Abstract
Cooperation among transcription factors is central for their ability to execute specific transcriptional programmes. The AP1 complex exemplifies a network of transcription factors that function in unison under normal circumstances and during the course of tumour development and progression. This Perspective summarizes our current understanding of the changes in members of the AP1 complex and the role of ATF2 as part of this complex in tumorigenesis.
Activator protein 1 (AP1)1,2 functions in almost all areas of eukaryotic cellular behaviour, from cell cycle proliferation and development to stress response and apoptosis. Indeed, AP1 is activated in response to a plethora of extracellular signals from cytokines and growth factors to stress and inflammation3,4. The expansive transcriptional repertoire executed by AP1 complexes is propagated from the diverse compositional array of homodimeric or heterodimeric combinations formed by members of the Jun, Atf, Fos and Maf transcription factor families (BOX 1). The dimeric combinations and transcriptional activity observed in vivo are largely influenced by the tissue-specific expression patterns of the individual proteins, and importantly by their specific activating mechanisms and post-translational modifications that facilitate their individual ability to dimerize with other basic leucine zipper (bZIP) domain proteins. This inherently diverse composition of AP1 complexes and their central role in transcriptional regulation places AP1 complexes at a functional epicenter for pathological signal relay in disease, particularly in the context of malignant cellular transformation in which AP1 proteins are often deregulated by oncoprotein signalling4–6. This Perspective describes the function and cooperation of Jun, Fos and Atf family members in tumour cells, and the emerging function of ATF2 as part of the dynamic AP1 complex.
Box 1. The Ap1 transcription factor complex.
The mammalian AP1 proteins are homodimers and heterodimers composed of proteins from the Jun (JUN, JUNB and JUND) and Fos (FOS, FOSB, FRA1 and FRA2) families, and the closely related activating transcription factor (Atf and Creb) subfamily and the Maf subfamily5. AP1 constituent proteins are structurally distinguished by a basic leucine zipper (bZIP) domain that is composed of leucine zipper and basic domains. It is through these domains that AP1 proteins dimerize and bind to DNA. These proteins are typically activated through phosphorylation by the indicated upstream kinases. The different AP1 dimers bind to DNA with varying affinities and differ in their transactivation efficiencies8,15. Jun proteins can form stable dimers that bind to the AP1 DNA recognition element 5′-TGAC/GTCA-3′ (also known as TPA response element (TRE)) based on their ability to mediate transcriptional induction in response to the phorbol ester tumour promoter TPA2,15. Atf proteins, conversely, form dimers that preferentially bind to cyclic AMP responsive elements (CRE; 5′-TGACGTCA-3′)15. AP1 proteins also dimerize efficiently with other transcription factors, including some that are not members of the bZIP family193.
JUN and the Jun family
JUN was originally identified as the normal cellular counterpart of the avian sarcoma (ASV17) viral Jun oncoprotein (v-jun)7. The Jun family consists of JUN, JUNB and JUND, and each protein has distinct characteristics. JUN is important for cell proliferation, survival and apoptosis, and accordingly mice lacking JUN die between day 12.5 and 13.5 of embryonal development owing to hepatic failure and heart defects8,9. Similarly, JUNB is essential for embryonal development, however, JUND is dispensable10. Although the JUN locus is not mutated in human cancer, it was recently shown to be a target of 1p32 amplifications in undifferentiated and aggressive human sarcomas11,12. Moreover, many human cancers exhibit overexpression of JUN and/or other Jun family members (TABLE 1), which in most cases, is the result of upstream oncogene activation13. There is now good evidence that JUN activation is a crucial contributing factor for transformation and tumorigenesis, rather than an indirect effect of oncogenesis.
Table 1. Deregulation of Jun family members in cancer.
| Observation | Tumour type | Method | Refs |
|---|---|---|---|
| JUN amplification and JUN overexpression | Liposarcoma | CGH, FISH, Real-time PCR and WB | 12 |
| Increased JUN expression | Non-small-cell lung cancer | IHC | 29,202 |
| Acute myeloid leukaemia | Microarray | 203 | |
| Colorectal neoplasm | IHC | 204 | |
| Pancreatic cancer | IHC | 205 | |
| Oral squamous cell carcinomas | IHC | 206 | |
| Increased levels of phosphorylated JUN | Astrocytoma | IHC | 207 |
| Invasive breast cancer | IHC | 146 | |
| Osteosarcoma | IHC | 208 | |
| Glioblastoma | IHC | 209 | |
| Melanoma | WB | 204 | |
| JUN and FOS overexpression | Prostate cancer | TMA | 175 |
| JUN and JUND overexpression | Breast cancer | TMA | 210 |
| JUN and JUNB overexpression | CD30+ lymphoma* | TMA and IHC | 174 |
| JUND overexpression | Primary cutaneous B cell lymphoma | IHC | 211 |
| JUNB and JUND overexpression | Colorectal adenocarcinoma | IHC and WB | 212 |
| Increased AP1 activity | Lung and bladder carcinoma | EMSA | 213 |
| Increased AP1 binding activity (FOS–JUNB) | Endometrial cancer | WB | 214 |
| Cervical cancer | EMSA and WB | 215 |
CGH, comparative genomic hybridization; EMSA, electrophoretic mobility shift assay; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; TMA, tumour microarray; WB, western blot.
Classic Hodgkin lymphoma, anaplastic large cell lymphoma, diffuse large B cell lymphoma.
JUN activation
JUN is an ‘immediate early gene’ and is responsive to mitogenic stimuli, as well as DNA damage and stress. JUN expression levels are tightly controlled by a combination of protein stability and a short mRNA half-life of 20–25 minutes, owing to an AU-rich RNA destabilizing element in the 3′-untranslated region. Post-translational modifications trigger a positive autoregulatory loop that involves the binding of AP1 dimers to a phorbol TPA response element (TRE; also known as a JUN1 site) and a cyclic AMP responsive element CRE (also known as a JUN2 site) in the JUN promoter resulting in increased transcription14. The AP1 dimers primarily involved in JUN transcription are JUN– FOS and JUN–ATF2 for TRE and CRE, respectively15. JUN transcription is also induced by SP1, nuclear factor-κB (NF-κB), ternary complex factors (TCFs), MEF2 or CCAAT-binding transcription factors16,17. ERK contributes to JUN transcription by activating FOS, TCFs and MEF217. The signals that trigger JUN transcription also activate RNA-binding proteins that increase JUN mRNA stability. In all cases, JUN exhibits a rapid, but transient upregulation, which effectively stimulates the transcription of genes important for entry into the G1 and S phases of the cell cycle such as cyclin D1 (REF. 11), cyclin A18 and cyclin E. However, JUN also controls anti-proliferative cell cycle regulators such as p53 (REFS 19,20), p21, INK4A13 and ARF19,21 (for an extensive review of JUN transcriptional targets see REF. 8; representative JUN targets are shown in Supplementary information S1 (table)). Despite the high degree of sequence homology shared between the three Jun proteins, they have distinct transactivation properties and biological effects, mainly attributed to the lower degree of conservation of the amino-terminal region (residues 1–95). Although JUN and JUND have strong transactivation activity, the transcriptional activity of JUNB is much weaker22, and unlike JUN, both JUNB and JUND can repress transcription22–24.
Consistent with its important role in cell cycle regulation, JUN levels and N-terminal phosphorylation (which is crucial for its activation) are cell cycle regulated25,26. Ectopic overexpression of JUN promotes cell growth in many cell lines27,28, whereas mouse fibroblasts lacking JUN and cancer cells expressing TAM67 — a dominant-negative form of JUN that lacks the transactivation domain encompassing amino acids 3–122 — exhibit severely impaired or inhibited proliferation, demonstrating the central role that JUN has in cell growth and tumorigenesis20,29–31. It should be noted that TAM67 generally inhibits AP1, regardless of the specific binding partner, owing to its promiscuous interaction with most AP1 proteins. The role of JUNB and JUND is more complex, and they can promote or inhibit growth under different conditions. In Jun-knockout mice Junb can rescue Jun-induced phenotypes in a dose-dependent manner32 and prevent p53 expression. These experiments suggest that, in the absence of Jun, Junb exhibits proliferative effects and its anti-proliferative activity requires the formation of a growth-inhibiting JUN–JUNB heterodimer. Similarly, Jund can suppress p53-induced senescence and apoptosis in fibroblasts19, although it has also been observed to function as a tumour suppressor10,13. It is thought that this dual role depends on an interaction with the tumour suppressor Menin33. However, most of the evidence indicates that JUND antagonizes JUN in cell growth regulation and transformation19. Notably, JUNB and JUND are regulated by different protein kinases than the kinases that regulate JUN. Therefore, stimuli that differentially activate JUNB and JUND regulatory kinases might dictate positive or negative effects on the interaction of JUNB or JUND with JUN and so on their corresponding response elements.
JUN function, stability34 and transactivation potential35 are crucially enhanced by phosphorylation of Ser63 and Ser73 by JUN N-terminal kinase (JNK)17, which docks with JUN primarily through the δ-domain (amino acids 34–60) of JUN8 (FIG. 1). Increasing evidence implicates differential regulation of JUN by JNK1 compared with JNK2, which may explain the variable binding affinity that JUN exhibits to JNK family members36,37. JNK-mediated phosphorylation can stimulate JUN transcriptional activity by promoting either interaction with basal transcriptional machinery or co-activators38–40, or by promoting dissociation of transcriptional repressor complexes containing histone deacetylase 3 (REF. 41). Therefore, JUN can regulate gene transcription through the regulation of transcriptional machinery including RNA polymerase II, as well as other co-activators or co-repressors, and chromatin structural changes. Although JNK phosphorylation sites on Ser63 and Ser73 are conserved, the Jun proteins differ markedly in their regulation by JNK42. JUNB has a JNK docking site, but the lack of phospho-acceptor residues prevents its phosphorylation by JNK. By contrast, JUND lacks an effective docking site, resulting in only weak phosphorylation by JNK42. These residues of JUND can still be phosphorylated by ERK1 and ERK2 (REF. 43). Whether these phosphorylations affect differential dimerization with other AP1 partners remains largely unexplored.
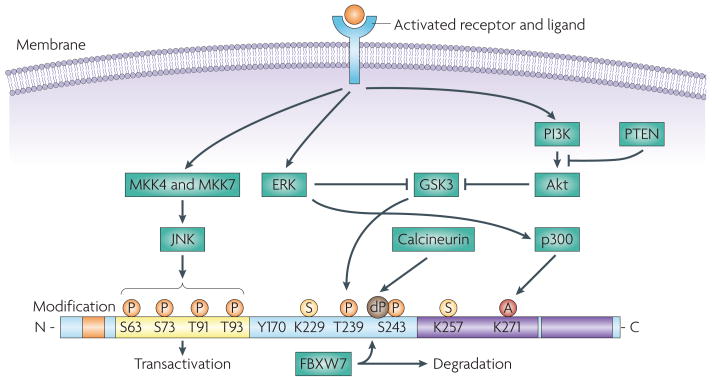
Figure 1. Structure and regulation of JUN.
JUN is encoded by a 3.34 kb intronless gene, located on chromosome 1 (1p32-p31) and results in the expression of a 334 amino acid protein product composed of four main domains, which are involved in DNA binding, transcription and dimerization197. JUN activity is regulated by post-translational modifications, which are largely controlled by components of the MAPK family of serine and threonine kinases, including JUN N-terminal kinase (JNK), ERK and p38 isoforms17. JUN is phosphorylated on Ser63 and Ser73 by JNK, increasing its stability and transactivation potential35. JNK also phosphorylates Thr91 and Thr93, which are required for DNA binding and activation of its transcriptional activity. JUN is subject to ubiquitylation and this requires phosphorylation at Thr239 by glycogen synthase kinase 3 (GSK3). GSK3 can target JUN only once Ser243 is phosphorylated. Phosphorylation on these sites is required for recruitment of the F-box and WD domain repeated 7 (FBXW7) ubiquitin ligase198. Inactivation of GSK3, owing to the activation of ERK and PI3K–Akt signalling cascades results in JUN stabilization44,45. The effect of GSK3 can be antagonized by the dephosphorylation of Ser243 by calcineurin199. JUN can be sumoylated on Lys257 and Lys229, which leads to a reduced transcriptional activity51. ERK induces the acetylation of the lysine residues in the JUN DNA binding region200, thereby increasing JUN transcriptional activity. Post-translational modifications are indicated as small coloured circles. The four domains are indicated as follows: the δ-domain is orange, the basic region (DNA binding) is blue, the transactivation domain is yellow and the leucine zipper is purple. A, acetylation; dP, dephosphorylation; P, phosphorylation; S, sumoylation.
JUN degradation
In most cells, JUN is a labile protein (with a half-life of approximately 2 hours), and its expression levels are tightly regulated by polyubiquitylation on multiple lysine residues and concomitant degradation by the 26S proteasome34 (FIG. 2). Stabilization of JUN occurs following inactivation of GSK3, attributable to ERK and PI3K–Akt signalling cascades44,45. Inhibition of GSK3-dependent phosphorylation of JUN on Ser243 prevents binding of the E3 ligase F-box and WD domain repeated 7 (FBXW7), which targets JUN for polyubiquitylation and proteasomal degradation46,47. Whether FBXW7 degrades active (N-terminally phosphorylated by JNK) or inactive JUN molecules, is controversial47,48. JUN stability is increased after phosphorylation by JNK, which promotes degradation of JUN under non-stressed conditions17,36,49,50. JUN is also subject to sumoylation, which reduces the transcriptional activity of the JUN–FOS heterodimer51. Interestingly, the SUMO protease, SENP1, increases JUN-mediated transcription through the desumoylation of the p300 CRD1 domain, offering an alternative mechanism for the regulation of JUN transcription52. Although ubiquitin and SUMO modifications were also identified for other members of the AP1 family, the role of these modifications remains largely unexplored. For example, JUNB seems to undergo ubiquitylation-mediated proteasomal degradation, although the ubiquitin ligase that controls this modification is still unknown53. JUNB was shown to be sumoylated in T cells, resulting in its transcriptional activation54. Although JUND is also ubiquitylated, the consequences of this modification are yet to be determined, as its ubiquitylation does not result in its degradation55.
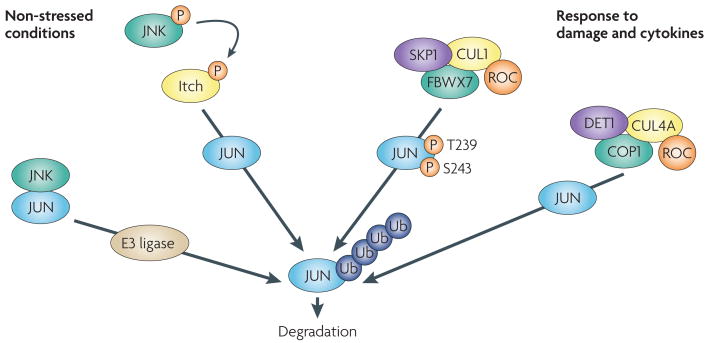
Figure 2. Mechanisms of JUN degradation.
Several mechanisms exist to limit JUN stablity. Under non-stressed conditions, JUN N-terminal kinase (JNK) is tightly bound to JUN, targeting JUN for ubiquitylation and degradation. Activation of JNK by stress results in JUN phosphorylation and dissociation from JNK, enhancing its protein stability, although phosphorylation on Ser63 and Ser73 promotes the F-box and WD domain repeated 7 (FBXW7)-mediated degradation of JUN48. As part of a feedforward mechanism, JNK also phosphorylates the E3 ligase Itch in T cells after stimulation, accelerating degradation of JUN and JUNB, independently of Ser63 and Ser73 phosphorylation34. Phosphorylation on JUN at Thr239 by glycogen synthase kinase 3 (GsK3) (FIG. 1) allows FBXW7 binding and ubiquitin-mediated degradation by a Skp1-Cullin1-F-box (SCF) complex. Following osmotic stress, MEKK1 can activate JUN N-terminal phosphorylation by activating MKK4, an upstream kinase for JNKs that can also function as a JUN E3 ubiquitin ligase, promoting its ubiquitin–proteasome-dependent degradation34 (not shown). De-etiolated 1 (DET1) contributes to JUN degradation by promoting the formation of a ubiquitin ligase complex containing DNA damage binding protein 1 (DDB1), cullin 4A (CUL4A), regulator of cullins 1 (ROC1) and constitutively photomorphogenic 1 (COP1)201. Other mechanisms modulating JUN ubiquitin-mediated degradation have been reported34. P, phosphorylation; Ub, ubiquitylation.
FOS and the Fos family
The Fos family of transcription factors is composed of FOS, FOSB, FOS-related antigen 1 (FRA1; also known as FOSL1) and FRA2 (also known as FOSL2). Apart from the classic bZIP domain and basic DNA binding domain in other AP1 proteins, FOS and FOSB also have strong transactivation domains, which FRA1 and FRA2 do not possess56. Fos family members can heterodimerize with JUN and some Atf family members, giving rise to complexes with different biochemical and transcriptional behaviour15. The negative charge of residues adjacent to the hydrophobic interphase of their leucine zipper electrostatically destabilizes Fos homodimers57 and favours the formation of JUN–FOS heterodimers, which exhibit increased stability, DNA-binding activity and greater transforming potential.
Fos activation and degradation
Like Jun family members, FOS and FOSB are immediate early genes expressed at low or undetectable levels in most cell types, with rapid and transient transcriptional activation following mitogenic stimuli or cellular stress58. Within minutes of growth factor stimulation and subsequent ERK activation, transcription of both genes is induced by ELK1, the cyclic AMP response element-binding protein (CREB) and serum-response factor (SRF). Although transcription of FRA1 and FRA2 also increases as a result of mitogenic stimulation through TRE, SRE, MYC and Atf sites, they are often expressed under non-stimulated conditions59. Similar to JUN, FRA1 transcription is partly autoregulated by an AP1 site60.
FOS activity and degradation are primarily regulated by phosphorylation. The major phospho-acceptor sites include Thr325, Thr331 and Ser374, which are phosphorylated by ERK, Ser362, phosphorylated by RSK1 and RSK2 (which are substrates of ERK), and Thr232, phosphorylated by an unknown kinase61. Transient activation of ERK alone results in Ser374 and Ser362 phosphorylation and stabilization of FOS, but these are insufficient to increase its transcriptional activity. Rather, these two modifications expose a docking site for ERK, which facilitates ERK-mediated phosphorylation of Thr331 and Thr325 that increases FOS transcriptional activity62. FOS is also phosphorylated by p38 at Thr232, Thr325, Thr331 and Ser374 in response to ultraviolet light treatment63. Unlike JUN, FOS is primarily degraded by the proteasome through ubiquitin-independent mechanisms. FOS degradation is differentially regulated by autonomous degrons at its N-terminal and carboxy-terminal ends. The activity of the C-terminal degron is reduced by phosphorylation of Ser362 and Ser374 (REF. 64). Similarly, FRA1 stabilization relies on the inhibition of a C-terminal degron by ERK-mediated phosphorylation of Ser252 and Ser265 (REF. 61). FOS shuttles between the nucleus and the cytoplasm owing to the presence of two nuclear localization signals. Dimerization with Jun proteins inhibits FOS nuclear export (notably, the strongest nuclear retention of FOS is observed when dimerized with JUN), thereby preventing the degradation of monomeric FOS in the cytoplasm65. Like other integral members of the AP1 transcriptional complex, Fos family members are reportedly deregulated in numerous human pathologies, and particularly in cancer (Supplemental information S2 (table)).
ATF2
ATF2 is one of 16 members of the Atf and Creb group of bZIP transcription factors that contribute to multiple cellular functions, from development to cellular responses to stresses, such as hypoxia or DNA damage response66–68. Although particularly enriched in brain tissue69, ATF2 is an ubiquitously expressed protein that is implicated in transcriptional control, chromatin remodelling and the DNA damage response70–72. Complete somatic loss of Atf2 results in postnatal lethality, whereas partial deregulation of ATF2 is implicated in cancer73–79.
ATF2 is located on chromosome 2q32 and comprises 12 exons, and in its full-length form, is translated into a protein 505 amino acids in length80. Like JUN and FOS, ATF2 is also characterized by a basic structural region and a leucine zipper domain that are crucial for AP1 homodimerization and heterodimerization81. ATF2 contains two canonical nuclear localization sequences (NLS) and one export sequence (NES) in its basic and leucine zipper regions, respectively. Its nuclear export has been shown to be CRM1-dependent82. Further complexity is added by tissue-specific expression of ATF2 splice variants, although to date studies evaluating the function of the splice variants have been limited (BOX 2).
Box 2. ATF2 splice variants.
Differential splicing or promoter usage in a tissue-specific manner can result in the expression of alternative splice isoforms of ATF2. Of the studied isoforms, most are ubiquitously expressed, with particular variants exhibiting tissue-specific enrichment. Studies on murine T cells revealed three isoforms (CRE-BP1, CRE-BP2 and CRE-BP3) of ATF2. The basic leucine zipper domain (bZIP) domain is conserved between these isoforms and variation between them resides mostly in their amino- and extreme carboxy-termini194,195, where ATF2 is frequently post-translationally modified and regulated85. Whereas CRE-BP2 lacks exons 1–7 and most of exon 12, CRE-BP2 varies from CRE-BP1 by an 8 amino acid substitution for the first 15 amino acids of CRE-BP1 only. This diversity suggests conservation of the transcription factor function between these isoforms, but variation in their regulation. ATF2-sm is an intriguing isoform of ATF2 that lacks all major bZIP functional domains and comprises the first and last two exons of full-length ATF2 only. It has been shown to be transcriptionally active, and exhibits polarized expression patterns in myometrial tissue and is differentially regulated before and during pregnancy and labour. Such differential expression patterns suggest that different ATF2 isoforms have tissue and temporal-specific functions, an idea that is supported by the finding that ATF2 and ATF2-sm transcriptionally regulate distinct subsets of genes196.
ATF2 phosphorylation
ATF2 is negatively regulated by intramolecular auto-inhibitory binding of its C-terminal DNA binding domain to its N-terminal activation domain83. This prevents ATF2 monomers from dimerizing with partner proteins during unstimulated (unstressed) conditions. Whether ATF2 monomers have a cellular function is unknown. In response to stress stimuli or cytokines, ATF2 is phosphorylated on Thr69 and/or Thr71 by either JNK or p38. Certain growth factors have also been shown to induce ERK-dependent phosphorylation of ATF2 on Thr71 followed by RALGDS–SRC–p38-dependent phosphorylation of Thr69 (REF. 84). In all cases, phosphorylation of these residues is required to de-repress ATF2 intramolecular inhibition allowing its homodimerization or heterodimerization with other members of the AP1 transcription factor family, such as JUN (Supplemental information S3 (table)), CREB, Fos and Fra85. The N-terminal phosphorylation of ATF2 and its dimerization, which facilitate ATF2 transcriptional function, also promote its ubiquitylation and degradation — a mechanism that limits ATF2 transcriptional output. Indeed, ATF2 mutants that are incapable of dimerization exhibit enhanced protein stability86–88. Phosphorylation of ATF2 at C-terminal Ser490 and Ser498 by ataxia-telangectasia mutated (ATM) is required for the contribution of ATF2 to the DNA damage response. ATM phosphorylation of ATF2 is important for the intra-S phase checkpoint following ionizing radiation (IR), essential for halting entry into the DNA replication phase of cell cycle. Furthermore, this phosphorylation was also found to promote ATF2 localization at irradiation-induced foci where it localizes with components of the DNA repair machinery, including MRE11, RAD50 and NBS1 (REF. 89). Another kinase shown to phosphorylate ATF2 on Ser121 is PKC, and this is essential for ATF2-mediated late-phase response to stress90.
ATF2 transcriptional targets
The basic DNA binding region of ATF2 homodimers exhibits binding specificity for CRE sequences, TGACGTCA91. However, depending on specific stimulus and cell type context, ATF2 can interact with other promoter elements including, but not limited to, other AP1 sequences, the proximal promoter of interferon-γ, stress-response element and the URE promoter. Like Jun and Fos family members, ATF2 dimerization with different partners significantly influences DNA binding specificity and affinity15,78,92,93, and ultimately the transcriptional outcome.
Although it is unknown whether ATF2 itself is cell cycle regulated, ATF2 does regulate cell cycle progression through the transcriptional control of several key genes, including RB1, cyclin A, cyclin D, GADD45A, GADD45B and maspin (TABLE 2). ATF2 can further enhance proliferation by promoting survival through regulation of Bcl2 expression in certain cell types94. ATF2 transcriptionally regulates a wide array of gene targets controlling other cellular pathways, ranging from Atf, Jun and Fos transcription factors, to extracellular, cytokine and intracellular signalling pathways (TABLE 2).
Table 2. ATF2 transcriptional targets.
| Gene | Context | Methods | Refs |
|---|---|---|---|
| Extracellular stimuli | |||
| IL8 | ATF2 recruits macroH2A nucleosome to repress IL8 transcription | HeLa and Namalwa B-cells, WB and Luc | 72 |
| Pdgfra | ATF2-null mice exhibit decreased PDGFRα expression levels that are rescued on re-expression of ATF2 | Mutant mice and CHO cells, WB and Luc | 73 |
| MMP2 | ATF2 activates MMP2 transcription | MCF10A cells and Luc | 78 |
| PLAU | ATF2 cooperatively induces PLAU in response to IL-1 and TPA | HepG2 cells, WB and Luc | 216 |
| Crip2 | TGFβ upregulation of CRP2 requires ATF2 | Rat and mouse vascular smooth muscle cells, WB and Luc | 217 |
| Psen1 | Presenilin 1 expression is reduced in Atf2-null keratinocytes | Murine keratinocytes and WB | 115 |
| SELE | ATF2 contributes to E-selectin induction during inflammatory response | Human endothelial cells, WB and Luc | 218 |
| TNF | Autoamplification of death signals are mediated by ATF2 | Jurkat and CEM cells, and WB | 219 |
| ATF3 | Radiation-activated ATM signalling induces ATF3 through ATF2 | Normal human diploid fibroblasts and WB | 220 |
| Nos2a | ATF2 mediates NOS2A upregulation in rat glial cells | C6 glial cells and Luc | 221 |
| IFNG | Jun–ATF2 dimers regulate IFNG transcription through proximal promoter | Jurkat cells, WB and Luc | 222 |
| Hspa5 | ATF2 and CREB mediate stress induction of HSPA5 | 9L rat brain tumour cells and WB | 223 |
| Th | ATF2 positively regulates tyrosine hydroxylase transcription | PC12D cells, WB and Luc | 224 |
| Cell cycle and transcription | |||
| CCND1 | p38 and ATF2 induce cyclin D1 in breast cancer in response to oestradiol and spermine | MCF7 cells, WB and Luc | 119 |
| Ccna1 | ATF2 mediates serum induction of cyclin A | Rat chondrosarcoma cells, WB and Luc | 114 |
| Rb1 | ATF2 modulates RB during skeletal growth | ATDC cells and primary chondrocytes, WB and Luc | 112 |
| PI5 | ATF2 binds directly to the PI5 promoter (which encodes maspin) | Mutant mice, MEFs, MCF7 cells, WB and Luc | 116 |
| GADD45A | ATF2 is recruited by OCT1 and NF1 to the GADD45A promoter to activate transcription | Mutant mice, MEFs, MCF7 cells, WB and Luc | 116 |
| PKC | PKC phosphorylates ATF2 to activate JUN transcription | F9 teratoma, MEFs, HeLa cells, WB and Luc | 90 |
| Ppargc1a | ATF2 mediates exercise-induced PGC1α upregulation | C2C12 myocytes, WB and Luc | 225 |
| Atf and Creb, Jun and Fos family genes | ATF2 transcriptionally regulates other members of the Atf and Creb, and Jun and Fos families, contingent on cell type and stimulus context | F9 teratoma, COS-1, MCF7 cells, WB and Luc | 4,82, 226 |
| Mapk14 | ATF2 negatively feeds back on p38 signalling through transcriptional induction of MAPK phosphatases | Mutant mouse tissue and cells, and WB | 68 |
| Cell death | |||
| Bcl2 | ATF2 controls Bcl2 promoter activation in chondrocytes | Mutant mice, WB and Luc | 94 |
| TRAIL | Auto-amplification of death signals are mediated by ATF2 | Jurkat and CEM cells, and WB | 219 |
| ACHE | ATF2 mediates hydrogen peroxide-induction of acetylcholinesterase | 293T cells, WB and Luc | 227 |
ACHE, acetylcholinesterase; ATM, ataxia-telangectasia mutated; CCND1, cyclin D1; CHO, Chinese hamster ovary; Crip2, cysteine-rich intestinal protein 2; Hspa5, heat shock protein A5; IFNG, interferon (IFN)-γ; IL8, interleukin-8; Luc, luciferase assay; MEF, mouse embryonic fibroblast; MMP2, matrix metalloproteinase 2; Nos2a, nitric oxide synthase 2A; Pdgfra, platelet derived growth factor A; PI5, protease inhibitor 5; PLAU, urinary palminogen activator; Ppargc1a, peroxisome proliferator-activated receptor-γ coactivator 1α; Psen1, presenilin 1; SELE, E selectin; TGFβ, transforming growth factor-β; Th, tyrosine hydroxylase; TNF, tumour necrosis factor; TRAIL, tumour necrosis factor-related apoptosis-inducing ligand; WB, western blot.
JUN, FOS and ATF2 in tumorigenesis
JUN overexpression in vitro is sufficient, in certain cases, to transform mammalian cells8. Consistently, loss of JUN decreases the incidence of papilloma outgrowth by abrogation of epidermal growth factor receptor (EGFR) signalling in skin subjected to the two-stage skin carcinogenesis protocol95. However, transformation of other cells, such as rat embryonic fibroblasts, require the presence of either additional oncogenes such as Ras and SRC or other AP1 components such as FRA1 (REFS 8,96). Despite its oncogenic potential in vitro, JUN overexpression in transgenic mice does not result in the development of tumours8,13. HRAS-induced transformation of immortalized mouse fibroblasts requires JUN expression, as transformation is suppressed in the absence of JUN or the presence of a dominant-negative JUN97. Fibroblasts with JUN Ala63 and Ala73 can be efficiently transformed by v-ras, but show reduced tumorigenicity in nude mice98. This is consistent with the ability of v-jun to contribute to cell transformation despite a lack of phosphorylation on the Ser63 and Ser73 sites. The expression of oncogenic HRAS can increase AP1 transcriptional activity by activating ERK and JNK, leading to increased expression of Fos proteins and N-terminal phosphorylation of JUN99. An alternative mechanism was recently proposed by Talotta et al.100, who showed that HRAS can trigger a positive AP1 feedback loop in solid tumours through promoting JUN–FRA1 heterodimer formation with subsequent upregulation of microRNA-21. As a result, miR-21 causes the downregulation of tumour suppressors and negative AP1 regulators, including the tumour suppressor PTEN and programmed cell death 4 (PDCD4)100.
Fos family proteins have oncogenic potential both in vitro and in vivo by regulating proliferation and transformation, angiogenesis, tumour invasion and metastasis101. Expression of FRA1 confers anchorage-independent growth in rat fibroblasts in vitro and promotes tumour development in athymic mice102. Similar to JUN, FOS overexpression correlates with tumour grade and adverse outcome in some cancers. Its overexpression alone is sufficient to transform chicken embryonic fibroblasts103, and its oncogenicity is linked to JUN, as immortalized fibroblasts expressing v-ras, v-fos and a non-JNK phosphorylatable JUN mutant showed reduced tumorigenicity in nude mice98. Transgenic expression of Fos promoted the transformation of chondroblasts and osteoblasts, resulting in chondrogenic and osteogenic tumour formation in mice104. Overexpression of FRA2 in mice can also induce tumour formation in the pancreas, thymus and lung4. Knockdown or dominant-negative mutants of FOS can abrogate transformation by upstream oncogenes, such as activated Ras105,106. However, in contrast to their oncogenic contributions, recent reports suggest a possible tumour suppressor role for the Fos family. For instance, ubiquitous FRA1 overexpression accelerates osteoblast differentiation and subsequent osteosclerosis107, whereas overexpression of FOS inhibits cell cycle progression, stimulating mouse hepatocyte cell death and strongly suppresses tumour formation in vivo108. Based on findings from human familial breast and ovarian cancer, one possible mechanism for the tumour-suppressor activity of FOS could be its potential involvement in the regulation of BRCA1 (REF. 109). The function of FOS in apoptosis might also influence its capacity to suppress tumour formation (see below). Together, these data highlight the functional duality of Fos family transcription factors and the importance of their tissue-specific context and resulting heterodimerization partners.
Evidence to date indicates that ATF2 can elicit tumour suppressor or oncogene activities in a cell- and tissue-dependent context4. For example, in melanoma, inhibition of ATF2 activity by ATF2 inhibitory peptides results in the suppression of tumorigenesis and metastasis, concomitant with sensitization of melanoma tumour cells to genotoxic stress in vitro and in vivo74,93,110,111. Consistent with its cell cycle regulatory role, increased expression of ATF2 increased cell proliferation in mouse cancer models112–114. By contrast, expression of transcriptionally inactive ATF2 in the presence of oncogene activation (such as Ras mutations) in non-melanoma skin cancers increases papilloma formation owing to the deregulated expression of genes that promote proliferation, such as CTNNB1 (REF. 115). In agreement with this, mammary tumour formation rates are accelerated in Atf2 heterozygous mice that also carry a mutant allele of Trp53 (REF. 116). Such results indicate that depending on the tissue type, ATF2 can elicit a tumour suppressive function, and that loss of ATF2 can cooperate with oncogenes and mutation of tumour suppressor genes to promote tumorigenesis115. As the loss of ATF2 alone does not induce tumour formation, but rather predisposes mouse models to more rapid onset and increased tumour incidence with additional genetic mutations, the functional loss of ATF2 might have a cooperative role as opposed to an initiator role in multistage tumorigenic processes75,115.
A phenomenon that might shed light on the divergent function of ATF2 is its differential subcellular localization. Immunohistochemical studies have demonstrated an upregulation and activation of ATF2 in the nuclear compartment in certain cancer types75,117–119. Furthermore, immunohistochemical analysis of patient-derived tumour tissue microarrays found enriched nuclear localization of ATF2 in advanced metastatic melanoma samples, which correlated with poor prognosis and survival120. By contrast, melanoma samples exhibiting strong cytoplasmic localization correlated with primary tumours and favourable prognosis. Analysis of tissue microarrays from patient-derived squamous and basal cell carcinoma samples revealed reduced nuclear levels and increased cytoplasmic levels of ATF2, further substantiating the idea that ATF2 transcriptional activity may be attenuated in non-melanoma and papillary tumours115. Recent studies using IR of prostate cancer cells showed that IR can induce cytoplasmic localization of ATF2, in contrast to its predominant nuclear localization during basal conditions121. Notably, cytoplasmic accumulation of ATF2 was associated with the appearance of a neuroendocrine-like (differentiation) phenotype. As ATF2 is known to promote differentiation in certain tissue contexts when dimerized with JUN, it is also possible that enhanced ATF2 binding with JUN might outcompete JUN binding with other factors, such as FOS and FRA2, both of which enhance cell cycle re-entry and progression122. Although the significance of the cytosolic localization of ATF2 is not known, its distinct distribution and activities probably depend on post-translational modification with available heterodimeric partners of the AP1 network (FIG. 3). Dimerization with JUN has been shown to promote nuclear import of ATF2 while monomeric ATF2 remained cytoplasmic, suggesting that monomeric forms of ATF2 in the cytoplasm have an alternative function82.
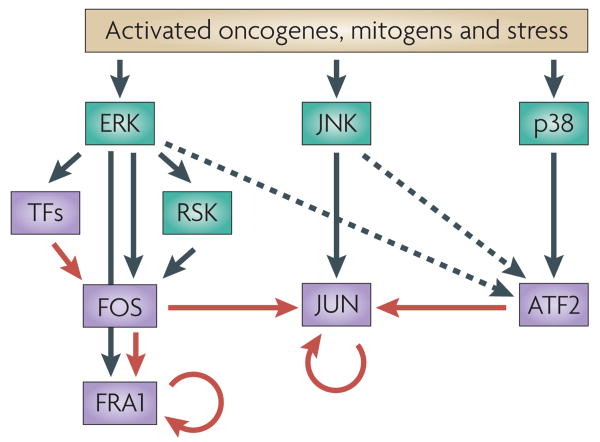
Figure 3. Network of AP1 signalling.
ERK, JUN N-terminal kinase (JNK) and p38 are predominantly responsible for the phosphorylation and activation of FOS, JUN and ATF2 respectively (black arrows) in response to stress, mitogens or oncogene activation. ERK and JNK also regulate FOS and JUN degradation, respectively (not depicted) and participate in ATF2 activation (dashed arrows). ERK also induces the transcription of FOS through the activation of non-AP1 transcription factors (TFs). Transcription of some AP1 proteins (that is, JUN and FRA1) is regulated by crosstalk among AP1 complexes (red arrows) as well as by autoregulation (circular red arrows).
AP1 in tumorigenesis
Chronic exposure to certain environmental or dietary carcinogens can promote tumorigenesis through the stimulation of a wide array of signalling pathways, ranging from inflammatory to pro-proliferative and survival pathways23, and carcinogens have been observed to induce or at least correlate with increased AP1 activity. Long-term exposure to tobacco smoke or nicotine, for instance, activates AP1 activity in mouse brain or epithelial cell lines, and specifically, FOS and JUN are upregulated in rat and hamster cell lines during chronic asbestos exposure123–126. Chronic ethanol exposure of human neuroblastoma cells enhances AP1 activity127. In several studies, AP1 activity is crucial for tumorigenesis, as inhibition of AP1 function by dominant-negative JUN mutants or AP1 decoys, for example, effectively inhibits tumour formation in vivo. Such studies have also enabled the identification of AP1 target genes involved in different aspects of carcinogenesis30,31,128,129. Interestingly, AP1 activity is reported to be upregulated in certain tumour cell lines that acquire drug resistance after chronic anti-oestrogen therapy or cisplatin treatment, suggesting the possibility that some chemotherapeutic agents, similar to long-term carcinogenic stimuli, can elicit AP1 activation that can facilitate tumour survival and render them refractory to long-term treatments130,131. Numerous studies have shown the importance of AP1 in tumorigenesis.
Invasion and metastasis
Extensive evidence suggests that JUN and other AP1 proteins coordinate multigene expression programmes required for invasive and metastatic behaviour (Supplementary information S1, S3 (tables)). For example, AP1 has consistently been linked to invasive properties of aggressive breast cancer132. Overexpression of JUN in MCF7 breast cancer cells increased tumour formation in nude mice, as well as motility, invasiveness and liver metastasis27. Enforced expression of JUN in human bronchial epithelial cells significantly increased cell viability and colony formation in soft agar, whereas expression of TAM67 inhibited their anchorage-independent growth29,133. Similarly, the treatment of oral squamous cell carcinoma cells with AP1 decoys attenuates their invasiveness134. Consistently, cells from conditional Jun-knockout mice exhibit increased cellular adhesion, stress fibre formation and reduced cellular migration, a phenotype that was reverted by addition of stem cell factor (SCF; a JUN target gene)135. Among genes that are regulated by JUN and may mediate these changes are genes encoding Stathmin, HMGA1 or cyclin A18,136,137. Other JUN-induced genes that may contribute to enhanced tumorigenesis are involved in cellular migration and invasion as well as inflammation (Supplementary information S1 (table)). These findings suggest that increased expression of JUN, as well as FOS (see below), may be involved in the acquisition of anchorage independence in the process of human carcinogenesis.
FOS family members transcriptionally regulate numerous genes involved in cell movement and invasiveness, such as the genes encoding matrix metalloproteinase 1 (MMP1), MMP3, cathepsin L and ezrin138,139. Therefore, it is not surprising that epithelial–mesenchymal transition can be induced by FOS140. As FRA1-containing complexes can activate transcription from both TRE and CRE elements, most of the genes shown to be activated by FOS have also been shown to be activated by FRA1 (REFS 141,142).
Several observations support a contribution of ATF2 to the regulation of cellular invasiveness and migration. For example, increased expression and phosphorylation of ATF2 correlates with increased tumour invasiveness in patients with extramammary Paget's disease143. Furthermore, in vitro studies with MCF10A cells demonstrate that ATF2 signalling driven by p38 mediates the transcription of MMP2, thereby influencing invasive migration78. Other members of the Atf and Creb family are also implicated in invasion and migration. For example, the expression of a dominant-negative CREB mutant impairs the invasiveness of MeWo melanoma cells144.
Angiogenesis
Activated JUN is predominantly found at the invasive front of tumours and is associated with replicating cells, microvessel density and vascular endothelial growth factor A (VEGFA) expression145. Targeting JUN by catalytic DNA molecules known as DNAzymes blocked endothelial cell proliferation, migration, chemoinvasion and tubule formation in mouse tumour models146. The same DNAzymes also suppressed the growth and angiogenesis of solid squamous cell carcinomas in severe combined immunodeficient (SCID) mice by inhibiting MMP2, MMP9, VEGFA and fibroblast growth factor 2 expression147. This is consistent with previous data showing that a transactivation domain deletion mutant of JUN attenuated the formation of squamous cell carcinoma148. JUN was also found to control expression of proliferin, an angiogenic placental hormone that also has a role in tumour angiogenesis149. Recently, it was shown that interleukin-7 (IL-7) promotes lymphangiogenesis in lung cancer by inducing VEGFD expression that is dependent on FOS–JUN dimers150.
Survival and apoptosis
The pro-apoptotic or anti-apoptotic function of JUN is cell type specific and dependent on both the type of external or internal stimuli and the potential JUN binding partners (Supplementary information S1 (table)). In part, the overexpression of JUN has been implicated in the induction of apoptosis in neurons151, endothelial152, myeloma cells153 and fibroblasts154, although the JUN targets have remained mostly unidentified. Activation of JNK and subsequent JUN phosphorylation has been associated to apoptotic cell death. However, Atf family members, such as ATF3, can cooperatively promote survival with JUN through the induction of heat shock protein 27 in neurons during injury, indicating that the heterodimerization of JUN with other AP1 factors can antagonize its pro-apoptotic functions.
Enhanced growth conferred by induction of JUN has been attributed not only to cell cycle alterations, but also to enhanced cell survival that is concomitant with the reduction of cell death. Among the mediators for cell survival signalling, JUN suppresses PTEN155, which inhibits cell growth through negative regulation of the Akt survival pathway. Eferl et al.156 showed that mice with a targeted disruption of Jun in hepatocytes156 presented with reduced liver tumour mass and higher survival rates than control mice in a chemically induced hepatocellular carcinoma model. JUN deficiency resulted in the accumulation of p53 and increased apoptosis without affecting the proliferation rate of these cells, confirming a role for JUN in cell survival. This function of JUN seems to be distinct from its role in proliferation, which also involves p53 (REF. 157). Therefore, JUN may promote tumorigenesis by antagonizing the proapoptotic and anti-proliferative activities of p53 through different mechanisms.
FOS also seems to have a pro-apoptotic function, as do FRA1 and FRA2. FOS mediates MYC-induced cell death, probably through the p38 MAPK pathway and the induction of CD95L and tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)158,159. Moreover, reduced levels of FOS, FRA1 or FRA2 might potentiate chemoresistance160–162. The proapoptotic downstream molecular targets of FOS are mostly poorly understood. Interestingly, JUN and FRA1 also induce the expression of the tumour suppressor ARF — a key link between oncogenic signalling and the p53 pathway — which resulted in the induction of growth arrest in primary mouse fibroblasts21.
In contrast to its pro-proliferative function, ATF2 has been found to mediate apoptosis in various circumstances, although specific signalling mechanisms remain largely understudied. For example, in chondrocytes, ATF2 and CREB1 can heterodimerize to directly regulate the Bcl2 promoter94. ATF2 was shown to mediate apoptosis in non-differentiated PC12 cells163.
With strong evidence to support the role of JUN in normal and tumour cell biology, and the observations that JUN can also induce apoptosis, under certain conditions111,154,164, it is likely that these diverse functions are dependent on the nature of its heterodimeric partners. For example, abrogation of ATF2 interaction with JNK sensitizes melanoma cells to anisomycin-induced cell death that is JUN- and JUND-dependent, suggesting that a pro-apoptotic JUN and/or JUND-containing AP1 complex is enriched in the absence of active ATF2 (REF. 111). In neurons, JUN dimerization to ATF2 promotes apoptosis through the transcription of harakiri, whereas this is abrogated by increases in FOS expression, probably because FOS competes with JUN for binding to ATF2 (REF. 165). Conversely, FOS–JUN dimers have been shown to induce apoptosis in prostate cancer cells by the transcriptional repression of the anti-apoptotic molecule CASP8 and FADD-like apoptosis regulator (FLIP)166. These data imply that perturbations in the fine balance between the tissue-specific compositions of AP1 dimers are sufficient to alter transcriptional programmes for either cell survival or cell death, and are likely to have a major role during tumorigenesis.
Stem cell self-renewal and differentiation
Several in vitro and in vivo studies have indicated that AP1 function is involved in stem cell and tumour cell self-renewal or differentiation. Whereas JUN is stabilized at the protein and mRNA levels during induced differentiation of teratocarcinoma and erythroid cells, respectively, other Jun family members, such as JUNB, can negatively regulate proliferation of long-term repopulating myeloid stem cells167,168. JUN is particularly implicated in hepatogenesis and cardiac development9,169, and consistent with this JUN and FOS are implicated in icariin-induced cardiomyocyte differentiation of mouse embryonic stem cells170. Downregulation of cyclin A during differentiation of human embryonic carcinoma cells depends on promoter depletion of ATF1 and ATF2 (REF. 113). By contrast, ATF2 also interacts with undifferentiated embryonic cell transcription factor 1 (UTF1), an important transcriptional co-activator during early embryogenesis that apparently enhances ATF2-dependent transcription in F9 embryonic carcinoma cells171, indicating specific AP1 transcriptional programme switches during differentiation.
Jun, Fos and ATF2 in human cancer
Many human cancers exhibit overexpression of JUN and/or other Jun family members (TABLE 1), which is predominantly the result of activation of upstream oncogenes, including Ras, BRAF and EGFR. Activating mutations of NRAS or BRAF, which occur in >70% of melanomas, super-activate ERK, driving increased expression of JUN by increasing its transcription and stability44. Moreover, inhibition of JUN function consistently attenuates the growth of various human tumour cell lines both in vitro and in mouse xenografts172. Consistent with the idea that JUN promotes tumorigenicity is its overexpression in some of the more aggressive CD30-positive lymphomas173,174. Similarly, increased JUN levels correlate with more advanced tumour stage and poor prognosis in prostate cancer175. In breast cancer, other altered pathways, including RB, VEGF and EGFR have been implicated in inducing increased JUN expression.
Interestingly, altered FOS expression in tumours depends on the tissue of origin. Its increased expression is associated with poor clinical outcomes in osteosarcoma and endometrial carcinoma, and loss of FOS expression is associated with tumour progression and adverse outcome in ovarian carcinoma and gastric carcinoma176,177. FRA1 overexpression is associated with diverse tumours, including thyroid, breast, lung, brain, nasopharyngeal, oesophageal, endometrial, prostate and colon carcinomas, along with glioblastomas and mesotheliomas, and so may hold prognostic value178 (Supplementary information S2 (table)).
Overexpression and activation (phosphorylation) of ATF2, altered subcellular localization and enhanced interaction with other AP1 proteins, in particular with oncogenic JUN, is observed in many cancer types and transformation models46,117. Such increased expression of ATF2 might be of diagnostic value in the clinic118. However, loss of ATF2 function is also observed in cancer. Although germline mutations in ATF2 are infrequent, mutations that inactivate ATF2 have been observed in certain cancer types such as lung and breast cancer, and neuroblastoma76,77.
The dynamic network of AP1 signalling
The diverse functions attributed to AP1 complexes have proved difficult to discern as each is dictated by the distinct heterodimeric combination that can be assembled from an array of potential complexes that these proteins can form. ATF2, for example, was reported to form 8 different complexes with other members of the Atf, Jun and Fos family, whereas JUN can form 15 different dimeric complexes. FOS was reported to form heterodimeric complexes with all Jun members, ATF2, ATF4, CREB1 and all Maf members. Differential dimerization between JUN, ATF2 and FOS with different family members is sufficient to alter their promoter-binding specificity, drastically changing the transcriptional capacity, protein stability and localization, and ultimately the transcriptional repertoire of these proteins179. Such dimerization and consequent functional differences are largely attributed to tissue- and cell type-specific expression levels of the individual AP1 proteins and the degree of activation of upstream pathways such as MAPK or SAPK pathways. For example, whereas JUN–ATF2180,181 and/or JUN–FOS dimers182 can promote proliferation in some cell types, in skin and breast cancer, ATF2 suppresses tumour outgrowth115,116. In addition to cell- and tissue-specific conditions, AP1 dimer composition is subject to influence by cell cycle progression and specific stimuli. For example, mitogenic stimulation upregulates FOS–JUN dimers, which are later displaced by FRA1 and FRA2 in accordance with the duration of ERK1 and ERK2 activity183.
Apart from variation in transcriptional activity that can be attributed to altered promoter binding, individual AP1 proteins (particularly those exhibiting weak transactivation potential, such as JUNB, JUND, FRA1 and FRA2) can function as transcriptional repressors, by competitively out-binding partners of transcriptionally active AP1 complexes165,184,185,186. This explains why some dimers activate, and others repress transcription through binding at the same DNA promoter sites187. Accordingly, as the composition of the AP1 complexes is paramount to their function, deregulation of this composition in favour of more oncogenic partnerships may account for the transcriptional alterations observed during tumorigenesis.
A revealing example of altered AP1 composition relevant for cancer is that observed during Ras transformation. Oncogenic Ras constitutively activates ERK and increases transcription of JUN, JUNB, FRA1 and FRA2, but not of FOS, increasing the population of JUN–FRA1 dimers and thus increasing AP1 activity188. Similarly, the adenoviral protein E1A alters AP1 composition by promoting ATF2–JUN dimer formation, resulting in strong activation of ATF2, but weak activation of ATF1 or CREB1 (REFS 189–191). The implication of this dynamic heterodimerization is exemplified by CREB1 dimerization with ATF2, which abrogates the ability of E1A to bind to ATF2, presumably suppressing that particular axis of E1A-mediated transformation189,192.
Collectively, these studies support the idea that manipulation of AP1 composition might have therapeutic applications in cancer treatment. The oncogenicity of JUN in the studies discussed above largely depends on its binding to ATF2, which is consistent with earlier studies showing that mutant v-jun with enhanced ATF2-binding capacity and mutant ATF2 that binds JUN with increased affinity can both enhance growth factor independence and tumorigenicity in vivo111,180. JUN–FOS and JUN–ATF2 dimers clearly have crucial roles in promoting tumorigenesis, however, the antagonistic effects of FOS- and ATF2-containing AP1 complexes implies that distinct cellular pathways are activated by each of these complexes. The complex changes in AP1 members at the transcriptional, translational and post-translational levels that enable their dynamic interchange during tumour development are still subject to intense investigation. The availability of genomic and proteomic technologies combined with the power of systems biology will, we hope, reveal the composition and therefore the mechanisms underlying this dynamic network in the near future.
Supplementary Material
Acknowledgments
We thank members of the Ronai laboratory for discussions and critical reading. We thank K. Wright for editorial assistance. Support by US National Cancer Institute (NCI) grants CA099961, CA051995, CA117927 (to Z. R.) and by Roemmers Foundation, The Harry J Lloyd Charitable Trust and ANPCyT (PICT-2007-01010) (to P.L.B.) is gratefully acknowledged. E.L. was supported by NCI grant T32 CA121929 and by the American Cancer Society (ACS), Illinois Division, Postdoctoral Fellowship, PF-09-112-01-GMC.
Footnotes
Competing interests statement: The authors declare no competing financial interests.
Databases: Entrez Gene: http://www.ncbi.nlm.nih.gov/gene
JUN
UniProtKB: http://www.uniprot.org
ATF2 | ATF3 | ATF4 | cathepsin L | CREB1 | cyclin A | ezrin | FBXW7 | FOS | FOSB | FRA1 | FRA2 | HMGA1 | JNK1 | JNK2 | JUNB | JUND | MMP1 | MMP2 | MMP3 | MMP9 | SENP1 | Stathmin | VEGFA
Pathway interaction Database: http://pid.nci.nih.gov/
ATF2
Further Information: Ze'ev Ronai's homepage: http://www.ronailab.net
Supplementary Information: See online article: S1 (table) | S2 (table) | S3 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Pablo Lopez-Bergami, Instituto de Biologia y Medicina Experimental, Vuelta de Obligado 2490, Buenos Aires1428, Argentina, pablo.bergami@gmail.com.
Eric Lau, Signal Transduction Program, Burnham Institute for Medical Research, La Jolla, CA 92037, USA, elau@burnham.org.
Ze'ev Ronai, Signal Transduction Program, Burnham Institute for Medical Research, La Jolla, CA 92037, USA.
References
- 1.Lee W, Mitchell P, Tjian R. Purified transcription factor AP1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 3.Wisdom R. AP1: One switch for many signals. Exp Cell Res. 1999;253:180–185. doi: 10.1006/excr.1999.4685. [DOI] [PubMed] [Google Scholar]
- 4.Eferl R, Wagner EF. AP1: a double-edged sword in tumorigenesis. Nature Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 5.Angel P, Karin M. The role of Jun, Fos and the AP1 complex in cell-proliferation and transformation. Biochem Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 6.Vlahopoulos SA, et al. The role of ATF-2 in oncogenesis. Bioessays. 2008;30:314–327. doi: 10.1002/bies.20734. [DOI] [PubMed] [Google Scholar]
- 7.Maki Y, Bos C, Davis C, Starbuck M, Vogt P. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci USA. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt PK. Jun, the oncoprotein. Oncogene. 2001;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- 9.Eferl R, et al. Functions of c-Jun in liver and heart development. J Cell Biol. 1999;145:1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 11.Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariani O, et al. JUN Oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11:361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Shaulian E, Karin M. AP1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 14.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 15.van Dam H, Castellazzi M. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 16.Sng JCG, Taniura H, Yoneda Y. A tale of early response genes. Biol Pharm Bull. 2004;27:606–612. doi: 10.1248/bpb.27.606. [DOI] [PubMed] [Google Scholar]
- 17.Whitmarsh AJ, Davis RJ. Regulation of transcription factor function by phosphorylation. Cell Mol Life Sci. 2000;57:1172–1183. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katabami M, et al. Cyclin A is a c-Jun target gene and is necessary for c-Jun-induced anchorage-independent growth in RAT1a cells. J Biol Chem. 2005;280:16728–16738. doi: 10.1074/jbc.M413892200. [DOI] [PubMed] [Google Scholar]
- 19.Weitzman JB, Fiette L, Matsuo K, Yaniv M. JunD protects cells from p53-dependent senescence and apoptosis. Mol Cell. 2000;6:1109–1119. doi: 10.1016/s1097-2765(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 20.Schreiber M. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ameyar-Zazoua M, et al. AP1 dimers regulate transcription of the p14/p19ARF tumor suppressor gene. Oncogene. 2005;24:2298–306. doi: 10.1038/sj.onc.1208424. [DOI] [PubMed] [Google Scholar]
- 22.Deng T, Karin M. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 1993;7:479–490. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal BB, Gehlot P. Inflammation and cancer: how friendly is the relationship for cancer patients? Curr Opin Pharmacol. 2009;9:351–369. doi: 10.1016/j.coph.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passegue E, Wagner EF. JunB suppresses cell proliferation by transcriptional activation of p16INK4a expression. EMBO J. 2000;19:2969–2979. doi: 10.1093/emboj/19.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryseck RP, Hirai SI, Yaniv M, Bravo R. Transcriptional activation of c-jun during the G0/G1 transition in mouse fibroblasts. Nature. 1988;334:535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- 26.Mayo MW, Steelman LS, McCubrey JA. Phorbol esters support the proliferation of a hematopoietic cell line by upregulating c-jun expression. Oncogene. 1994;9:1999–2008. [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Critical role of c-Jun overexpression in liver metastasis of human breast cancer xenograft model. BMC Cancer. 2007;7:145. doi: 10.1186/1471-2407-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X, et al. Blockade of AP1 activity by dominant-negative TAM67 can abrogate the oncogenic phenotype in latent membrane protein 1-positive human nasopharyngeal carcinoma. Mol Carcinog. 2007;46:901–911. doi: 10.1002/mc.20319. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu Y, et al. Growth inhibition of non-small cell lung cancer cells by AP1 blockade using a cJun dominant-negative mutant. Br J Cancer. 2008;98:915–922. doi: 10.1038/sj.bjc.6604267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Q, et al. The AP1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene. 2008;27:366–377. doi: 10.1038/sj.onc.1210643. [DOI] [PubMed] [Google Scholar]
- 31.Suto R, et al. Dominant-negative mutant of c-Jun gene transfer: a novel therapeutic strategy for colorectal cancer. Gene Therapy. 2004;11:187–193. doi: 10.1038/sj.gt.3302158. [DOI] [PubMed] [Google Scholar]
- 32.Passegue E, Jochum W, Behrens A, Ricci R, Wagner EF. JunB can substitute for Jun in mouse development and cell proliferation. Nature Genet. 2002;30:158–166. doi: 10.1038/ng790. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal SK, et al. Transcription factor JunD, deprived of menin, switches from growth suppressor to growth promoter. Proc Natl Acad Sci USA. 2003;100:10770–10775. doi: 10.1073/pnas.1834524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laine A, Ronai Z. Ubiquitin chains in the ladder of MAPK signaling. Sci STKE. 2005;281:re5. doi: 10.1126/stke.2812005re5. [DOI] [PubMed] [Google Scholar]
- 35.Dérijard B, et al. JNK1: A protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 36.Sabapathy K, et al. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 37.Sabapathy K, Wagner EF. JNK2: a negative regulator of cellular proliferation. Cell Cycle. 2004;3:1520–1523. doi: 10.4161/cc.3.12.1315. [DOI] [PubMed] [Google Scholar]
- 38.Lively TN, Ferguson HA, Galasinski SK, Seto AG, Goodrich JA. c-Jun binds the N terminus of Human TAFII250 to derepress RNA polymerase II transcription in vitro. J Biol Chem. 2001;276:25582–25588. doi: 10.1074/jbc.M100278200. [DOI] [PubMed] [Google Scholar]
- 39.Franklin CC, McCulloch AV, Kraft AS. In vitro association between the Jun protein family and the general transcription factors, TBP and TFIIB. Biochem J. 1995;305:967–974. doi: 10.1042/bj3050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karin M, Liu ZG, Zandi E. AP1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 41.Weiss C, et al. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 2003;22:3686–3695. doi: 10.1093/emboj/cdg364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 43.Gallo A, et al. Menin uncouples Elk-1, JunD and c-Jun phosphorylation from MAP kinase activation. Oncogene. 2002;21:6434–6445. doi: 10.1038/sj.onc.1205822. [DOI] [PubMed] [Google Scholar]
- 44.Lopez-Bergami P, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhoumik A, Ronai Z. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle. 2008;7:2341–2345. doi: 10.4161/cc.6388. [DOI] [PubMed] [Google Scholar]
- 47.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 49.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 50.Fuchs SY, Dolan L, Davis RJ, Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- 51.Bossis G, et al. Down-regulation of c-Fos/c-Jun AP1 dimer activity by sumoylation. Mol Cell Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng J, Perkins ND, Yeh ET. Differential regulation of c-Jun-dependent transcription by SUMO-specific proteases. J Biol Chem. 2005;280:14492–14498. doi: 10.1074/jbc.M412185200. [DOI] [PubMed] [Google Scholar]
- 53.Farras R, Bossis G, Andermarcher E, Jariel-Encontre I, Piechaczyk M. Mechanisms of delivery of ubiquitylated proteins to the proteasome: new target for anti-cancer therapy? Crit Rev Oncol Hematol. 2005;54:31–51. doi: 10.1016/j.critrevonc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Garaude J, et al. SUMOylation regulates the transcriptional activity of JunB in T lymphocytes. J Immunol. 2008;180:5983–5990. doi: 10.4049/jimmunol.180.9.5983. [DOI] [PubMed] [Google Scholar]
- 55.Musti AM, Treier M, Peverali FA, Bohmann D. Differential regulation of c-Jun and JunD by ubiquitin-dependent protein degradation. Biol Chem. 1996;377:619–624. doi: 10.1515/bchm3.1996.377.10.619. [DOI] [PubMed] [Google Scholar]
- 56.Tulchinsky E. Fos family members: regulation, structure and role in oncogenic transformation. Histol Histopathol. 2000;15:921–928. doi: 10.14670/HH-15.921. [DOI] [PubMed] [Google Scholar]
- 57.Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- 58.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 59.Kovary K, Bravo R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adiseshaiah P, Peddakama S, Zhang Q, Kalvakolanu DV, Reddy SP. Mitogen regulated induction of FRA-1 proto-oncogene is controlled by the transcription factors binding to both serum and TPA response elements. Oncogene. 2005;24:4193–4205. doi: 10.1038/sj.onc.1208583. [DOI] [PubMed] [Google Scholar]
- 61.Basbous J, Jariel-Encontre I, Gomard T, Bossis G, Piechaczyk M. Ubiquitin-independent- versus ubiquitin-dependent proteasomal degradation of the c-Fos and Fra-1 transcription factors: is there a unique answer? Biochimie. 2008;90:296–305. doi: 10.1016/j.biochi.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 62.Pellegrino MJ, Stork PJ. Sustained activation of extracellular signal-regulated kinase by nerve growth factor regulates c-fos protein stabilization and transactivation in PC12 cells. J Neurochem. 2006;99:1480–1493. doi: 10.1111/j.1471-4159.2006.04250.x. [DOI] [PubMed] [Google Scholar]
- 63.Tanos T, et al. Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP1 response to UV light. J Biol Chem. 2005;280:18842–18852. doi: 10.1074/jbc.M500620200. [DOI] [PubMed] [Google Scholar]
- 64.Basbous J, Chalbos D, Hipskind R, Jariel-Encontre I, Piechaczyk M. Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Mol Cell Biol. 2007;27:3936–3950. doi: 10.1128/MCB.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malnou CE, et al. Heterodimerization with Jun family members regulates c-Fos nucleocytoplasmic traffic. J Biol Chem. 2007;282:31046–31059. doi: 10.1074/jbc.M702833200. [DOI] [PubMed] [Google Scholar]
- 66.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 67.Bhoumik A, Lopez-Bergami P, Ronai Z. ATF2 on the double — activating transcription factor and DNA damage response protein. Pigment Cell Res. 2007;20:498–506. doi: 10.1111/j.1600-0749.2007.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breitwieser W, et al. Feedback regulation of p38 activity via ATF2 is essential for survival of embryonic liver cells. Genes Dev. 2007;21:2069–2082. doi: 10.1101/gad.430207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takeda J, et al. Expression of the CRE-BP1 transcriptional regulator binding to the cyclic AMP response element in central nervous system, regenerating liver, and human tumors. Oncogene. 1991;6:1009–1014. [PubMed] [Google Scholar]
- 70.Kim HS, Choi ES, Shin JA, Jang YK, Park SD. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem. 2004;279:42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- 71.Bruhat A, et al. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 2007;35:1312–1321. doi: 10.1093/nar/gkm038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agelopoulos M, Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006;25:4843–4853. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maekawa T, et al. Mouse ATF-2 null mutants display features of a severe type of meconium aspiration syndrome. J Biol Chem. 1999;274:17813–17819. doi: 10.1074/jbc.274.25.17813. [DOI] [PubMed] [Google Scholar]
- 74.Papassava P, et al. Overexpression of activating transcription factor-2 is required for tumor growth and progression in mouse skin tumors. Cancer Res. 2004;64:8573–8584. doi: 10.1158/0008-5472.CAN-03-0955. [DOI] [PubMed] [Google Scholar]
- 75.Zoumpourlis V, et al. High levels of phosphorylated c-Jun, Fra-1, Fra-2 and ATF-2 proteins correlate with malignant phenotypes in the multistage mouse skin carcinogenesis model. Oncogene. 2000;19:4011–4021. doi: 10.1038/sj.onc.1203732. [DOI] [PubMed] [Google Scholar]
- 76.Woo IS, Kohno T, Inoue K, Ishii S, Yokota J. Infrequent mutations of the activating transcription factor-2 gene in human lung cancer, neuroblastoma and breast cancer. Int J Oncol. 2002;20:527–531. [PubMed] [Google Scholar]
- 77.Maekawa T, et al. Reduced levels of ATF-2 predispose mice to mammary tumors. Mol Cell Biol. 2007;27:1730–1744. doi: 10.1128/MCB.01579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song H, Ki SH, Kim SG, Moon A. Activating transcription factor 2 mediates matrix metalloproteinase-2 transcriptional activation induced by p38 in breast epithelial cells. Cancer Res. 2006;66:10487–10496. doi: 10.1158/0008-5472.CAN-06-1461. [DOI] [PubMed] [Google Scholar]
- 79.Reimold AM, et al. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature. 1996;379:262–265. doi: 10.1038/379262a0. [DOI] [PubMed] [Google Scholar]
- 80.Ozawa K, Sudo T, Soeda E, Yoshida MC, Ishii S. Assignment of the human CREB2 (CRE-BP1) gene to 2q32. Genomics. 1991;10:1103–1104. doi: 10.1016/0888-7543(91)90210-6. [DOI] [PubMed] [Google Scholar]
- 81.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 82.Liu H, et al. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO J. 2006;25:1058–1069. doi: 10.1038/sj.emboj.7601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li XY, Green MR. Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev. 1996;10:517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- 84.Ouwens DM, et al. Growth factors can activate ATF2 via a two-step mechanism: phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 2002;21:3782–3793. doi: 10.1093/emboj/cdf361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gupta S, Campbell D, Derijard B, Davis RJ. Transcription factor ATF2: regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 86.Firestein R, Feuerstein N. Association of activating transcription factor 2 (ATF2) with the ubiquitin-conjugating enzyme hUBC9. Implication of the ubiquitin/proteasome pathway in regulation of ATF2 in T cells. J Biol Chem. 1998;273:5892–5902. doi: 10.1074/jbc.273.10.5892. [DOI] [PubMed] [Google Scholar]
- 87.Fuchs SY, Ronai Z. Ubiquitination and degradation of ATF2 are dimerization dependent. Mol Cell Biol. 1999;19:3289–3298. doi: 10.1128/mcb.19.5.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fuchs SY, Tappin I, Ronai Z. Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation. J Biol Chem. 2000;275:12560–12564. doi: 10.1074/jbc.275.17.12560. [DOI] [PubMed] [Google Scholar]
- 89.Bhoumik A, et al. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol Cell. 2005;18:577–587. doi: 10.1016/j.molcel.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamasaki T, Takahashi A, Pan J, Yamaguchi N, Yokoyama KK. Phosphorylation of activation transcription factor-2 at serine 121 by protein kinase C controls c-Jun-mediated activation of transcription. J Biol Chem. 2009;284:8567–8581. doi: 10.1074/jbc.M808719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 92.Kerppola TK, Curran T. Selective DNA bending by a variety of bZIP proteins. Mol Cell Biol. 1993;13:5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ronai Z, et al. ATF2 confers radiation resistance to human melanoma cells. Oncogene. 1998;16:523–531. doi: 10.1038/sj.onc.1201566. [DOI] [PubMed] [Google Scholar]
- 94.Ma Q, et al. Activating transcription factor 2 controls Bcl-2 promoter activity in growth plate chondrocytes. J Cell Biochem. 2007;101:477–487. doi: 10.1002/jcb.21198. [DOI] [PubMed] [Google Scholar]
- 95.Zenz R, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 96.Shaulian E, Karin M. AP1 as a regulator of cell life and death. Nature Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 97.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Behrens A, Jochum W, Sibilia M, Wagner EF. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19:2657–2663. doi: 10.1038/sj.onc.1203603. [DOI] [PubMed] [Google Scholar]
- 99.Binetruy B, Smeal T, Karin M. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature. 1991;351:122–127. doi: 10.1038/351122a0. [DOI] [PubMed] [Google Scholar]
- 100.Talotta F, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 101.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 102.Bergers G, Graninger P, Braselmann S, Wrighton C, Busslinger M. Transcriptional activation of the fra-1 gene by AP1 is mediated by regulatory sequences in the first intron. Mol Cell Biol. 1995;15:3748–3758. doi: 10.1128/mcb.15.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jenuwein T, Muller R. Structure-function analysis of fos protein: a single amino acid change activates the immortalizing potential of v-fos. Cell. 1987;48:647–657. doi: 10.1016/0092-8674(87)90243-1. [DOI] [PubMed] [Google Scholar]
- 104.Sunters A, McCluskey J, Grigoriadis AE. Control of cell cycle gene expression in bone development and during c-Fos-induced osteosarcoma formation. Dev Genet. 1998;22:386–397. doi: 10.1002/(SICI)1520-6408(1998)22:4<386::AID-DVG8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 105.Ledwith BJ, Manam S, Kraynak AR, Nichols WW, Bradley MO. Antisense-fos RNA causes partial reversion of the transformed phenotypes induced by the c-Ha-ras oncogene. Mol Cell Biol. 1990;10:1545–1555. doi: 10.1128/mcb.10.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Olive M, et al. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 107.Jochum W, et al. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nature Med. 2000;6:980–984. doi: 10.1038/79676. [DOI] [PubMed] [Google Scholar]
- 108.Mikula M, et al. The proto-oncoprotein c-Fos negatively regulates hepatocellular tumorigenesis. Oncogene. 2003;22:6725–6738. doi: 10.1038/sj.onc.1206781. [DOI] [PubMed] [Google Scholar]
- 109.Graves ML, Zhou L, MacDonald G, Mueller CR, Roskelley CD. Regulation of the BRCA1 promoter in ovarian surface epithelial cells and ovarian carcinoma cells. FEBS Lett. 2007;581:1825–1833. doi: 10.1016/j.febslet.2007.03.072. [DOI] [PubMed] [Google Scholar]
- 110.Abbas S, et al. Preclinical studies of celastrol and acetyl isogambogic acid in melanoma. Clin Cancer Res. 2007;13:6769–6778. doi: 10.1158/1078-0432.CCR-07-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bhoumik A, Jones N, Ronai Z. Transcriptional switch by activating transcription factor 2-derived peptide sensitizes melanoma cells to apoptosis and inhibits their tumorigenicity. Proc Natl Acad Sci USA. 2004;101:4222–4227. doi: 10.1073/pnas.0400195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vale-Cruz DS, Ma Q, Syme J, LuValle PA. Activating transcription factor-2 affects skeletal growth by modulating pRb gene expression. Mech Dev. 2008;125:843–856. doi: 10.1016/j.mod.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 113.Nakamura T, et al. Down-regulation of the cyclin A promoter in differentiating human embryonal carcinoma cells is mediated by depletion of ATF-1 and ATF-2 in the complex at the ATF/CRE site. Exp Cell Res. 1995;216:422–430. doi: 10.1006/excr.1995.1053. [DOI] [PubMed] [Google Scholar]
- 114.Beier F, Taylor AC, LuValle P. Activating transcription factor 2 is necessary for maximal activity and serum induction of the cyclin A promoter in chondrocytes. J Biol Chem. 2000;275:12948–12953. doi: 10.1074/jbc.275.17.12948. [DOI] [PubMed] [Google Scholar]
- 115.Bhoumik A, et al. Suppressor role of activating transcription factor 2 (ATF2) in skin cancer. Proc Natl Acad Sci USA. 2008;105:1674–1679. doi: 10.1073/pnas.0706057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maekawa T, et al. ATF-2 controls transcription of Maspin and GADD45α genes independently from p53 to suppress mammary tumors. Oncogene. 2008;27:1045–1054. doi: 10.1038/sj.onc.1210727. [DOI] [PubMed] [Google Scholar]
- 117.Chen SY, et al. Overexpression of phosphorylated-ATF2 and STAT3 in cutaneous squamous cell carcinoma, Bowen's disease and basal cell carcinoma. J Dermatol Sci. 2008;51:210–215. doi: 10.1016/j.jdermsci.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 118.Knippen S, et al. Expression and prognostic value of activating transcription factor 2 (ATF2) and its phosphorylated form in mammary carcinomas. Anticancer Res. 2009;29:183–189. [PubMed] [Google Scholar]
- 119.Lewis JS, et al. Activation of cyclin D1 by estradiol and spermine in MCF-7 breast cancer cells: a mechanism involving the p38 MAP kinase and phosphorylation of ATF-2. Oncol Res. 2005;15:113–128. doi: 10.3727/096504005776367924. [DOI] [PubMed] [Google Scholar]
- 120.Berger AJ, et al. Subcellular localization of activating transcription factor 2 in melanoma specimens predicts patient survival. Cancer Res. 2003;63:8103–8107. [PubMed] [Google Scholar]
- 121.Deng X, et al. Ionizing radiation induces prostate cancer neuroendocrine differentiation through interplay of CREB and ATF2: implications for disease progression. Cancer Res. 2008;68:9663–9670. doi: 10.1158/0008-5472.CAN-08-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Daury L, et al. Opposing functions of ATF2 and Fos-like transcription factors in c-Jun-mediated myogenin expression and terminal differentiation of avian myoblasts. Oncogene. 2001;20:7998–8008. doi: 10.1038/sj.onc.1204967. [DOI] [PubMed] [Google Scholar]
- 123.Chu M, Guo J, Chen CY. Long-term exposure to nicotine, via ras pathway, induces cyclin D1 to stimulate G1 cell cycle transition. J Biol Chem. 2005;280:6369–6379. doi: 10.1074/jbc.M408947200. [DOI] [PubMed] [Google Scholar]
- 124.Manna SK, et al. Long term environmental tobacco smoke activates nuclear transcription factor-κB, activator protein-1 and stress responsive kinases in mouse brain. Biochem Pharmacol. 2006;71:1602–1609. doi: 10.1016/j.bcp.2006.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heintz NH, Janssen YM, Mossman BT. Persistent induction of c-fos and c-jun expression by asbestos. Proc Natl Acad Sci USA. 1993;90:3299–3303. doi: 10.1073/pnas.90.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Janssen YM, Heintz NH, Marsh JP, Borm PJ, Mossman BT. Induction of c-fos and c-jun proto-oncogenes in target cells of the lung and pleura by carcinogenic fibers. Am J Respir Cell Mol Biol. 1994;11:522–530. doi: 10.1165/ajrcmb.11.5.7946382. [DOI] [PubMed] [Google Scholar]
- 127.Fried U, Kotarsky K, Alling C. Chronic ethanol exposure enhances activating protein-1 transcriptional activity in human neuroblastoma cells. Alcohol. 2001;24:189–195. doi: 10.1016/s0741-8329(01)00151-3. [DOI] [PubMed] [Google Scholar]
- 128.Matthews CP, et al. Dominant-negative activator protein 1 (TAM67) targets cyclooxygenase-2 and osteopontin under conditions in which it specifically inhibits tumorigenesis. Cancer Res. 2007;67:2430–2438. doi: 10.1158/0008-5472.CAN-06-0522. [DOI] [PubMed] [Google Scholar]
- 129.Young MR, et al. Transgenic mice demonstrate AP1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Astruc ME, Chabret C, Bali P, Gagne D, Pons M. Prolonged treatment of breast cancer cells with antiestrogens increases the activating protein-1-mediated response: involvement of the estrogen receptor. Endocrinology. 1995;136:824–832. doi: 10.1210/endo.136.3.7867590. [DOI] [PubMed] [Google Scholar]
- 131.Brozovic A, et al. Long-term activation of SAPK/JNK, p38 kinase and fas-L expression by cisplatin is attenuated in human carcinoma cells that acquired drug resistance. Int J Cancer. 2004;112:974–985. doi: 10.1002/ijc.20522. [DOI] [PubMed] [Google Scholar]
- 132.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 133.Maeno K, et al. Altered regulation of c-jun and its involvement in anchorage-independent growth of human lung cancers. Oncogene. 2005;25:271–277. doi: 10.1038/sj.onc.1209018. [DOI] [PubMed] [Google Scholar]
- 134.Shiratsuchi T, Ishibashi H, Shirasuna K. Inhibition of epidermal growth factor-induced invasion by dexamethasone and AP1 decoy in human squamous cell carcinoma cell lines. J Cell Physiol. 2002;193:340–348. doi: 10.1002/jcp.10181. [DOI] [PubMed] [Google Scholar]
- 135.Katiyar S, Jiao X, Wagner E, Lisanti MP, Pestell RG. Somatic excision demonstrates that c-Jun induces cellular migration and invasion through induction of stem cell factor. Mol Cell Biol. 2007;27:1356–1369. doi: 10.1128/MCB.01061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hommura F, et al. HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. Mol Cancer Res. 2004;2:305–314. [PubMed] [Google Scholar]
- 137.Kinoshita I, et al. Identification of cJun-responsive genes in Rat-1a cells using multiple techniques: Increased expression of stathmin is necessary for cJun-mediated anchorage-independent growth. Oncogene. 2003;22:2710–2722. doi: 10.1038/sj.onc.1206371. [DOI] [PubMed] [Google Scholar]
- 138.Jooss KU, Muller R. Deregulation of genes encoding microfilament-associated proteins during Fos-induced morphological transformation. Oncogene. 1995;10:603–608. [PubMed] [Google Scholar]
- 139.Westermarck J, et al. Activation of fibroblast collagenase-1 expression by tumor cells of squamous cell carcinomas is mediated by p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase-2. Cancer Res. 2000;60:7156–7162. [PubMed] [Google Scholar]
- 140.Reichmann E, et al. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 1992;71:1103–1116. doi: 10.1016/s0092-8674(05)80060-1. [DOI] [PubMed] [Google Scholar]
- 141.Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 2005;24:1434–1444. doi: 10.1038/sj.onc.1208312. [DOI] [PubMed] [Google Scholar]
- 142.Ramos-Nino ME, Scapoli L, Martinelli M, Land S, Mossman BT. Microarray analysis and RNA silencing link fra-1 to cd44 and c-met expression in mesothelioma. Cancer Res. 2003;63:3539–3545. [PubMed] [Google Scholar]
- 143.Chen SY, et al. Concordant overexpression of phosphorylated ATF2 and STAT3 in extramammary Paget's disease. J Cutan Pathol. 2009;36:402–408. doi: 10.1111/j.1600-0560.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 144.Jean D, Bar-Eli M. Regulation of tumor growth and metastasis of human melanoma by the CREB transcription factor family. Mol Cell Biochem. 2000;212:19–28. [PubMed] [Google Scholar]
- 145.Vleugel MM, Greijer AE, Bos R, van der Wall E, van Diest P. J c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer Hum Pathol. 2006;37:668–674. doi: 10.1016/j.humpath.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 146.Zhang G, et al. Effect of deoxyribzymes targeting c-Jun on solid tumor growth and angiogenesis in rodents. J Natl Cancer Inst. 2004;96:683–696. doi: 10.1093/jnci/djh120. [DOI] [PubMed] [Google Scholar]
- 147.Zhang G, et al. Squamous cell carcinoma growth in mice and in culture is regulated by c-Jun and its control of matrix metalloproteinase-2 and -9 expression. Oncogene. 2006;25:7260–7266. doi: 10.1038/sj.onc.1209726. [DOI] [PubMed] [Google Scholar]
- 148.Bowden GT, Schneider B, Domann R, Kulesz-Martin M. Oncogene activation and tumor suppressor gene inactivation during multistage mouse skin carcinogenesis. Cancer Res. 1994;54:1882–1885. [PubMed] [Google Scholar]
- 149.Toft DJ, Rosenberg SB, Bergers G, Volpert O, Linzer DIH. Reactivation of proliferin gene expression is associated with increased angiogenesis in a cell culture model of fibrosarcoma tumor progression. Proc Natl Acad Sci USA. 2001;98:13055–13059. doi: 10.1073/pnas.231364798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ming J, Zhang Q, Qiu X, Wang E. Interleukin 7/interleukin 7 receptor induce c-Fos/c-Jun-dependent vascular endothelial growth factor-D up-regulation: a mechanism of lymphangiogenesis in lung cancer. Eur J Cancer. 2009;45:866–873. doi: 10.1016/j.ejca.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 151.Ham J, et al. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 152.Wang N, et al. c-Jun triggers apoptosis in human vascular endothelial cells. Circ Res. 1999;85:387–393. doi: 10.1161/01.res.85.5.387. [DOI] [PubMed] [Google Scholar]
- 153.Podar K, et al. Up-regulation of c-Jun inhibits proliferation and induces apoptosis via caspase-triggered c-Abl cleavage in human multiple myeloma. Cancer Res. 2007;67:1680–1688. doi: 10.1158/0008-5472.CAN-06-1863. [DOI] [PubMed] [Google Scholar]
- 154.Bossy-Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hettinger K, et al. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2006;14:218–229. doi: 10.1038/sj.cdd.4401946. [DOI] [PubMed] [Google Scholar]
- 156.Eferl R, et al. Liver Tumor development: c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 157.Stepniak E, et al. c-Jun/AP1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20:2306–2314. doi: 10.1101/gad.390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kalra N, Kumar V. c-Fos is a mediator of the c-myc-induced apoptotic signaling in serum-deprived hepatoma cells via the p38 mitogen-activated protein kinase pathway. J Biol Chem. 2004;279:25313–25319. doi: 10.1074/jbc.M400932200. [DOI] [PubMed] [Google Scholar]
- 159.Siegmund D, et al. Fas-associated death domain protein (FADD) and caspase-8 mediate up-regulation of c-Fos by Fas ligand and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) via a FLICE inhibitory protein (FLIP)-regulated pathway. J Biol Chem. 2001;276:32585–32590. doi: 10.1074/jbc.M100444200. [DOI] [PubMed] [Google Scholar]
- 160.Kustikova O, et al. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol Cell Biol. 1998;18:7095–7105. doi: 10.1128/mcb.18.12.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shirsat NV, Shaikh SA. Overexpression of the immediate early gene fra-1 inhibits proliferation, induces apoptosis, and reduces tumourigenicity of c6 glioma cells. Exp Cell Res. 2003;291:91–100. doi: 10.1016/s0014-4827(03)00346-x. [DOI] [PubMed] [Google Scholar]
- 162.Jochum W, Passegue E, Wagner EF. AP1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 163.Leppa S, Eriksson M, Saffrich R, Ansorge W, Bohmann D. Complex functions of AP1 transcription factors in differentiation and survival of PC12 cells. Mol Cell Biol. 2001;21:4369–4378. doi: 10.1128/MCB.21.13.4369-4378.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Behrens A, Sibilia M, Wagner EF. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nature Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 165.Yuan Z, et al. Opposing roles for ATF2 and c-Fos in c-Jun-mediated neuronal apoptosis. Mol Cell Biol. 2009 doi: 10.1128/MCB.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li W, Zhang X, Olumi AF. MG-132 sensitizes TRAIL-resistant prostate cancer cells by activating c-Fos/c-Jun heterodimers and repressing c-FLIPL. Cancer Res. 2007;67:2247–2255. doi: 10.1158/0008-5472.CAN-06-3793. [DOI] [PubMed] [Google Scholar]
- 167.Adunyah SE, Chander R, Barner VK, Cooper RS, Copper RS. Regulation of c-jun mRNA expression by hydroxyurea in human K562 cells during erythroid differentiation. Biochim Biophys Acta. 1995;1263:123–132. doi: 10.1016/0167-4781(95)00079-v. [DOI] [PubMed] [Google Scholar]
- 168.Santaguida M, et al. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15:341–352. doi: 10.1016/j.ccr.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hilberg F, Aguzzi A, Howells N, Wagner EF. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 170.Wo Y, Zhu D, Yu Y, Lou Y. Involvement of NF-κB and AP1 activation in icariin promoted cardiac differentiation of mouse embryonic stem cells. Eur J Pharmacol. 2008;586:59–66. doi: 10.1016/j.ejphar.2008.02.080. [DOI] [PubMed] [Google Scholar]
- 171.Okuda A, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 1998;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rapp UR, Troppmair J, Beck T, Birrer MJ. Transformation by Raf and other oncogenes renders cells differentially sensitive to growth inhibition by a dominant negative c-jun mutant. Oncogene. 1994;9:3493–3498. [PubMed] [Google Scholar]
- 173.Mathas S, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-κB. EMBO J. 2002;21:4104–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Drakos E, et al. c-Jun expression and activation are restricted to CD30+ lymphoproliferative disorders. Am J Surg Pathol. 2007;31:447–453. doi: 10.1097/01.pas.0000213412.25935.e4. [DOI] [PubMed] [Google Scholar]
- 175.Ouyang X, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68:2132–2144. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 176.Mahner S, et al. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br J Cancer. 2008;99:1269–1275. doi: 10.1038/sj.bjc.6604650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jin SP, et al. Prognostic significance of loss of c-fos protein in gastric carcinoma. Pathol Oncol Res. 2007;13:284–289. doi: 10.1007/BF02940306. [DOI] [PubMed] [Google Scholar]
- 178.Young MR, Colburn NH. Fra-1 a target for cancer prevention or intervention. Gene. 2006;379:1–11. doi: 10.1016/j.gene.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 179.Ramirez-Carrozzi V, Kerppola T. Asymmetric recognition of nonconsensus AP1 sites by Fos-Jun and Jun-Jun influences transcriptional cooperativity with NFAT1. Mol Cell Biol. 2003;23:1737–1749. doi: 10.1128/MCB.23.5.1737-1749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Huguier S, Baguet J, Perez S, van Dam H, Castellazzi M. Transcription factor ATF2 cooperates with v-Jun to promote growth factor-independent proliferation in vitro and tumor formation in vivo. Mol Cell Biol. 1998;18:7020–7029. doi: 10.1128/mcb.18.12.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.van Dam H, et al. Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 1998;12:1227–1239. doi: 10.1101/gad.12.8.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sunters A, Thomas DP, Yeudall WA, Grigoriadis AE. Accelerated cell cycle progression in osteoblasts overexpressing the c-fos proto-oncogene: induction of cyclin A and enhanced CDK2 activity. J Biol Chem. 2004;279:9882–9891. doi: 10.1074/jbc.M310184200. [DOI] [PubMed] [Google Scholar]
- 183.Chalmers CJ, Gilley R, March HN, Balmanno K, Cook SJ. The duration of ERK1/2 activity determines the activation of c-Fos and Fra-1 and the composition and quantitative transcriptional output of AP1. Cell Signal. 2007;19:695–704. doi: 10.1016/j.cellsig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 184.De Cesare D, et al. Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 185.Lemaigre FP, Ace CI, Green MR. The cAMP response element binding protein, CREB, is a potent inhibitor of diverse transcriptional activators. Nucleic Acids Res. 1993;21:2907–2911. doi: 10.1093/nar/21.12.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rutberg SE, et al. CRE DNA binding proteins bind to the AP1 target sequence and suppress AP1 transcriptional activity in mouse keratinocytes. Oncogene. 1999;18:1569–1579. doi: 10.1038/sj.onc.1202463. [DOI] [PubMed] [Google Scholar]
- 187.Chiu R, Angel P, Karin M. JunB differs in its biological properties from, and is a negative regulator of c-Jun. Cell. 1989;59:979–986. doi: 10.1016/0092-8674(89)90754-x. [DOI] [PubMed] [Google Scholar]
- 188.Mechta F, Lallemand D, Pfarr CM, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997;14:837–847. doi: 10.1038/sj.onc.1200900. [DOI] [PubMed] [Google Scholar]
- 189.Flint KJ, Jones NC. Differential regulation of three members of the ATF/CREB family of DNA-binding proteins. Oncogene. 1991;6:2019–2026. [PubMed] [Google Scholar]
- 190.Hagmeyer BM, Angel P, van Dam H. Modulation of AP1/ATF transcription factor activity by the adenovirus-E1A oncogene products. Bioessays. 1995;17:621–629. doi: 10.1002/bies.950170708. [DOI] [PubMed] [Google Scholar]
- 191.Pospelova TV, et al. E1A+ cHa-ras transformed rat embryo fibroblast cells are characterized by high and constitutive DNA binding activities of AP1 dimers with significantly altered composition. Gene Expr. 1999;8:19–32. [PMC free article] [PubMed] [Google Scholar]
- 192.Abdel-Hafiz HA, Chen CY, Marcell T, Kroll DJ, Hoeffler JP. Structural determinants outside of the leucine zipper influence the interactions of CREB and ATF-2: interaction of CREB with ATF-2 blocks E1a-ATF-2 complex formation. Oncogene. 1993;8:1161–1174. [PubMed] [Google Scholar]
- 193.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 194.Kara CJ, Liou HC, Ivashkiv LB, Glimcher LH. A cDNA for a human cyclic AMP response element-binding protein which is distinct from CREB and expressed preferentially in brain. Mol Cell Biol. 1990;10:1347–1357. doi: 10.1128/mcb.10.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Georgopoulos K, Morgan BA, Moore DD. Functionally distinct isoforms of the CRE-BP DNA-binding protein mediate activity of a T-cell-specific enhancer. Mol Cell Biol. 1992;12:747–757. doi: 10.1128/mcb.12.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bailey J, Europe-Finner GN. Identification of human myometrial target genes of the c-Jun NH2-terminal kinase (JNK) pathway: the role of activating transcription factor 2 (ATF2) and a novel spliced isoform ATF2-small. J Mol Endocrinol. 2005;34:19–35. doi: 10.1677/jme.1.01608. [DOI] [PubMed] [Google Scholar]
- 197.Ransone LJ, Verma IM. Nuclear proto-oncogenes fos and jun. Annu Rev Cell Biol. 1990;6:539–557. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 198.Wei W, Jin J, Schlisio S, Harper JW, Kaelin JWG. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 199.Huang CC, et al. Calcineurin-mediated dephosphorylation of c-Jun Ser-243 is required for c-Jun protein stability and cell transformation. Oncogene. 2007;27:2422–2429. doi: 10.1038/sj.onc.1210888. [DOI] [PubMed] [Google Scholar]
- 200.Wang YN, Chen YJ, Chang WC. Activation of extracellular signal-regulated kinase signaling by epidermal growth factor mediates c-Jun activation and p300 recruitment in keratin 16 gene expression. Mol Pharmacol. 2006;69:85–98. doi: 10.1124/mol.105.016220. [DOI] [PubMed] [Google Scholar]
- 201.Wertz IE, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 202.Szabo E, Riffe ME, Steinberg SM, Birrer MJ, Linnoila RI. Altered cJun expression: an early event in human lung carcinogenesis. Cancer Res. 1996;56:305–315. [PubMed] [Google Scholar]
- 203.Rangatia J, et al. Elevated c-Jun expression in acute myeloid leukemias inhibits C/EBPα DNA binding via leucine zipper domain interaction. Oncogene. 2003;22:4760–4764. doi: 10.1038/sj.onc.1206664. [DOI] [PubMed] [Google Scholar]
- 204.Zhang W, Hart J, McLeod HL, Wang HL. Differential expression of the AP1 transcription factor family members in human colorectal epithelial and neuroendocrine neoplasms. Am J Clin Path. 2005;124:11–19. doi: 10.1309/T1H2Y2CHWY7PD2BN. [DOI] [PubMed] [Google Scholar]
- 205.Tessari G, et al. The expression of proto-oncogene c-jun in human pancreatic cancer. Anticancer Res. 1999;19:863–867. [PubMed] [Google Scholar]
- 206.Acay RR, Santos Ed, Machado de Sousa SO. Correlation between c-Jun and human papillomavirus in oral premalignant and malignant lesions. Oral Oncology. 2008;44:698–702. doi: 10.1016/j.oraloncology.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 207.Assimakopoulou M, Varakis J. AP1 and heat shock protein 27 expression in human astrocytomas. J Cancer Res Clin Oncol. 2001;127:727–732. doi: 10.1007/s004320100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Papachristou DJ, Batistatou A, Sykiotis GP, Varakis I, Papavassiliou AG. Activation of the JNK-AP1 signal transduction pathway is associated with pathogenesis and progression of human osteosarcomas. Bone. 2003;32:364–371. doi: 10.1016/s8756-3282(03)00026-7. [DOI] [PubMed] [Google Scholar]
- 209.Assimakopoulou M, Kondyli M, Gatzounis G, Maraziotis T, Varakis J. Neurotrophin receptors expression and JNK pathway activation in human astrocytomas. BMC Cancer. 2007;7:202. doi: 10.1186/1471-2407-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Langer S, et al. Jun and Fos family protein expression in human breast cancer: correlation of protein expression and clinicopathological parameters. Eur J Gynaecol Oncol. 2006;27:345–352. [PubMed] [Google Scholar]
- 211.Mao X, Orchard G. Abnormal AP1 protein expression in primary cutaneous B-cell lymphomas. Br J Dermatol. 2008;159:145–151. doi: 10.1111/j.1365-2133.2008.08579.x. [DOI] [PubMed] [Google Scholar]
- 212.Wang H, Birkenbach M, Hart J. Expression of Jun family members in human colorectal adenocarcinoma. Carcinogenesis. 2000;21:1313–1317. [PubMed] [Google Scholar]
- 213.Linardopoulos S, et al. Human lung and bladder carcinoma tumors as compared to their adjacent normal tissue have elevated AP1 activity associated with the retinoblastoma gene promoter. Anticancer Res. 1993;13:257–262. [PubMed] [Google Scholar]
- 214.Bamberger AM, Milde-Langosch K, Rossing E, Goemann C, Loning T. Expression pattern of the AP1 family in endometrial cancer: correlations with cell cycle regulators. J Cancer Res Clin Oncol. 2001;127:545–550. doi: 10.1007/s004320100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Prusty BK, Das BC. Constitutive activation of transcription factor AP1 in cervical cancer and suppression of human papillomavirus (HPV) transcription and AP1 activity in HeLa cells by curcumin. Int J Cancer. 2005;113:951–960. doi: 10.1002/ijc.20668. [DOI] [PubMed] [Google Scholar]
- 216.Cirillo G, et al. Role of distinct mitogen-activated protein kinase pathways and cooperation between Ets-2, ATF-2, and Jun family members in human urokinase-type plasminogen activator gene induction by interleukin-1 and tetradecanoyl phorbol acetate. Mol Cell Biol. 1999;19:6240–6252. doi: 10.1128/mcb.19.9.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lin DW, et al. Transforming growth factor beta up-regulates cysteine-rich protein 2 in vascular smooth muscle cells via activating transcription factor 2. J Biol Chem. 2008;283:15003–15014. doi: 10.1074/jbc.M801621200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Read MA, et al. Tumor necrosis factorα-induced E-selectin expression is activated by the nuclear factor-kappaB and c-JUN N-terminal kinase/p38 mitogen-activated protein kinase pathways. J Biol Chem. 1997;272:2753–2761. doi: 10.1074/jbc.272.5.2753. [DOI] [PubMed] [Google Scholar]
- 219.Herr I, et al. Autoamplification of apoptosis following ligation of CD95-L, TRAIL and TNF-α. Oncogene. 2000;19:4255–4262. doi: 10.1038/sj.onc.1203776. [DOI] [PubMed] [Google Scholar]
- 220.Kool J, et al. Induction of ATF3 by ionizing radiation is mediated via a signaling pathway that includes ATM, Nibrin1, stress-induced MAPkinases and ATF-2. Oncogene. 2003;22:4235–4242. doi: 10.1038/sj.onc.1206611. [DOI] [PubMed] [Google Scholar]
- 221.Bhat NR, Feinstein DL, Shen Q, Bhat AN. p38 MAPK-mediated transcriptional activation of inducible nitric-oxide synthase in glial cells. Roles of nuclear factors, nuclear factor κB, cAMP response element-binding protein, CCAAT/enhancer-binding protein-β, and activating transcription factor-2. J Biol Chem. 2002;277:29584–29592. doi: 10.1074/jbc.M204994200. [DOI] [PubMed] [Google Scholar]
- 222.Penix LA, et al. The proximal regulatory element of the interferon-γ promoter mediates selective expression in T cells. J Biol Chem. 1996;271:31964–31972. doi: 10.1074/jbc.271.50.31964. [DOI] [PubMed] [Google Scholar]
- 223.Chen KD, Hung JJ, Huang HL, Chang MD, Lai YK. Rapid induction of the Grp78 gene by cooperative actions of okadaic acid and heat-shock in 9L rat brain tumor cells — involvement of a cAMP responsive element-like promoter sequence and a protein kinase A signaling pathway. Eur J Biochem. 1997;248:120–129. doi: 10.1111/j.1432-1033.1997.t01-1-00120.x. [DOI] [PubMed] [Google Scholar]
- 224.Suzuki T, Yamakuni T, Hagiwara M, Ichinose H. Identification of ATF-2 as a transcriptional regulator for the tyrosine hydroxylase gene. J Biol Chem. 2002;277:40768–40774. doi: 10.1074/jbc.M206043200. [DOI] [PubMed] [Google Scholar]
- 225.Akimoto T, et al. Exercise stimulates Pgc-1α transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 226.Kawasaki H, et al. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 1998;12:233–45. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang JY, et al. The JNK/AP1/ATF2 pathway is involved in H2O2-induced acetylcholinesterase expression during apoptosis. Cell Mol Life Sci. 2008;65:1435–1445. doi: 10.1007/s00018-008-8047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.