Abstract
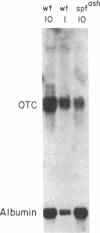
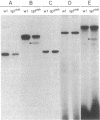
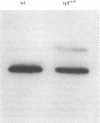
Ornithine transcarbamylase (ornithine carbamoyltransferase; carbamoyl-phosphate:L-ornithine carbamoyltransferase, EC 2.1.3.3) is a mitochondrial matrix enzyme of the mammalian urea cycle. The X chromosome-linked spfash mutation in the mouse causes partial ornithine transcarbamylase deficiency and has served as a model for the human disease. We show here that the spfash mutation is a guanine to adenine transition of the last nucleotide of the fourth exon of the ornithine transcarbamylase gene. This nucleotide change produces two remarkably different effects. First, this transition causes ornithine transcarbamylase mRNA deficiency because the involved exon nucleotide plays a part in the recognition of the adjacent splice donor site. As a result of the mutation, ornithine transcarbamylase pre-mRNA is spliced inefficiently both at this site and at a cryptic splice donor site 48 bases into the adjacent intron. Second, two mutant proteins are translated from these mRNAs. From the correctly spliced mRNA, the transition results in a change of amino acid 129 from arginine to histidine. This missense substitution has no discernable effect on mitochondrial import, subunit assembly, or enzyme activity. On the other hand, the elongated mRNA resulting from mis-splicing is translated into a dysfunctional ornithine transcarbamylase subunit elongated by the insertion of 16 amino acid residues.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Batshaw M. L., Hyman S. L., Coyle J. T., Robinson M. B., Qureshi I. A., Mellits E. D., Quaskey S. Effect of sodium benzoate and sodium phenylacetate on brain serotonin turnover in the ornithine transcarbamylase-deficient sparse-fur mouse. Pediatr Res. 1988 Apr;23(4):368–374. doi: 10.1203/00006450-198804000-00006. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Briand P., Francois B., Rabier D., Cathelineau L. Ornithine transcarbamylase deficiencies in human males. Kinetic and immunochemical classification. Biochim Biophys Acta. 1982 May 21;704(1):100–106. doi: 10.1016/0167-4838(82)90136-4. [DOI] [PubMed] [Google Scholar]
- Briand P., Miura S., Mori M., Cathelineau L., Kamoun P., Tatibana M. Cell-free synthesis and transport of precursors of mutant ornithine carbamoyltransferases into mitochondria. Biochim Biophys Acta. 1983 Nov 8;760(3):389–397. doi: 10.1016/0304-4165(83)90379-3. [DOI] [PubMed] [Google Scholar]
- Cavard C., Grimber G., Dubois N., Chasse J. F., Bennoun M., Minet-Thuriaux M., Kamoun P., Briand P. Correction of mouse ornithine transcarbamylase deficiency by gene transfer into the germ line. Nucleic Acids Res. 1988 Mar 25;16(5):2099–2110. doi: 10.1093/nar/16.5.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- DeMars R., LeVan S. L., Trend B. L., Russell L. B. Abnormal ornithine carbamoyltransferase in mice having the sparse-fur mutation. Proc Natl Acad Sci U S A. 1976 May;73(5):1693–1697. doi: 10.1073/pnas.73.5.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Hata A., Tsuzuki T., Shimada K., Takiguchi M., Mori M., Matsuda I. Structure of the human ornithine transcarbamylase gene. J Biochem. 1988 Feb;103(2):302–308. doi: 10.1093/oxfordjournals.jbchem.a122265. [DOI] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Kalousek F., Hendrick J. P., Rosenberg L. E. Two mitochondrial matrix proteases act sequentially in the processing of mammalian matrix enzymes. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7536–7540. doi: 10.1073/pnas.85.20.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek F., Orsulak M. D., Rosenberg L. E. Newly processed ornithine transcarbamylase subunits are assembled to trimers in rat liver mitochondria. J Biol Chem. 1984 May 10;259(9):5392–5395. [PubMed] [Google Scholar]
- Kraus J. P., Hodges P. E., Williamson C. L., Horwich A. L., Kalousek F., Williams K. R., Rosenberg L. E. A cDNA clone for the precursor of rat mitochondrial ornithine transcarbamylase: comparison of rat and human leader sequences and conservation of catalytic sites. Nucleic Acids Res. 1985 Feb 11;13(3):943–952. doi: 10.1093/nar/13.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. C., Wu X. W., Gibbs R. A., Cook R. G., Muzny D. M., Caskey C. T. Generation of cDNA probes directed by amino acid sequence: cloning of urate oxidase. Science. 1988 Mar 11;239(4845):1288–1291. doi: 10.1126/science.3344434. [DOI] [PubMed] [Google Scholar]
- Lee C. S., Curtis D., McCarron M., Love C., Gray M., Bender W., Chovnick A. Mutations affecting expression of the rosy locus in Drosophila melanogaster. Genetics. 1987 May;116(1):55–66. doi: 10.1093/genetics/116.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Sollner-Webb B., Cleveland D. W. Surprising S1-resistant trimolecular hybrids: potential complication in interpretation of S1 mapping analyses. Mol Cell Biol. 1985 Oct;5(10):2842–2846. doi: 10.1128/mcb.5.10.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake A., Takayanagi M., Yamamoto S., Nakajima H., Mori M. Ornithine transcarbamylase deficiency in spf and spf-ash mice: genes, mRNAs and mRNA precursors. Biochem Biophys Res Commun. 1987 Aug 14;146(3):1064–1070. doi: 10.1016/0006-291x(87)90755-8. [DOI] [PubMed] [Google Scholar]
- Qureshi I. A., Letarte J., Ouellet R. Ornithine transcarbamylase deficiency in mutant mice I. Studies on the characterization of enzyme defect and suitability as animal model of human disease. Pediatr Res. 1979 Jul;13(7):807–811. doi: 10.1203/00006450-197907000-00003. [DOI] [PubMed] [Google Scholar]
- Rosenberg L. E., Kalousek F., Orsulak M. D. Biogenesis of ornithine transcarbamylase in spfash mutant mice: two cytoplasmic precursors, one mitochondrial enzyme. Science. 1983 Oct 28;222(4622):426–428. doi: 10.1126/science.6623083. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Scherer S. E., Veres G., Caskey C. T. The genetic structure of mouse ornithine transcarbamylase. Nucleic Acids Res. 1988 Feb 25;16(4):1593–1601. doi: 10.1093/nar/16.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres G., Gibbs R. A., Scherer S. E., Caskey C. T. The molecular basis of the sparse fur mouse mutation. Science. 1987 Jul 24;237(4813):415–417. doi: 10.1126/science.3603027. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]