Abstract
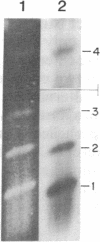
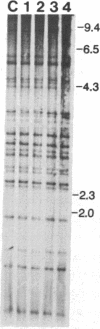
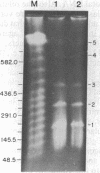
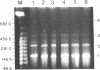
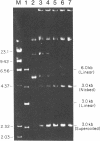
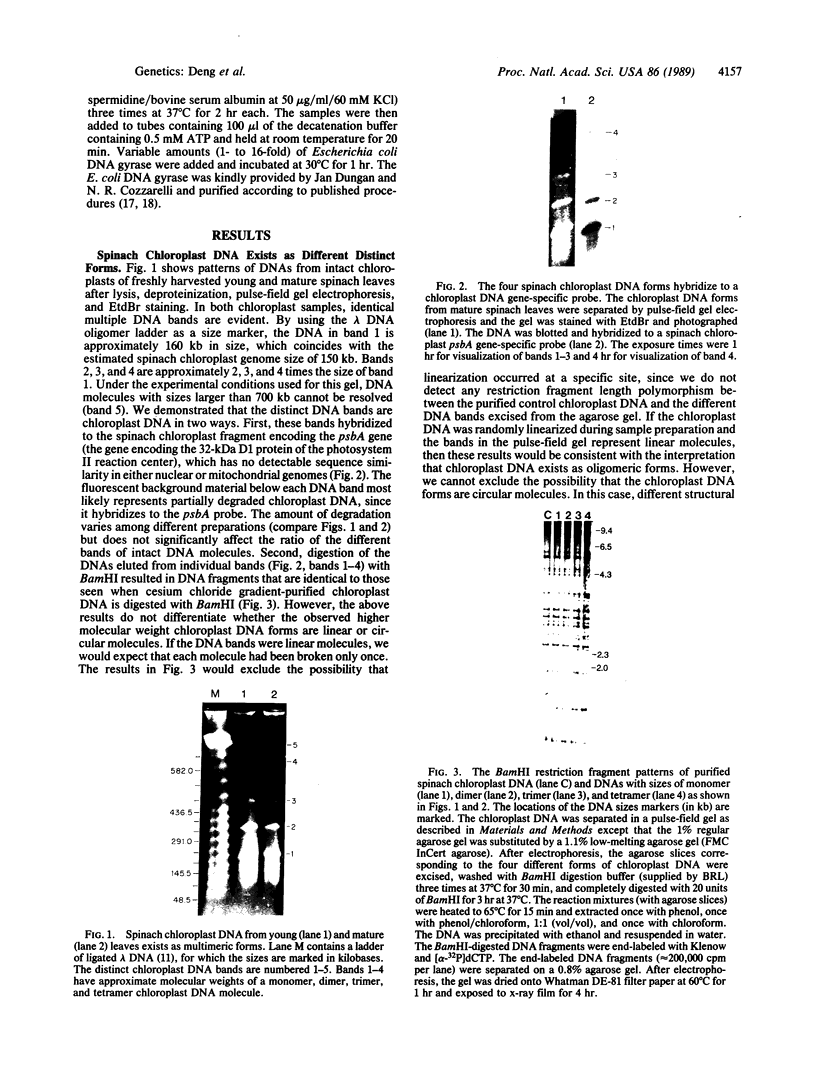
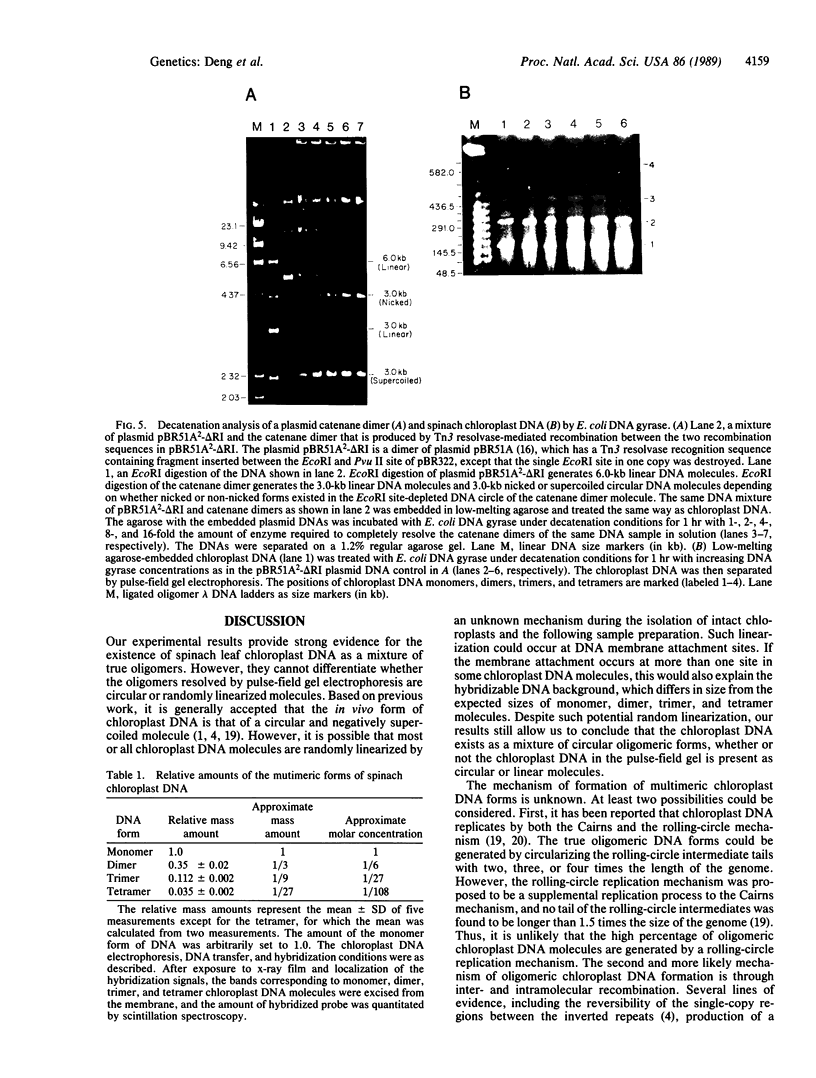
Chloroplast DNA conformation was analyzed by pulse-field gel electrophoresis. We found that spinach leaf chloroplast DNA molecules exist in at least four distinct forms with the apparent molecular weights of monomer, dimer, trimer, and tetramer. Two-dimensional gel analysis of DNA after UV nicking and in the presence of ethidium bromide indicates that they are not isomers that differ in superhelical density. DNA gyrase decatenation analysis demonstrates that the majority of the DNA molecules are oligomers rather than catenanes. The relative amounts of monomer, dimer, trimer, and tetramer forms, quantitated by molecular hybridization, are 1, 1/3, 1/9, and 1/27, respectively, and do not change during leaf maturation. The possible mechanisms of chloroplast DNA oligomer formation are discussed.
Keywords: spinach, chloroplast DNA, in vivo DNA forms, DNA oligomers, leaf maturation
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988 Jun 10;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987 May 8;49(3):379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Kolodner R. D., Tewari K. K. Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature. 1975 Aug 28;256(5520):708–711. doi: 10.1038/256708a0. [DOI] [PubMed] [Google Scholar]
- Kolodner R. D., Tewari K. K. Denaturation mapping studies on the circular chloroplast deoxyribonucleic acid from pea leaves. J Biol Chem. 1975 Jul 10;250(13):4888–4895. [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Inverted repeats in chloroplast DNA from higher plants. Proc Natl Acad Sci U S A. 1979 Jan;76(1):41–45. doi: 10.1073/pnas.76.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. The molecular size and conformation of the chloroplast DNA from higher plants. Biochim Biophys Acta. 1975 Sep 1;402(3):372–390. doi: 10.1016/0005-2787(75)90273-7. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980 May;20(1):245–254. doi: 10.1016/0092-8674(80)90252-4. [DOI] [PubMed] [Google Scholar]
- Medgyesy P., Fejes E., Maliga P. Interspecific chloroplast recombination in a Nicotiana somatic hybrid. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6960–6964. doi: 10.1073/pnas.82.20.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter R., Cozzarelli N. R. Escherichia coli DNA gyrase. Methods Enzymol. 1983;100:171–180. doi: 10.1016/0076-6879(83)00053-1. [DOI] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Vollrath D., Davis R. W. Resolution of DNA molecules greater than 5 megabases by contour-clamped homogeneous electric fields. Nucleic Acids Res. 1987 Oct 12;15(19):7865–7876. doi: 10.1093/nar/15.19.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Bohnert H. J., Whitfeld P. R., Bottomley W. Nucleotide sequence of the gene for the M(r) 32,000 thylakoid membrane protein from Spinacia oleracea and Nicotiana debneyi predicts a totally conserved primary translation product of M(r) 38,950. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7699–7703. doi: 10.1073/pnas.79.24.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]