [Google Scholar]
- Engel S. A., Rumelhart D. E., Wandell B. A., Lee A. T., Glover G. H., Chichilnisky E.-J., and Shadlen M. N., “fMRI of human visual cortex,” Nature 369, 525 (1994). 10.1038/369525a0 [DOI] [PubMed] [Google Scholar]
- DeYoe E. A., Bandettini P., Neitz J., Miller D., and Winans P., “Functional magnetic resonance imaging (fMRI) of the human brain,” J. Neurosci. Methods 54, 171–187 (1994). 10.1016/0165-0270(94)90191-0 [DOI] [PubMed] [Google Scholar]
- Warnking J., Dojat M., Guérin-Dugué A., Delon-Martin C., Olympieff S., Richard N., Chéhikian A., and Segebarth C., “fMRI retinotopic mapping—step by step,” Neuroimage 17, 1665–1683 (2002). 10.1006/nimg.2002.1304 [DOI] [PubMed] [Google Scholar]
- Sereno M. I., Dale A. M., Reppas J. B., Kwong K. K., Belliveau J. W., Brady T. J., Rosen B. R., and Tootell R. B. H., “Borders of multiple visual areas in humans revealed by functional magnetic-resonance-imaging,” Science 268(5212), 889–893 (1995). 10.1126/science.7754376 [DOI] [PubMed] [Google Scholar]
- Culver J. P., Ntziachristos V., Holboke M. J., and Yodh A. G., “Optimization of optode arrangements for diffuse optical tomography: a singular-value analysis,” Opt. Lett. 26(10), 701–703 (2001). 10.1364/OL.26.000701 [DOI] [PubMed] [Google Scholar]
- Azizi L., Zarychta K., Ettori D., Tinet E., and Tualle J.-M., “Ultimate spatial resolution with diffuse optical tomography,” Opt. Express 17(14), 12132–12144 (2009). 10.1364/OE.17.012132 [DOI] [PubMed] [Google Scholar]
- Custo A., Wells W. M. I., Barnett A. H., Hillman E. M. C., and Boas D. A., “Effective scattering coefficient of the cerebral spinal fluid in adult head models for diffuse optical imaging,” Appl. Opt. 45(19), 4747–4755 (2006). 10.1364/AO.45.004747 [DOI] [PubMed] [Google Scholar]
- Dehghani H., White B. R., Zeff B. W., Tizzard A., and Culver J. P., “Depth sensitivity and image reconstruction analysis of dense imaging arrays for mapping brain function with diffuse optical tomography,” Appl. Opt. 48(10), D137–D143 (2009). 10.1364/AO.48.00D137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saager R. and Berger A., “Measurement of layer-like hemodynamic trends in scalp and cortex: implications for physiological baseline suppression in functional near-infrared spectroscopy,” J. Biomed. Opt. 13(3), 034017 (2008). 10.1117/1.2940587 [DOI] [PubMed] [Google Scholar]