Abstract
The antimicrobial activity of chitosan and chitosan derivatives has been well established. However, although several mechanisms have been proposed, the exact mode of action is still unclear. Here we report on the investigation of antibacterial activity and the antibacterial mode of action of a novel water-soluble chitosan derivative, arginine-functionalized chitosan, on the gram-negative bacteria Pseudomonas fluorescens and Escherichia coli. Two different arginine-functionalized chitosans (6% arginine-substituted and 30% arginine-substituted) each strongly inhibited P. fluorescens and E. coli growth. Time-dependent killing efficacy experiments showed that 5000 mg L-1 of 6% substituted and 30% substituted chitosan-arginine killed 2.7 logs and 4.5 logs of P. fluorescens, and 4.8 logs and 4.6 logs of E. coli in 4 h, respectively. At low concentrations, the 6% substituted chitosan-arginine was more effective in inhibiting cell growth even though the 30% substituted chitosan-arginine appeared to be more effective in permeabilizing the cell membranes of both P. fluorescens and E. coli. Studies using fluorescent probes, 1-N-phenylnaphthylamine (NPN), nile red (NR) and propidium iodide (PI), and field emission scanning electron microscopy (FESEM) suggest that chitosan-arginine's antibacterial activity is, at least in part, due to its interaction with the cell membrane, in which it increases membrane permeability.
Keywords: chitosan derivative, antibacterial action, fluorescent probe, P. fluorescens, E. coli
1. Introduction
Chitosan is a carbohydrate biopolymer derived from deacetylation of chitin, the main component of crustacean (e.g., shrimp, crab, lobster) exoskeletons. Chitin's abundance is second only to cellulose among polysaccharides found on Earth [1]. Chitosan is medically important due to its biological properties, such as antimicrobial activity, haemostatic activity, anti-tumor activity, acceleration of wound healing, tissue-engineering scaffolds, and promise for drug delivery [2]. It is also biodegradable and biocompatible, with low toxicity to mammalian cells [1]. Bacteria are not known to develop chitosan resistance [3].
Chitosan's antimicrobial activity has been well documented. It displays a broad spectrum of antibacterial activity against both gram-positive and gram-negative bacteria, with minimum inhibitory concentrations (MICs) reported to range from 100 to 10 000 mg L-1 against gram-negative bacteria [4], and from 100 to 1 250 mg L-1 against gram-positive bacteria [5-8].
Chitosan's antimicrobial activities are thought to be affected by chemical, physical and biological factors that include chitosan concentration, molecular weight, degree of deacetylation, pH, temperature, salinity, divalent cations, chitosan solvent, suspending medium, and bacterial growth phase [1,8-20]. Because chitosan and its derivatives have been tested under widely varied conditions, it is hard to compare chitosan's antibacterial effect among results obtained by different researchers.
The exact mode of action of chitosan is still not fully understood, although several mechanisms have been proposed for its antimicrobial activity [1,21]. The key feature of chitosan is thought to be its positive charge of the amino groups (−NH3+) at the C-2 positions in the glucose monomer when the pH is lower than its pKa (∼ 6.3). This forms a polycationic structure that can interact with the anionic compounds and macromolecular structures of bacteria [4,22]. This charge interaction can alter bacterial surface morphology, which either increases membrane permeability causing leakage of intracellular substances (e.g., proteins including lactate dehydrogenase, nucleic acids and glucose), or decreases membrane permeability preventing nutrient transport [14,15,17,23, 24]. The bulk of evidence supports increased membrane permeability and disruption of cell membranes [3,4,12,14,15,24]. It has also been postulated that positively charged chitosan binds with cellular DNA following chitosan penetration into the cells, thereby inhibiting transcription [1,17].
Researchers have applied multiple techniques to investigate chitosan's antibacterial mode of action, including outer membrane (OM) permeability assays (1-N-phenylnaphthylamine (NPN) uptake, SDS-promoted cell lysis), an inner membrane (IM) permeability assay (β-galactosidase activity), lipopolysaccharide (LPS) release, intracellular constituents leakage (OD260nm, OD280nm, SDS-PAGE), scanning electron microscopy, transmission electron microscopy, and atomic force microscopy [3,4,12,14,15,23-31].
The use of chitosan is limited because of its insolubility at neutral pH. Therefore, much effort has been made to prepare functionalized chitosan derivatives that are soluble in water at physiological pH. Chemically modified chitosans include a maltose-chitosan derivative [19] and the proprietary arginine-functionalized chitosan that we tested in this study [32]. Arginine-functionalized chitosan is a chitosan derivative modified with arginine groups to different degrees of substitution. It is highly soluble in water owing to the high pKa of the guanidinium side chain of arginine (pKa = 12.48), rendering it positively charged in neutral pH environments.
Here we report on the antibacterial activity and the antibacterial mode of action of arginine-functionalized chitosan on model gram-negative bacteria, Pseudomonas fluorescens and Escherichia coli. Fluorescence spectroscopy and electron microscopy were used to evaluate cellular effects in the target bacteria. Three fluorescence probes, 1-N-phenylnaphthylamine (NPN), nile red (NR), and propidium iodide (PI), were used in the study. NPN is a hydrophobic probe widely used to assess cell membrane permeability. NR stains, and is sensitive to, neutral lipids in cells. PI, a DNA intercalator, is used to indicate cell death. The fluorescence spectra of these probe molecules are sensitive to their surroundings, reflecting the slight change of their excited state under different environment, as detailed in the discussion section.
2. Materials and methods
2.1 Chemicals and bacteria
Proprietary arginine-functionalized chitosans (6% arginine-substituted and 30% arginine -substituted) were provided by BioStar West, Claremont CA. Trypticase Soy Broth (TSB) (Difco™, BD Company), Luria-Bertani Broth (LB) (Difco™, BD Company), Cation-Adjusted Muller Hinton Broth (MH II broth) (BBL™, BD Company), and 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) were purchased from Fisher Scientific; NPN, NR, PI, tannic acid, and poly-L-lysine from Sigma; ethanol, glutaraldehyde, paraformaldehyde, osmium tetroxide, and hexamethyldisilazane (HMDS) from Electron Microscopy Science; PBS buffer from Invitrogen. Both bacteria were from ATCC.
2.2 Bacteria overnight culture growth
E. coli (ATCC # 25922) and P. fluorescens (ATCC # 700830) were routinely cultivated in LB or TSB at 37 °C with shaking overnight.
2.3 Evaluation of chitosan-arginine's antibacterial activity
2.3.1 Bacterial growth inhibition
Overnight cultures of E. coli and P. fluorescens were inoculated into fresh LB to ∼ 106 cells ml-1 and were mixed with a series of concentrations of chitosan-arginine (0 mg L-1 – 512 mg L-1). Optical density (OD) at 595 nm was monitored at 37°C using a Thermomax Microplate Reader for 24 h. The value of OD595nm reported is the average of triplicate samples. Statistical analysis was performed for data points collected at 24 h by two-way ANOVAs using Minitab-15 software.
2.3.2 MICs and minimum bactericidal concentrations (MBCs) determination
MICs were determined by microtiter broth dilution method, following the guidelines in the literature [33]. Briefly, inocula of P. fluorescens and E. coli were prepared by adjusting overnight culture to containing 2×105 cells ml-1 in MH II broth. Aliquots of 50 μl inoculum were mixed with 50 μl of serial two-fold dilutions of 6%-substituted and 30%-substituted chitosan-arginine in MH II broth in a 96-well plate. The plate was incubated with shaking at 37°C for 18 h. MIC was defined as the lowest concentration of chitosan-arginine where no growth was observed by microscopic examination. After 18 h incubation, 10 μl mixtures from wells with no growth were spread on agar plates for MBC determination. MBC was defined as the lowest concentration of chitosan-arginine where no colony growth was observed on agar plates after 48 h incubation at 37°C. The MIC/MBC determinations were carried out in triplicates, with two independent experiments performed.
2.3.3 Time-dependent killing efficacy
Briefly, overnight cultures of P. fluorescens and E. coli were adjusted in MH II broth to contain 107 cells ml-1 and mixed with 5000 mg L-1 of 6% substituted and 30% substituted chitosan-arginine. The mixtures were incubated at 37°C with shaking and aliquots were withdrawn to perform colony count every 30 min for 4 h.
2.4 Fluorescent probe-permeability assays
Overnight cultures of E. coli and P. fluorescens were centrifuged at 1000× g for 10 min. The supernatant was discarded. Bacterial pellets were re-suspended in 5 mM HEPES (pH 5.3 ± 0.1) to OD600nm ∼ 0.2 for fluorescence probe assays. All assays were performed at room temperature.
All fluorescence measurements were done on a spectrofluorometer (Photon Technology International) with a xenon lamp as the excitation source. The slit widths were set to 4 nm for both the excitation and the emission monochromators.
For each assay with respective probes, 3 ml of bacterial suspension was first mixed with either NPN (1.1 mg/ml in acetone), NR (1 mg/ml in methanol) or PI (2.5 mg/ml in water) to a final probe concentrations of 2.2 μg/ml for NPN, 10 μg/ml for NR, and 17 μg/ml for PI. Fluorescence measurements were then taken. Next, nanopure water (for controls) or chitosan-arginine (final concentration of 50 mg L-1) was added to the mixture. The mixture was thoroughly stirred before fluorescence measurements were taken again. There was typically a 3-minute lapse between the addition of chitosan-arginine and the following fluorescence measurement for NPN and NR. Fluorescence intensity of both excitation and emission peaks was monitored over time. For PI, fluorescence intensity was taken every ten minutes for up to 2 h, or hourly for up to 8 h and at 24 h.
2.5 Electron Microscopy
Overnight E. coli and P. fluorescens cultures were washed once with 0.85% NaCl solution, then re-suspended in 5 mM HEPES (pH 5.3 ± 0.1) to OD600nm ∼ 0.4. After 3-h incubation with or without 100 mg L-1 chitosan-arginine at 37°C, E. coli and P. fluorescens cells were washed once with PBS and fixed (2% paraformaldehyde and 1% glutaraldehyde in pH 7.3 ± 0.1 PBS). Cells were then spread onto glass cover-slips pre-treated with poly-L-lysine, post-fixed in 1% osmium tetroxide, post-stained in 1% tannic acid, dehydrated in a series of increasing ethanol concentrations (70% – 100%), and air-dried in HMDS. Samples were sputter-coated with platinum using a K950X EmiTech coater and imaged at 80 kV using an S-800 Hitachi field emission scanning electron microscope.
3. Results
3.1 Evaluation of antibacterial activity
Chitosan-arginine affected growth of P. fluorescens (Figure 1A). Statistical analysis using two-way ANOVA showed the effects of concentration (p = 0.057), degree of arginine substitution (6% vs. 30%, p = 0.019) and interactive effects (p < 0.001). 6%- and 30%- substituted chitosan-arginine showed concentration-dependent antibacterial effects on P. fluorescens (Fig. 1A). At 24 h, 6%-substituted chitosan-arginine significantly inhibited P. fluorescens growth at concentrations ≥ 64 mg L-1. 30%-substituted chitosan-arginine inhibited P. fluorescens growth at concentrations ≥ 128 mg L-1. Chitosan-arginine also inhibited E. coli growth (Figure 1B); both the concentration and the degree of arginine substitution showed significant effects (p = 0.005 and p = 0.005, respectively) as well as interactive effects (p < 0.001). At 24 h, both 6% and 30%-substituted chitosan-arginine significantly inhibited E. coli growth at concentrations ≥ 32 mg L-1. The significant interactions indicate that the effects of concentration differ depending on the degree of arginine substitution on the chitosan.
Fig 1.
Fig 1 (A-B) – The growth of P. fluorescens (A) and E. coli (B) in the presence of a series of concentrations of 6%-substituted and 30%-substituted chitosan-arginine, after 24 h incubation.
MICs and MBCs of 6% substituted and 30% substituted chitosan-arginine against P. fluorescens and E. coli were 2.5 mg mL-1, 5.0 mg mL-1, or higher (Table 1). The results of time-dependent killing efficacy experiments showed that 5000 mg L-1 of 6% substituted and 30% substituted chitosan-arginine killed 2.7 logs and 4.5 logs of P. fluorescens, and 4.8 logs and 4.6 logs of E. coli in 4 h, respectively (Figure 2).
Table 1.
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of 6%-substituted and 30%-substituted chitosan-arginine against P. fluorescens and E. coli.
| Bacteria | MIC (mg mL-1) | MBC (mg mL-1) | ||
|---|---|---|---|---|
| 6%-substituted | 30%-substituted | 6%-substituted | 30%-substituted | |
| P. fluorescens ATCC 700830 | 2.5 | > 5.0 | 2.5 | > 5.0 |
| E. coli ATCC 25922 | 5.0 | 2.5 | > 5.0 | 2.5 |
Fig 2.
Time-dependent killing efficacy of 5000 mg L-1 of 6% substituted and 30% substituted chitosan-arginine against P. fluorescens (A) and E. coli (B). Each data point is the average of two independent experiments and bar is the standard deviation.
3.2 Fluorescent probe-permeability
3.2.1 Effect of chitosan-arginine on probe fluorescence
For every fluorescent probe, control experiments were carried out by adding chitosan-arginine into HEPES solution containing only the probe molecules, without the presence of bacteria. No change was observed for either the fluorescence spectra or the fluorescence intensity of the respective probes (data not shown).
3.2.2 NPN
The change of NPN fluorescence upon the addition of 6%- or 30%- substituted chitosan-arginine is shown in Figure 3. The intensity increased in P. fluorescens within one minute and in E. coli within a few minutes. Fluorescence intensity of NPN in P. fluorescens and E. coli remained stable in the absence of 6%- or 30%- substituted chitosan-arginine (Fig. 3).
Fig 3.
Fig 3 (A-B) – The uptake of NPN probe by P. fluorescens (A) and E. coli (B) over time, with the addition of 50 mg L-1 6%- and 30%- substituted chitosan-arginine.
3.2.3 Nile Red
Figure 4 shows the excitation/emission (Ex/Em) spectra of NR, before and after the addition of 6%- and 30%- substituted chitosan-arginine, in P. fluorescens and E. coli. The Ex/Em wavelengths were 584 nm/654 nm in P. fluorescens and E. coli. The addition of 6%- or 30%- substituted chitosan-arginine to P. fluorescens quickly shifted NR's Ex/Em to 548 nm/638 nm (Fig. 4A). On the other hand, the addition of 6%- and 30%- substituted chitosan-arginine to E. coli shifted NR's Ex/Em wavelengths gradually over time, to the final positions of 542 nm / 618∼624 nm shown in Fig. 4B.
Fig 4.
Fig 4 (A-D) – The fluorescence spectra of (Nile Red) NR before and after the addition of 50 mg L-1 6%- substituted chitosan-arginine to P. fluorescens (A) or E. coli (B). The uptake of Nile Red (NR) probe by P. fluorescens (C) or E. coli (D) over time with the addition of 50 mg L-1 6%- and 30%- substituted chitosan-arginine.
NR's fluorescence intensity in both P. fluorescens and E. coli increased after the addition of 6%- and 30%- substituted chitosan-arginine (Fig. 4C and 4D). The rate of NR band shifting in E. coli (data not shown) and intensity increase in P. fluorescens and E. coli was greater in the case of 30%-substituted chitosan-arginine than that of 6%-substituted chitosan-arginine. Fluorescence intensity of NR in P. fluorescens and E. coli remained stable in the absence of 6%- or 30%- substituted chitosan-arginine (Fig. 4C and 4D).
3.2.4 PI
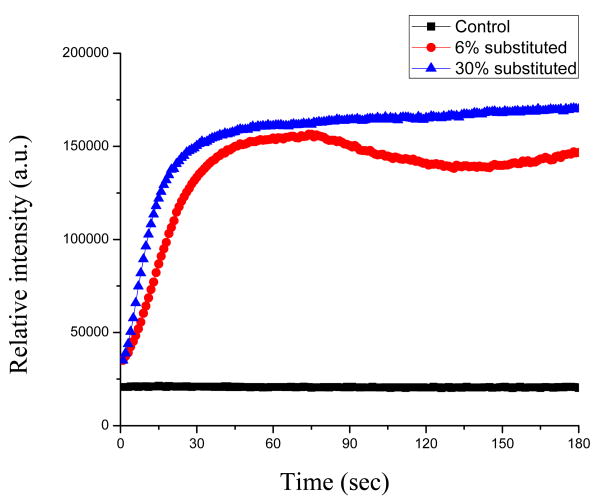
The uptake of PI probe over time after the addition of 6%- or 30%- substituted chitosan-arginine by P. fluorescens and E. coli, as indicated by the increase of PI's fluorescence intensity, is shown in Figure 5 and Figure 6, respectively. The increase in fluorescence intensity of PI occurred almost immediately upon 6%- or 30%- substituted chitosan-arginine addition to P. fluorescens (Fig. 5). The fluorescence intensity increased dramatically within one minute for 6%-substituted chitosan-arginine, and even sooner for 30%-substituted chitosan-arginine. Thereafter, the fluorescence intensity approached a plateau and remained stable.
Fig 5.
The uptake of PI probe by P. fluorescens with the addition of 50 mg L-1 6%- and 30%- substituted chitosan-arginine.
Fig 6.
Fig 6 (A-C) – The fluorescence spectra of PI before and after the addition of 50 mg L-1 6%- substituted chitosan-arginine to E. coli (A). The uptake of PI probe by E. coli in a period of 2 h (B) and up to 24 h (C) with the addition of 50 mg L-1 6%- and 30%- substituted chitosan-arginine.
The addition of 6%- or 30%- substituted chitosan-arginine to E. coli shifted excitation and emission bands as well as increased PI intensity over time (Fig. 6). PI's Ex/Em wavelengths were 466 nm/626 nm in the presence of E. coli alone. The addition of chitosan-arginine shifted PI's Ex/Em to 524 nm/612 nm, i.e., excitation maximum shifted to red and emission maximum shifted to blue (Fig. 6A). The fluorescence intensity increased most dramatically within the first 2-3 h after chitosan-arginine addition, and eventually approached a maximum. The fluorescence intensity increase was faster in the case of 30%-substituted chitosan-arginine than with 6%-substituted chitosan-arginine (Fig. 6B and 6C). Fluorescence intensity of PI in P. fluorescens (Fig. 5) and E. coli (Fig. 6B and 6C) remained stable in the absence of 6%- or 30%- substituted chitosan-arginine.
3.3 SEM
Figure 7 shows SEM images of E. coli cells treated with 100 mg L-1 6%-substituted and 30%-substituted chitosan-arginine under both high magnification (Fig. 7A–7C) and low magnification (Fig. 7D–7F). After 3 h treatment by either 6%-substituted or 30%-substituted chitosan-arginine, E. coli cells tended to aggregate (Fig. 7E and 7F) and were found to be covered by an amorphous substance (Fig. 7B and 7C). No obviously damaged or lysed cells were observed in either 6%- or 30%- substituted chitosan-arginine-treated samples. Figure 8 shows analogous SEM images of P. fluorescens cells treated with either 6%- or 30%- substituted chitosan-arginine under both high magnification (Fig. 8A–8C) and low magnification (Fig. 8D–8F). After 3 h incubation, P. fluorescens cells appeared to be covered by an amorphous substance (Fig. 8B and 8C). Again, no obviously damaged or lysed cells were observed in either 6%- or 30%-substituted chitosan-arginine-treated samples.
Fig 7.

Fig 7 (A-F) – SEM of E. coli after incubation with 100 mg L-1 chitosan-arginine for 3 h. Controls (A and D), cells treated with 6%-substituted chitosan-arginine (B and E), and cells treated with 30%-substituted chitosan-arginine (C and F).
Fig 8.

Fig 8 (A-F) – SEM of P. fluorescens after incubation with 100 mg L-1 chitosan-arginine for 3 h. Controls (A and D), cells treated with 6%-substituted chitosan-arginine (B and E), and cells treated with 30%-substituted chitosan-arginine (C and F).
4. Discussion
Chitosan was thought to have antibacterial activities only at acidic pH because of its poor solubility at pH > 6.5. Some chitosan derivatives' antibacterial activities were proven to be stronger at acidic pH than those at neutral pH [21]; in fact, Sudarshan et al. reported that chitosan had no antibacterial activity at pH 7 [31]. Our results using arginine functionalized chitosan, soluble at neutral pH, indicated that both 6% -and 30%-substituted chitosan-arginine significantly inhibited P. fluorescens and E. coli growth in 24 hours at concentrations ≥ 128 mg L-1 for P. fluorescens and ≥ 32 mg L-1 for E. coli under neutral pH (Fig. 1). In addition, 5000 mg L-1 of 6% substituted and 30% substituted chitosan-arginine killed 2.7 logs and 4.5 logs of P. fluorescens, and 4.8 logs and 4.6 logs of E. coli in 4 h, respectively (Fig. 2). Because the bacterial strains and the experimental conditions in this study are different from those in the literature, it is difficult to directly compare chitosan-arginine's antibacterial effect with results obtained by other researchers for other chitosan derivatives.
The gram-negative bacterial outer membrane (OM) contains polyanionic lipopolysaccharide stabilized by divalent cations, such as Mg2+ and Ca2+ [34]. The OM serves as an effective permeability barrier to restrict macromolecules and hydrophobic substances from entering or leaving bacterial cells [35]. The cation-binding sites of LPS are critical to OM integrity. However, cationic substances, known as membrane permeabilizers, e.g., polymyxin and aminoglycosides, can compete with divalent cations to bind with LPS, thereby disorganizing the OM structure [34].
Chitosan is a random copolymer of glucosamine and acetyl glucosamine. Arginine functionalized chitosans used in this study are obtained by the addition of a single arginine residue via formation of a stable peptide bond of the arginine carboxylic acid with the amine on the glucosamine. The reported % functionalization is reflected as the % of free amines on the molecule that have been functionalized with a single arginine. The resulting cationic polymer is soluble and polycationic at neutral pH, due to the higher pKa (12.48) of the guanidinium side chain relative to that of the amine on the chitosan. In comparison, unmodified chitosan is positively charged only at acidic pH. It may therefore be reasonable to propose that chitosan-arginine has a similar antibacterial mode of action to that of other cationic membrane permeabilizers such as polyethyleneimine (PEI) [36]. NPN is a hydrophobic fluorescence probe widely used to assess cell membrane permeability since its quantum yield increases greatly in hydrophobic environments compared to aqueous environments [37]. Normally, NPN is excluded from the intact bacterial cell membrane lipid bilayer by the OM barrier. When the OM structure is damaged, however, NPN can partition into the hydrophobic interior of the OM or plasma membrane, leading to a dramatic increase of its fluorescence. Therefore, the increase of NPN fluorescence intensity can be used as an indicator for increased cell membrane permeability.
The reported wavelengths for using NPN uptake to assess membrane permeabilization of chitosan range from 340 nm to 355 nm for excitation, and from 405 nm to 430 nm for emission [4,15,23,38]. In our experiments, NPN's Ex/Em wavelength maxima were 340 nm/462 nm in HEPES and shifted to 348/406 nm in P. fluorescens and E. coli. This is very close to the report by Loh et al., which found a shift of Ex/Em wavelengths from 340 nm/460 nm to 350nm/420nm in P. aeruginosa after the addition of gentamicin, a polycationic aminoglycoside antibiotic that is well known to increase membrane permeability [37].
The addition of 6%- or 30%- substituted chitosan-arginine increased NPN fluorescence intensity in P. fluorescens almost immediately and that in E. coli in a few minutes (Fig. 3), suggesting that chitosan-arginine increases the permeability of the outer membranes of these model gram-negative bacteria very effectively. Similar observations have been reported by other researchers for chitosan-treated E. coli,P. aeruginosa, and Salmonella typhimurium [4,15,23]. However, as Helander et al. pointed out, chitosan-induced NPN uptake with unmodified chitosan was effective only at acidic pH [4]. In contrast, chitosan-arginine induces NPN uptake at neutral pH and it is likely that NPN uptake occurs via a similar mechanism upon exposure to either modified or unmodified cationic chitosan polymers. The foremost functional consequence and advantage of an arginine-modified chitosan is the novel property of having a poly-cationic polymer at neutral, physiological pH.
Nile red is a hydrophobic probe typically used to localize and quantify neutral lipid droplets in cells. It is almost non-fluorescent in polar environments such as water, whereas in nonpolar environments, such a lipid droplets or bilyers, its fluorescence intensity is greatly enhanced and its excitation and emission wavelengths display a significant blue shift [39]. NR's Ex/Em wavelengths were 584 nm/654 nm in P. fluorescens and E. coli. The addition of 6%- and 30%- substituted chitosan-arginine to P. fluorescens and E. coli resulted in a blue shift of NR's Ex/Em wavelengths, and a dramatic increase of NR's fluorescence intensity (Fig. 4). These shifts suggested that the environment of NR becomes less polar after the addition of chitosan-arginine. Again, this can be explained by proposing that the NR's diffusion through the permeabilized cell outer membrane is increased by chitosan-arginine.
The rate of NR fluorescence intensity increase in P. fluorescens and E. coli, and the Ex/Em band shifting in E. coli, with the addition of 6%- and 30%- substituted chitosan-arginine, was greater upon addition of the 30%- than for 6%- substituted chitosan-arginine, suggesting that 30%- substituted chitosan-arginine was more effective as a membrane permeabilizer.
PI is a nucleic acid probe indicating cell wall permeability often associated with cell death. Once bound with nucleic acids, PI's quantum yield increases 20-30-fold. Moreover, its excitation peak red shifts 30-40 nm, while its emission peak blue shifts ∼15 nm [39]. We observed that PI's Ex/Em peaks (466 nm/626 nm) in E. coli only were similar to those in buffer solution. As expected, the addition of chitosan-arginine to E. coli shifted PI's Ex/Em to 524 nm/612 nm (Fig. 6A). This indicated that PI bound to nucleic acids only after the addition of chitosan-arginine.
On the other hand, PI's initial Ex/Em peaks in P. fluorescens were already at 524 nm/612 nm (data not shown). The addition of chitosan-arginine did not further shift PI's Ex/Em bands. This could be because a few PI molecules already bound to nucleic acids in P. fluorescens even before the chitosan-arginine was added. Since bound PI molecules have a much higher quantum yield, their spectral behavior likely dominated the spectrum. Nevertheless, the fluorescence intensity of PI increased dramatically and almost immediately after the chitosan-arginine addition (Fig. 5), suggesting an increased presence of PI molecules in P. fluorescens cells and binding to DNA shortly after chitosan-arginine addition.
The addition of 6%- or 30%- substituted chitosan-arginine to E. coli led to a slow increase of PI fluorescence intensity over time (Fig. 6B and 6C), which may indicate that membrane permeability changed gradually over a rather long period of several hours. The fluorescence intensity increased most significantly 2-3 h after chitosan-arginine addition, eventually approaching a plateau. The fluorescence intensity increase was more pronounced and faster for 30%-substituted chitosan-arginine than for the 6% substituted chitosan-arginine, again suggesting that the 30%-substituted chitosan-arginine was more effective as a membrane permeabilizer.
Although 6%-substituted chitosan-arginine was more effective than the 30%-substituted chitosan-arginine in inhibiting bacterial growth (Fig. 1), at low concentrations 30%-substituted chitosan-arginine appeared to be more effective than 6%-substituted chitosan-arginine in permeabilizing the cell membranes of both P. fluorescens and E. coli to NR and PI. This can be understood if one considers that the initial site of action of chitosan-arginine is the bacterial outer membrane. Since 30%-substituted chitosan-arginine is less viscous than 6%-substituted chitosan-arginine, it may contact the outer membrane and permeabilize it more quickly. After chitosan permeabilized the bacterial membranes, it is likely that it was the consequences of this permeabilization as well as the additional intracellular effects of chitosan-arginine that led to growth inhibition.
We observed cell aggregation for both E. coli and P. fluorescens immediately after addition of chitosan-arginine in the SEM images (data not shown), and for E. coli after the 3 h treatment with chitosan-arginine at low magnification (1 000×) (Fig. 7E and 7F). This was similar to Didenko's report on cell aggregation of Klebsiella and Staphylococcus after incubation with chitosan for 24 h [3]. The aggregation of bacterial cells under the influence of chitosan (in our case, chitosan-arginine) might indeed have contributed to growth inhibition. Under the high magnification (50 000× for E. coli and 30 000× for P. fluorescens) of SEM, it was noted that E. coli and P. fluorescens cells incubated with chitosan-arginine for 3 h became covered by an amorphous substance (Fig. 7B-7C and 8B-8C), similar to that of chitosan-treated algae reported previously [40].
Our SEM observations were consistent with some previous reports on chitosan experiments. Helander observed that, under TEM, after 1 h treatment with chitosan at room temperature, E. coli cell surfaces were covered with an additional layer of a vesicular structure which made the cell envelope appear thickened. Interestingly, the plasma membrane was not affected [4]. Chung also showed the adsorption of chitosan on E. coli cell surface after 4 h in their TEM images [25]. Kim noticed that there was no significant morphological change in chitosan derivative-treated E. coli cells [27]. We thus conclude that E. coli cells remained unlysed after the chitosan-arginine treatment. On the other hand, some researchers have reported much stronger effects of chitosan on bacterial cells in their TEM or SEM studies than we observed with chitosan-arginine. Didenko showed that chitosan disorganized both the cell wall and plasma membrane of Klebsiella and Staphylococcus [3]. Kumar et al. demonstrated pore formation in chitosan-treated Bacillus cereus and E. coli cells [24,28]. Furthermore, Moon reported lysed Staphylococcus aureus cells [29], and Choi reported Actinobacillus actinomycetemcomitans cells degraded into irregular bleb-like structures [12], both after only 30 min treatment with chitosan-oligosaccharide.
As we noted earlier, it is reasonable to conclude from our data that the initial site of action of chitosan-arginine is the outer membrane. However, we argue that chitosan's effect might not be as dramatic as lysing bacteria cells within a short period of time, for example, 30 min, as reported by Moon or Choi [12,29]. Helander showed that the interaction between chitosan and E. coli did not involve the release of LPS or other membrane lipids [4]. Raafat et al. observed that chitosan treatment of Staphylococcus simulans 22 cells did not lead to cell wall lysis and that the cell membrane remained intact [30]. Our data supported the notion that chitosan-arginine's effect on bacteria was not as violent as that of known bactericides, e.g., penicillins and cephalosporins. Rather, chitosan permeabilized bacterial envelopes and affected plasma membranes [15], which likely leads to the leakage of small intracellular substances, or allows other substances such as hydrophobic macromolecules to enter into cells, eventually killing the bacteria. It is worthwhile to point out that, although our fluorescent probe and SEM experiments were carried out at pH 5.3 (Fig. 7 and Fig. 8), images from further SEM experiments indicated that the surface morphology of E. coli and P. fluorescens cells appeared similar at pH 7.0 upon chitosan-arginine treatment (data not shown), which suggested that there was no significant difference between cells treated with chitosan-arginine at acidic and neutral pH. Thus, chitosan-arginine appeared to be similarly effective in neutral pH environment and acidic pH environment.
In summary, the results of our fluorescence and SEM studies on the antibacterial mode of action of chitosan-arginine against gram-negative bacteria were consistent and complementary. They combined to support the idea that the initial action of chitosan-arginine's, and presumably chitosan's, antibacterial activity is due to the interaction of this polycation with cell membranes, specifically increasing the cell membrane permeability. Further studies to investigate how chitosan damages the plasma membrane, whether self-promoted uptake of chitosan takes place, and whether there is a leakage of intracellular substances, are appropriate. The answers to these questions should help us further improve our understanding of the exact mode of action of chitosan-arginine's/chitosan's antibacterial activity. The antibacterial property of chitosan-arginine in neutral pH environment and its improved solubility in aqueous media will broaden the scope of applications for the chitosan derivatives.
Acknowledgments
We thank Dr. Kevin Kirk for helpful discussions on statistical analysis. This work is funded by Hawaii Chitopure under U.S. Army Medical Research Acquisition Activity Contract #W81XWH-05-1-0504, Process Development and Chemical Modification of Medical Grade Chitosan, and partially supported by Grant Number RR-016480 from NCRR of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lim SH, Hudson SM. Review of chitosan and its derivatives as antimicrobial agents and their uses as textile chemicals. J Macromol Sci Part C – Polymer Reviews. 2003;C43:223–269. [Google Scholar]
- 2.Burkatovskaya M, Tegos GP, Swietlik E, Demidova TN, Castano AP, Hamblin MR. Use of chitosan bandage to prevent fatal infections developing from highly contaminated wounds in mice. Biomaterials. 2006;27:4157–4164. doi: 10.1016/j.biomaterials.2006.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didenko LV, Gerasimenko DV, Konstantinova ND, Silkina TA, Avdienko ID, Bannikova GE, et al. Ultrastructural study of chitosan effects on Klebsiella and Staphylococci. Bull Exp Biol Med. 2005;140:343–347. doi: 10.1007/s10517-005-0489-6. [DOI] [PubMed] [Google Scholar]
- 4.Helander IM, Nurmiaho-Lassila EL, Ahvenainen R, Rhoades J, Roller S. Chitosan disrupts the barrier properties of the outer membrane of gram-negative bacteria. Int J Food Microbiol. 2001;71:235–244. doi: 10.1016/s0168-1605(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 5.Bae K, Jun EJ, Lee SM, Paik DI, Kim JB. Effect of water-soluble reduced chitosan on Streptococcus mutans, plaque regrowth and biofilm vitality. Clin Oral Invest. 2006;10:102–107. doi: 10.1007/s00784-006-0038-3. [DOI] [PubMed] [Google Scholar]
- 6.Jeon YJ, Park PJ, Kim SK. Antimicrobial effect of chitooligosaccharides produced by bioreactor. Carbohydr Polym. 2001;44:71–76. [Google Scholar]
- 7.Kumar ABV, Varadaraj MC, Lalitha RG, Tharanathan RN. Low molecular weight chitosans: preparation with the aid of papain and characterization. Biochimica et Biophysica Acta. 2004;1670:137–146. doi: 10.1016/j.bbagen.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 8.No HK, Park NY, Lee SH, Meyers SP. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int J Food Microbiol. 2002;74:65–72. doi: 10.1016/s0168-1605(01)00717-6. [DOI] [PubMed] [Google Scholar]
- 9.Anas A, Paul S, Jayaprakash NS, Philip R, Bright Singh IS. Antimicrobial activity of chitosan against vibrios from freshwater prawn Macrobrachium rosenbergii larval rearing systems. Dis Aqu Org. 2005;67:177–179. doi: 10.3354/dao067177. [DOI] [PubMed] [Google Scholar]
- 10.Chen YL, Chou CC. Factors affecting the susceptibility of Staphylococcus aureus CCRC 12657 to water soluble lactose chitosan derivative. Food Microbiol. 2005;22:29–35. [Google Scholar]
- 11.Chen YM, Chung YC, Wang LW, Chen KT, Li SY. Antibacterial properties of chitosan in waterborne pathogen. J Environ Sci and Health Part A – Toxic/Hazardous Substances & Environmental Engineering. 2002;A37(7):1379–1390. doi: 10.1081/ese-120005993. [DOI] [PubMed] [Google Scholar]
- 12.Choi BK, Kim KY, Yoo YJ, Oh SJ, Choi JH, Kim CY. In vitro antimicrobial activity of a chitooligosaccharide mixture against Actinobacillus actinomycetemcomitans and Streptococcus mutans. Int J Antimicrob Agents. 2001;18:553–557. doi: 10.1016/s0924-8579(01)00434-4. [DOI] [PubMed] [Google Scholar]
- 13.Chung YC, Wang HL, Chen YM, Li SL. Effect of abiotic factors on the antibacterial activity of chitosan against waterborne pathogens. Bioresour Technol. 2003;88:179–184. doi: 10.1016/s0960-8524(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 14.Hwang JK, Kim HJ, Yoon SJ, Pyun YR. Bactericidal activity of chitosan on E. coli. In: Chen RH, Chen HC, editors. Advances in Chitin Science III. Taiwan: Rita Advertising Co Ltd; 1998. pp. 340–344. [Google Scholar]
- 15.Liu H, Du Y, Wang X, Sun L. Chitosan kills bacteria through cell membrane damage. Int J Food Microbiol. 2004;95:147–155. doi: 10.1016/j.ijfoodmicro.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, Chen XG, Park HJ, Liu CG, Liu CS, Meng XH, et al. Effect of MW and concentration of chitosan on antibacterial activity of Escherichia coli. Carbohydr Polym. 2006;64:60–65. [Google Scholar]
- 17.Tsai GJ, Su WH. Antibacterial activity of shrimp chitosan against Escherichia coli. J Food Prot. 1999;62:239–243. doi: 10.4315/0362-028x-62.3.239. [DOI] [PubMed] [Google Scholar]
- 18.Tsai GJ, Hwang SP. In vitro and in vivo antibacterial activity of shrimp chitosan against some intestinal bacteria. Fish Sci. 2004;70:675–681. [Google Scholar]
- 19.Yang TC, Li CF, Chou CC. Cell age, suspending medium and metal ion influence the susceptibility of Escherichia coli O157:H7 to water-soluble maltose chitosan derivative. Int J Food Microbiol. 2007;113:258–62. doi: 10.1016/j.ijfoodmicro.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Zheng LY, Zhu JF. Study on antimicrobial activity of chitosan with different molecular weights. Carbohydr Polym. 2003;54:527–530. [Google Scholar]
- 21.Rabea EI, Badawy MET, Stevens CV, Smagghe G, Steurbaut W. Chitosan as antimicrobial agent: applications and mode of action. Biomacromolecules. 2003;4:1457–1465. doi: 10.1021/bm034130m. [DOI] [PubMed] [Google Scholar]
- 22.Jeon YJ, Kim SK. Effect of antimicrobial activity by chitosan oligosaccharide N-conjugated with asparagine. J Microbiol Biotechnol. 2001;11:281–286. [Google Scholar]
- 23.Je JY, Kim SK. Chitosan derivatives killed bacteria by disrupting the outer and inner membrane. J Agric Food Chem. 2006;54:6629–6633. doi: 10.1021/jf061310p. [DOI] [PubMed] [Google Scholar]
- 24.Kumar ABV, Varadaraj MC, Gowda LR, Tharanathan RN. Low molecular weight chitosans – Preparation with the aid of pronase, characterization and their bactericidal activity towards Bacillus cereus and Escherichia coli. Biochem Biophy Acta. 2007;1770:495–505. doi: 10.1016/j.bbagen.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Chung YC, Su YP, Chen CC, Jia G, Wang HL, Wu JCG, et al. Relationship between antibacterial activity of chitosan and surface characteristics of cell wall. Acta Pharmacol Sin. 2004;25(7):932–936. [PubMed] [Google Scholar]
- 26.Eaton P, Fernandes JC, Pereira E, Pintado ME, Malcata FX. Atomic force microscopy study of the antibacterial effects of chitosans on Escherichia coli and Staphylococcus aureus. Ultramicroscopy. 2008;108:1128–1134. doi: 10.1016/j.ultramic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Kim CH, Kim SY, Choi KS. Synthesis and antibacterial activity of water-soluble chitin derivatives. Polym Adv Technol. 1997;8:319–325. [Google Scholar]
- 28.Kumar ABV, Varadaraj MC, Gowda LR, Tharanathan RN. Characterization of chito-oligosaccharides prepared by chitosanolysis with the aid of papain and pronase, and their bactericidal action against Bacillus cereus and Escherichia coli. Biochem J. 2005;391:167–175. doi: 10.1042/BJ20050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon JS, Kim HK, Koo HC, Joo YS, Nam HM, Park YH, et al. The antibacterial and immunostimulative effect of chitosan-oligosaccharides against infection by Staphylococcus aureus isolated from bovine mastitis. Appl Microbiol Biotechnol. 2007;75(5):989–998. doi: 10.1007/s00253-007-0898-8. [DOI] [PubMed] [Google Scholar]
- 30.Raafat D, Bargen KV, Haas A, Sahl HG. Insight into the mode of action of chitosan as an antibacterial compound. Appl Environ Microbiol. 2008;74:3764–3773. doi: 10.1128/AEM.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudarshan NR, Hoover DG, Knorr D. Antibacterial action of chitosan. Food Biotech. 1992;6:257–272. [Google Scholar]
- 32.Baker SM, Wiesmann WP, Ryan SJ. Chitosan-derivative compounds and methods of controlling microbial populations. US Application 11/657,382 and PCT application PCT/US07/02078. 2007 Jan 24; filed.
- 33.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(S1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 34.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hancock REW. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 36.Helander IM, Alakomi HL, Latva-Kala K, Koski P. Polyethyleneimine is an effective permeabilizer of Gram-negative bacteria. Microbiol. 1997;143:3193–3199. doi: 10.1099/00221287-143-10-3193. [DOI] [PubMed] [Google Scholar]
- 37.Loh B, Grant C, Hancock REW. Use of the fluorescent probe 1-n-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:546–551. doi: 10.1128/aac.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helander IM, Mattila-Sandholm T. Fluorometric assessment of gram-negative bacteria permeabilization. J Appl Microbiol. 2000;88:213–219. doi: 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- 39.Haugland RP. Handbook of Fluorescent Probes and Research Products. 10th. Eugene, OR: Molecular Probes; 2005. Section 8.1 Nucleic acid stains and Section 13.5 Other nonpolar and amphiphilic probes; pp. 328pp. 630–631. [Google Scholar]
- 40.Cuero RG. Antimicrobial action of exogenous chitosan. EXS. 1999;87:315–333. doi: 10.1007/978-3-0348-8757-1_23. [DOI] [PubMed] [Google Scholar]