Abstract
Recent work has greatly contributed to the understanding of the biology and biochemistry of RecQ4. It plays an essential non-enzymatic role in the formation of the CMG complex, and thus replication initiation, by means of its Sld2 homologous domain. The helicase domain of RecQ4 has now been demonstrated to possess 3'-5' DNA helicase activity, like the other members of the RecQ family. The biological purpose of this activity is still unclear, but helicase-dead mutants are unable to restore viability in the absence of wildtype RecQ4. This indicates that RecQ4 performs a second role, which requires helicase activity and is implicated in replication and DNA repair. Thus, it is clear that two helicases, RecQ4 and Mcm2-7, are integral to replication. The nature of the simultaneous involvement of these two helicases remains to be determined, and possible models will be proposed.
Keywords: DNA replication, RecQ helicase, Sld2, Rothmund-Thomson Syndrome, RAPADILINO, Baller-Gerold Syndrome
Introduction
RecQ helicases are involved in various aspects of DNA metabolism, particularly replication and recombination. All prokaryotes and eukaryotes possess at least one member of the family, and some plants have as many as seven. Five RecQ helicases have been identified in humans and mice, and three in Drosophila. Within a single species, multiple family members may exhibit a level of parallel functionality (reviewed in Seki et al., 2008). This mitigates the impact of defects in any one RecQ helicase. Even so, mutations in three of the five RecQ helicases in humans (Blm, Wrn, and RecQ4) lead to clinical syndromes characterized by increased genetic instability, indicating that each of these plays a unique and important role (reviewed in Bohr, 2008). Of these, the least understood is RecQ4. Recently much work has been done to elucidate its biological and biochemical significance, pointing to an essential role for RecQ4 in replication. This paper will review what is currently known about RecQ4 and argue that it is a replicative helicase, acting alongside Mcm2-7. Because it is not intuitive that there should be two helicases at a single replication fork, models of how this may occur will also be proposed.
The Health Impact of RecQ4
RecQ4 was originally identified in 1998 in a human genome search for RecQ helicases (Kitao et al., 1998), and a year later was found to be associated with the rare type II Rothmund-Thomson syndrome (RTS) (Kitao et al., 1999). RTS is characterized by a wide array of defects, including cataracts, sparse hair, poikiloderma, growth deficiencies, various skeletal abnormalities and a greater propensity for osteosarcomas (reviewed in Vennos et al., 1992). The clinical impact of mutations in RecQ4 was later expanded to include the even rarer RAPADILINO (named for an acronym of the diagnostic symptoms) (Siitonen et al., 2003) and Baller-Gerold (Van Maldergem et al., 2006) syndromes. RTS, RAPADILINO and Baller-Gerold syndrome have a significant overlap of symptoms (Fig. 1), all occurring in tissues with a high proliferation rate. The absence of a strong correlation between specific mutations in RecQ4 and specific phenotypes (reviewed in Larizza et al., 2006 and Siitonen et al., 2009) has led some to propose that the three syndromes be reclassified as a single one (Van Maldergem et al., 2006).
Figure 1.
The symptoms of the RecQ4 associated Rothmund-Thomson (RTS), RAPADILINO, and Baller-Gerold (BGS) Syndromes. This is a Venn diagram of the symptoms of these diseases, showing which are unique and which are shared between syndromes. If these three are in fact a single syndrome, the key phenotypes would include growth deficiency, facial dysmorphia, and gastrointestinal disturbance. Adapted by permission from BMJ Publishing Group Limited. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. Van Maldergem et al. J. Med. Genet. 43, 148–152, 2006.
The wide spectrum of phenotypes associated with mutations in RecQ4 has been mirrored in studies using transgenic mice. Mouse RecQ4 knockouts were embryonic lethal (Ichikawa et al., 2002). Simply truncating the enzyme in the middle of the centrally located helicase domain showed no embryonic lethality, though only 5% survival was observed two weeks after birth. Mice that reached adulthood showed phenotypes similar to those found in RecQ4 associated syndromes, including smaller size, skin abnormalities, and hair discoloration (Hoki et al., 2003). Truncating the enzyme just before the helicase domain (rather than within it) significantly reduced morbidity, giving a survival rate of 84%. Phenotypes associated with RecQ4 associated diseases were still present, though they were different from those found in mice with RecQ4 truncated within the helicase domain (Mann et al., 2005). Thus three different genotypic defects in murine RecQ4 result in three very different phenotypic patterns.
The wide variation and heterozygosity of mutations in humans makes it difficult to derive enzymatic information about RecQ4 based on phenotypes, but no such difficulty exists with mice. Comparisons of human and murine RecQ4 are particularly relevant because of the high conservation of the enzyme between the two species (63.4% overall sequence identity, and 85.7% similarity) (Ohhata et al., 2000). Such comparisons show that RecQ4 plays an essential role in development, as its complete absence leads to embryonic lethality. This explains the extreme rarity of RecQ4 associated syndromes in humans, as severe mutations may result in spontaneous early miscarriage, and so are never observed. The helicase domain itself is not essential for viability, but its partial presence is more harmful than its absence. This suggests that the role played by the helicase domain is one for which there exist redundancies. In the absence of the helicase domain, these act to ensure the survival of the cell, albeit with a reduced efficiency leading to those phenotypes of RecQ4 associated syndromes that are less severe. In the presence of a partial or non-functional RecQ4 helicase domain such redundancies are unable to act. This results in the higher morbidity and more severe symptoms of RTS and other RecQ4 associated syndromes.
The above phenotypic analysis of mutant mice and RTS, RAPADILINO, and Baller-Gerold Syndrome patients demonstrates that RecQ4 plays two roles, though it does little to indicate the nature of those roles. One role is essential and does not require the helicase domain. The other is performed by the helicase domain, but either is non-essential, or can be performed by another enzyme in the absence of this domain. As will be discussed below, evidence indicates that both roles are involved in DNA replication, and that the latter role may also be involved in DNA repair.
RecQ4's Role in the Replication
The first direct evidence for RecQ4's involvement in replication came in 2005, when two groups independently observed sequence homology between Sld2 in Saccharomyces cerevisiae and the N-terminus of xRTS, the RecQ4 homolog in Xenopus (Fig. 2) (Matsuno et al., 2006; Sangrithi et al., 2005). Sld2 is one of two essential targets for S-phase cyclin dependent kinase (CDK), the other being Sld3. Phosphorylation of Sld2 and Sld3 by S-phase CDK leads them to interact with Dpb11 (Tanaka et al., 2007b; Zegerman and Diffley, 2007). These interactions are essential for the initiation of DNA replication in S. cerevisiae (reviewed in Tanaka et al., 2007 and Araki, 2009; for a more comprehensive review of the roles of these proteins in the initiation of replication see Sclafani and Holzen, 2007). RecQ4 is the only known metazoan homolog of Sld2 and is, by virtue of this comparison, strongly implicated in replication.
Figure 2.
Alignment of RecQ4 from various species with Sld2 and the other RecQ helicases in Homo sapiens and Saccharomyces cerevisiae. The RecQ helicases are aligned based on their shared RecQ helicase domain (in blue). The N-terminus of RecQ4 is the less conserved between species than the helicase domain and C-terminus. Note the large insertions in both the Sld2 homologous domain (in green). Unlike most RecQ family members, RecQ4 does not have an RQC (RecQ C-terminal) domain (in orange) or an HRDC (Helicase and Rnase D C-terminal) domain (in yellow).
This implication is borne out by data from Xenopus and Drosophila model systems, as well as from human tissue culture cells. The N-terminus of xRTS (including the Sld2 homologous domain, but not the helicase domain) is necessary for replication initiation in Xenopus oocyte extract. It is also necessary for chromatin binding by DNA Polymerase α (Matsuno et al., 2006). Supplementing xRTS-depleted Xenopus oocyte extract with the N-terminus of human RecQ4 restores replication to ~20% of wildtype levels (Sangrithi et al., 2005). The fact that restoration is only partial suggests that the helicase domain also plays a role in replication. Similarly, Drosophila with null or hypomorphic expression of RecQ4 are severely defective in normal premitotic replication. Hypomorphic Drosophila are also defective in endoreplication (genome synthesis without subsequent mitosis, leading to the polyploidy found in salivary glands) and chorion gene amplification (nested synthesis of the chorion gene, allowing for subsequent rapid production of the protein during egg shell generation). These defects occur during initiation, not elongation (Wu et al., 2008; Xu et al., 2009b). In human tissue culture cells, RecQ4 is associated with replication origins only during late G1-phase and S-phase, and depletion of RecQ4 inhibits cell proliferation and DNA synthesis (Thangavel et al., 2010). Work in Xenopus, Drosophila and human systems thus confirms that RecQ4 plays a role in replication initiation, similar to Sld2.
In yeast, as mentioned earlier, it is known that Sld2's role in replication is mediated by Dpb11 and associated with Sld3 (reviewed in Tanaka et al., 2007a and Sclafani and Holzen, 2007). However, evidence is not clear about whether the interactions between Dpb11 and phosphorylated Sld2 and Sld3 are conserved in metazoans. There is no known metazoan homolog of Sld3. It is not clear that Sld2's interaction with Dpb11 is paralleled by the interaction of RecQ4 with the known homologues of Dpb11, which include TopBP1 in humans, Cut5 in Xenopus, and Mus101 in Drosophila (reviewed in Garcia et al., 2005). In Xenopus oocyte extract, Cut5 and the N-terminus of xRTS co-immunoprecipitate independent of xRTS phosphorylation (Matsuno et al., 2006). Therefore, unlike Sld2 and Dpb11, RecQ4 does not need to be phosphorylated to interact with Cut5. And although xRTS is necessary for replication, it does not mediate the replication origin's interaction with either Cut5 or the CMG (Cdc45; Mcm2-7; GINS) complex, as Cut5 and the components of the CMG complex load onto chromatin even in xRTS-depleted oocyte extract (Sangrithi et al., 2005). Data from human tissue culture cells suggests that the key interaction is not between RecQ4 and TopBP1/Cut5/Mus101, but rather between RecQ4 and Mcm10. RecQ4 co-immunoprecipitates with Mcm10 (Xu et al., 2009a), and both RecQ4 and Mcm10 are necessary for the formation of the CMG complex (Im et al., 2009). In contrast, TopBP1 neither co-immunoprecipitates with RecQ4, nor is necessary for the stable formation of the CMG complex (Im et al., 2009; Xu et al., 2009a). It is possible that the way RecQ4-Mcm10 stabilizes the CMG complex is by causing GINS to associate with Mcm2-7 and Cdc45 (Xu et al., 2009a). Thus RecQ4-Mcm10 mediates the binding of the components of the CMG complex to each other, rather than to the replication origin. The differing data from Xenopus and human systems with respect to the interaction of RecQ4 and TopBP1/Cut5 may reflect either species-specific differences, or subtle dynamics in the formation of the metazoan replication complex. Regardless, it is clear that RecQ4 is central to the formation of the replication complex, and that it is loaded on the origin prior to Polymerase α or RPA (Matsuno et al., 2006; Sangrithi et al., 2005).
From the discussion above, it is clear that the N-terminal Sld2 domain of RecQ4 plays a necessary role in replication initiation. This role, if fully corresponding to that of Sld2, is nonenzymatic, and the presence of the domain is crucial for proper function. Absence of the domain leads to the early developmental lethality seen in RecQ4 null mutants. However, one cannot disregard the replicative importance of the other domains of RecQ4, particularly the helicase domain. Both the Sld2 domain and the helicase domain are required for viability in Drosophila, though the C-terminus is not essential (Xu et al., 2009b). More precisely, point mutants inactivating the helicase domain are unable to restore replication in either xRTS-depleted Xenopus oocyte extract or Drosophila null mutants (Capp et al., 2009; Sangrithi et al., 2005). The inability of these otherwise intact proteins to restore replication or viability indicates that RecQ4's helicase activity is also important for replication. Therefore, there must be a second role for which enzymatic activity is necessary.
RecQ4's Role in Repair
Data suggest that RecQ4 may also be involved in DNA damage repair, but this is considerably cloudier than that concerning its involvement in replication. In one study, cell lines derived from RTS patients show sensitivity similar to wildtype cell lines when subjected to a variety of DNA damaging agents, including those that induce DNA double-strand breaks (DSB), oxidative damage, inter-strand crosslinks, as well as those that interfere with replication, such as hydroxyurea (Cabral et al., 2008). However, this is the only study reporting that the loss of RecQ4 has no effect on sensitivity to such agents. Another study, using similar approaches, instead found increased sensitivity of RTS cell lines to agents that interfere with replication, and wildtype sensitivity to those that induce DSBs (Jin et al., 2008). Such data are consistent with RecQ4 being involved in the restart of stalled replication forks. While normal human cells undergo significant S-phase arrest after treatment with hydroxyurea or UV irradiation, RTS cells and T-293 cells with RecQ4 knocked down by shRNA have been shown to not enter such an arrest (Park et al., 2006). RecQ4 may thus play a role in the signaling of cell cycle arrest during DNA damage response. There is also some evidence that RecQ4 is involved in response to oxidative damage, which would suggest that it is part of the base excision repair pathway. RTS cells and those with RecQ4 transiently knocked down by siRNA have been shown by one group to be hypersensitive to oxidative damage (Schurman et al., 2009). The enzyme can also relocalize to the nucleolus in response to oxidative damage (Woo et al., 2006), indicating that the intracellular localization of RecQ4 may be regulated by DNA damage response. In human fibroblasts after induction of DSBs, the proportion of RecQ4 nuclear foci colocalizing with promyelotic leukemia protein was reduced, in favor of associating with Rad51 (Petkovic et al., 2005). Though this shift was relatively minor, it still indicates a response to DSB induction. Taking these results together, it is difficult to form a consistent model concerning RecQ4 and DNA damage repair. There is relative consensus that RecQ4 is involved somehow in replication arrest and repair of oxidative damage. It is more likely that this involvement is due to the helicase domain than to the Sld2 domain, but one also cannot eliminate the possibility of an undiscovered activity in the C-terminus being responsible. Certainly, much remains to be done to determine RecQ4's involvement in DNA damage repair.
The Localization and Regulation of RecQ4
It is generally essential that DNA be replicated once and only once during the life of a cell. Accordingly, many of the proteins involved in replication are tightly regulated in terms of expression, cellular localization, and enzymatic activity (reviewed in Remus and Diffley, 2009). RecQ4 is no exception. Both the expression and localization of RecQ4 are closely regulated, and it is possible that the activity of the enzyme is as well.
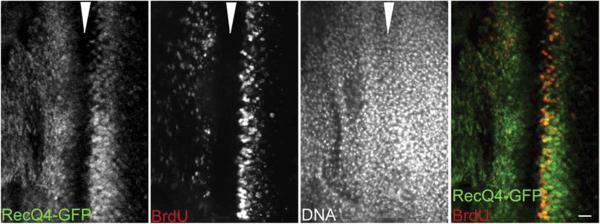
Drosophila has proved to be a useful model system for examination of the expression and localization patterns of RecQ4 (Wu et al., 2008). Cell cycle dependent expression patterns can be observed using the synchronized cell cycle progression found in the eye imaginal disc during Drosophila development. The eye imaginal disc has a morphogenic furrow (see arrow on Fig. 3), in which the cells are all paused in G1. Anterior to the furrow (left of the arrow), cells are asynchronously cycling. Posterior to it, the cells enter S-phase at precisely the same time, as indicated by BrdU incorporation, and proceed through the following mitotic wave in lockstep before entering into a non-replicative state. Throughout this process, RecQ4 expression is observed in asynchronously cycling cells (anterior to the furrow) and in mitotic cells (posterior to the furrow). It is notably absent within the morphogenic furrow itself. This means that RecQ4 must be destroyed prior to entrance into G1, only to be re-synthesized when replication is about to occur. Once the cells enter the extended non-replicative state (to the far right of the arrow), RecQ4 again diminishes, indicating degradation (Wu et al., 2008). It is consistent with this pattern that p53 represses expression of RecQ4 during G1 in human fibroblasts (Sengupta et al., 2005). Thus RecQ4 expression is tightly regulated by mechanisms addressing both its synthesis and degradation, confining its presence to S-phase and shortly thereafter.
Figure 3.
RecQ4 expression in the eye imaginal disc of Drosophila is coincident with DNA replication. Although the cell density (as seen by DAPI staining of the DNA) is constant, synthesis (as seen by BrdU incorporation) occurs only in two mitotic waves, separated by a morphogenic furrow (arrowhead) of cells in extended G1. The first, anterior to the morphogenic furrow (left of the arrowhead), is asynchronous. The second one, immediately posterior to the morphogenic furrow (right of the arrowhead), is synchronous. RecQ4 is absent upon entrance in to the extended G1 phase of the morphogenic furrow, but is again expressed with the beginning of synthesis. This indicates that RecQ4 is expressed only during S-phase, and that in other phases it is repressed and/or degraded. Scale bar: 10μm. This figure originally appeared in Wu et al., 2008. This research was originally published in Developmental Biology. Wu et al. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev. Biol. 2008; 323:130–142. © Elsevier.
The localization of RecQ4 within the cell is also closely regulated. Early Drosophila embryos do not have separate cells, though they do have distinct membrane-contained nuclei. The nuclei all proceed synchronously through the cell cycle (albeit lacking G1 and G2 phases) until cellularization occurs, after which the cell cycle lengthens and synchrony is lost (Foe et al., 1993). As expected for an enzyme involved in replication, RecQ4 is associated with the chromatin during interphase (Wu et al., 2008). During metaphase and anaphase, the nuclear membranes break down and RecQ4 is dispersed to the cytoplasm. After the membrane re-forms during telophase, RecQ4 is again found associated with the chromatin. This localization pattern is continued even after the formation of discrete cells (Wu et al., 2008). Work using human tissue culture cells has defined within the N-terminus a nuclear localization sequence, a nuclear retention sequence (Burks et al., 2007), and a lysine-rich nucleolar localization sequence (Woo et al., 2006) (Fig. 2). Deletion of the nuclear retention sequence results in localization to both the nucleus and the cytoplasm (Burks et al., 2007), and has been correlated to the occurrence of osteosarcomas and lymphomas among RTS and RAPADILINO patients (Siitonen et al., 2009). Acetylation by p300 of the lysines within the nucleolar localization sequence also causes RecQ4 to be excluded from the nucleus (Dietschy et al., 2009), suggesting a means of regulating the cell-cycle dependent localization of RecQ4.
Besides acetylation, there are several other post-translational modifications of RecQ4 that may also regulate the enzyme. The C-terminus of RecQ4 has been shown to coimmunoprecipitate with poly(ADP-Ribose) Polymerase-1, and to be an in vitro substrate for poly-ADP ribosylation (Woo et al., 2006). Although RecQ4 has also been shown to form stable interactions with UBR1 and UBR2 (ubiquitin E3 ligases of the N-end rule pathway, which targets proteins for degradation by the proteasome), it is neither ubiquitylated nor rapidly degraded (Yin et al., 2004). As mentioned earlier, it is clear that Sld2 depends on being phosphorylated for functionality. But while it is known that RecQ4 can serve as a substrate for phosphorylation in vitro, it has not been established that this actually occurs in vivo. Biochemical identification and characterization of these and other as yet to be discovered post-translational modifications will be a critical area for future investigation.
The Biochemistry of RecQ4
As seen above, comparison of the data from RTS patients and mouse, Xenopus, and Drosophila model systems leads to the prediction of a second role for RecQ4, which can be traced to enzymatic activity from the helicase domain. However, it has proven difficult to biochemically assess the helicase activity of RecQ4. In 2004, RecQ4 was first isolated by immunoprecipitating the endogenous enzyme from HeLa cells. This enzyme was unable to act as a helicase or DNA translocase, though DNA-stimulated ATP hydrolysis was detected (Yin et al., 2004). In 2006, human RecQ4 was expressed in E. coli, and purified by several ion exchange and affinity chromatography steps. Helicase activity was not observed, though both DNA-stimulated ATP hydrolysis and DNA annealing activities were (Macris et al., 2006). This was not expected, as each of the four other human RecQ helicases had demonstrated 3'-5' helicase activity (reviewed in Seki et al., 2008; Bohr, 2008; and Bachrati and Hickson, 2008), and these all share with RecQ4 a well conserved helicase domain. Then, in 2009, eleven years after the initial identification of RecQ4, three groups independently reported the observation of helicase activity from the enzyme.
The first of these also isolated human RecQ4 from E. coli (Xu and Liu, 2009). Like previous work, they observed annealing and ATP hydrolysis. Helicase activity, detected by the separation of radio-labeled oligonucleotide from its cold complement, was only observed in the presence of an excess cold oligonucleotide, which prevented the complementary strand from re-annealing to the newly liberated radio-labeled oligonucleotide. The necessity of such a trap was attributed to RecQ4's innate annealing activity rendering the helicase activity undetectable. Helicase activity was determined to proceed in a 3'-5' direction, like the other RecQ helicases, but also to act on blunt-ended substrates, which is uncommon in the family. In an unexpected turn, this helicase activity was mapped to both the helicase domain and the Sld2 domain, the latter contributing most of the activity. The Sld2 domain was found to interact with ATP using both spin column assays and UV crosslinking. This suggests the presence of completely new ATP binding motifs. The helicase domain was shown to be competent for robust helicase activity when part of a RecQ4-RecQ5 chimera, indicating that the weakness of its helicase activity is due to inhibition by the rest of the RecQ4 enzyme, rather than to any qualities inherent to the domain itself (Xu and Liu, 2009).
The second group to observe helicase activity from RecQ4 purified the human enzyme using the baculoviral expression system (Suzuki et al., 2009). They observed 3'-5' helicase activity in the absence of a reaction product trap. RecQ4 was found to efficiently displace a 17 basepair-long oligonucleotide annealed to single-stranded circular DNA, but could not unwind a similar 37 basepair region. This gives some sense of the step-size of the enzyme, and indicates that it may not be processive. Further analysis showed maximal helicase activity between pH 8 and pH 10, at 5mM ATP, and at MgCl2 concentrations of 8mM and above. All of these assays used either oligonucleotides annealed to single-stranded circular DNA, or blunt-ended linear DNA with an internal single-stranded region. As such, beyond determining the direction of helicase activity, substrate preference was not characterized (Suzuki et al., 2009).
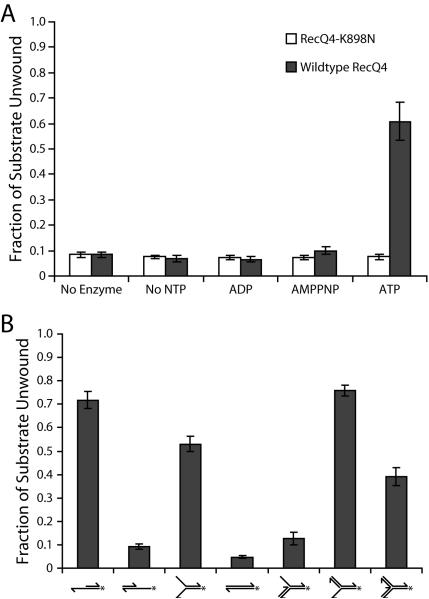
The most recent group to examine the biochemistry of RecQ4 used the baculoviral expression system to express Drosophila RecQ4, and purified it with two affinity chromatography steps, followed by a glycerol density gradient (Capp et al., 2009). Enzyme thus obtained was more than 96% pure, and demonstrated DNA-stimulated ATP hydrolysis, annealing, DNA binding, and helicase activity. RecQ4 formed a stable complex with single-stranded DNA in the presence of AMPPNP. This was not observed with completely duplex DNA, and there is a specific requirement for AMPPNP as a cofactor. The absence of cofactor, or the substitution of ADP for AMPPNP, abolishes the tight binding to single-stranded DNA. The annealing activity of RecQ4 was inhibited by AMPPNP, possibly due to the trapping of single-stranded DNA in this stable protein/DNA complex. Robust helicase activity was observed in the absence of any reaction product trap (Fig. 4A). RecQ4 could unwind duplex regions as long as 30 basepairs, and required the single-stranded DNA 3' of the region to be separated (Fig. 4B). Neither a 60 basepair blunt-ended DNA substrate, nor a substrate with single-stranded DNA 5' of a 30 basepair duplex region could be unwound. This confirms the 3'-5' directionality of the enzyme. Further characterization of substrate preference found these same trends when looking at fork-based oligonucleotide substrates. Thus, a fork substrate with a duplex 5' branch and a single-stranded 3' branch (but not the reverse) was unwound. Interestingly, a fork substrate with two duplex branches could be unwound, though this is attributed to transiently exposed single-stranded regions at the fork junction. Helicase activity and AMPPNP-dependent DNA binding were both derived exclusively from the helicase domain, because a point mutant inactivating ATP binding by the domain abolished helicase activity (Fig. 4A), DNA binding, and inhibition of annealing by AMPPNP. An active helicase domain was determined to be necessary for viability, as the point mutant could not rescue RecQ4-null flies (Capp et al., 2009).
Figure 4.
Analysis of the helicase activity substrate preference of Drosophila RecQ4. (A) RecQ4 (shaded bars) unwinds the 3' extension substrate in the presence of ATP. This activity is due to the helicase domain of RecQ4 because the helicase inactive mutant RecQ4-K898N (open bars) does not unwind the 3' extension substrate. (B) Quantification of RecQ4 activity on substrates. RecQ4 acts as a helicase only on substrates with a 3' single-stranded region, with the exception of the 3-way duplex junction substrate (far right). Error bars in these experiments indicate standard deviation (n =3). This research was originally published in the Journal of Biological Chemistry. Capp et al. Drosophila RecQ4 Has a 3'-5' DNA Helicase Activity That Is Essential for Viability. J. Biol. Chem. 2009; 284:30845–30852. © the American Society for Biochemistry and Molecular Biology.
It has now been established that RecQ4 is a 3'-5' DNA helicase that also possesses annealing activity. This is in keeping with the general biochemistry of RecQ helicase family members. Helicase activity is clearly more robust when the enzyme is expressed in insect cells with baculovirus vectors than when it is expressed in E. coli, implying that post-translational modifications and/or eukaryotic-specific protein folding mechanisms are important for activation. It remains to be determined whether or which particular post-translational modifications are responsible for the observed difference in helicase activities. It is not clear why the initial isolation of RecQ4 from HeLa cells did not demonstrate helicase activity. However, because the potential effect of antibody on enzymatic activities was not examined, it is possible that the use of immunoprecipitation in purification resulted in inhibition of helicase activity. The tracking of helicase activity to the Sld2 domain of human RecQ4 is unprecedented, as no enzymatic activity was predicted to come from this region. Because a reaction product trap was required to observe the activity, it perhaps could be better described as strand-exchange activity (Xu and Liu, 2009). Sld2 domain-derived helicase/strand-exchange activity was not found in Drosophila RecQ4, but this may simply be reflective of differences between species. This alternate source of helicase activity could explain why humans and mice entirely lacking the helicase domain are still viable. Nonetheless, the demonstration of helicase activity by the helicase domain provides a source of enzymatic activity consistent with the predicted second role for RecQ4. It remains to be determined what the biological nature of this role is.
Discussion: The Dual Roles of RecQ4
The biological and biochemical data, when taken together, suggest two essential roles for RecQ4 during replication. The first of these roles is in the loading and licensing of the replication machinery, and is dependent upon the N-terminal Sld2 domain. The nature of the second role is less clear, but may occur during the elongation phase of replication, and requires an active helicase domain. RecQ4 is tightly regulated by a number of mechanisms: expression level through degradation before entrance into G1 (Wu et al., 2008), and repression by p53 during G1 (Sengupta et al., 2005); intracellular localization by acetylation of the nucleolar localization sequence (Dietschy et al., 2009); and possibly helicase activity by an as yet undetermined post-translational modification. Therefore, it is critical for the appropriate function of RecQ4 that the protein is expressed only during the correct time, at the correct level, and in the correct specific cellular locale.
The best understood role of RecQ4 is that which is played by the Sld2 domain. This understanding is largely based on comparison with the function of Sld2 in S. cerevisiae. The similarity is instructive but not authoritative, due to the more complicated nature of replication in metazoans. Like Sld2 in S. cerevisiae, RecQ4 is involved in the construction of the replicative complex. When preparing for replication during G1, the origin recognition complex and Mcm2-7 bind the origin, forming the pre-replicative complex (pre-RC). RecQ4 and Mcm10 (along with Ctf4) interact with the pre-RC at the beginning of S-phase, causing it to form the CMG complex with Cdc45 and GINS (Im et al., 2009; Xu et al., 2009a). This complex then interacts with Polymerase α and RPA as replication is initiated. TopBP1/Cut5 plays some role in this process and can interact with RecQ4. However, unlike Dpb11 and Sld2, it is able to bind chromatin independently of RecQ4 (Matsuno et al., 2006). If the interaction between TopBP1/Cut5 and RecQ4 is important to replication, it is not in the same manner as the interaction between Dpb11 and Sld2. In fact, it is possible that the functional homolog of the Sld2-Dpb11 interaction is that of RecQ4 and Mcm10. Phosphorylation by metazoan S-CDKs probably regulates or modulates this process, but this also remains to be determined. The role played by RecQ4's Sld2 domain in the assembly of the replicative complex is absolutely essential, and one for which there is no “backup” redundant system. It is failure to perform at least this role that renders RecQ4 knockout mutants early embryonic lethal (the small amount of development in such mutants being attributed to maternal loading of functional RecQ4).
RecQ4's Sld2 domain, however, cannot be responsible for the symptoms of RecQ4 associated syndromes. Patients of these syndromes are able to replicate DNA and generally have RecQ4 mutations that occur outside the Sld2 domain. Such mutations may affect the protein sequence outside the Sld2 domain, or may occur in the promoter and only affect expression level, but either way the Sld2 domain remains intact. Therefore, the phenotypes seen in RecQ4 associated syndromes and in RecQ4-truncated mouse models are due to defects occurring in the rest of the enzyme. The most obvious potential source of these defects is the helicase domain.
We suggest that RecQ4 plays a second important role in replication, one which involves the active helicase domain. This role may also be applied to DNA damage repair. Work in mouse and fly model systems indicate that an inactivated or partially present helicase domain results in lethality. On the other hand, the complete absence of the helicase domain in transgenic mice only slightly reduces viability, though developmental abnormalities do occur. One can envision that the role played by the helicase domain may not be essential, but, when inactive, it interferes with the normal mechanics of replication, which results in defects of varying severity depending on the extent of interference. If the helicase domain is entirely absent, it provides neither benefit nor detriment, and so defects are less severe. A more specific variation of this model is that RecQ4's helicase domain would have an essential role that could also be performed by a redundant system. Such a system would not be able to compete with the helicase domain for binding, and so the presence of an inactive helicase domain would hinder its ability to act, leading to more severe defects. When employed, the redundancy would show reduced efficiency compared to RecQ4, which would result in an increased incidence of replication defects, but not annul replication altogether. It must be noted that the redundant system could come from within RecQ4 itself. If helicase activity attributed to the Sld2 domain is confirmed, this could serve in the absence of the helicase domain. In this case, the lethality of helicase-inactivated point mutants from Xenopus and Drosophila would imply that such an internal redundancy was chiefly characteristic of mammalian RecQ4.
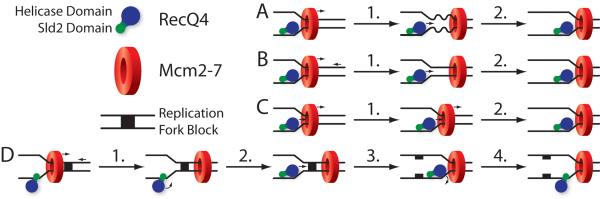
The question remains, what is the functional role for a helicase other than the Mcm2-7 complex at the replication fork? One possibility is that Mcm2-7 and RecQ4 work in concert to unwind DNA, in an active-passive pairing. Active helicases use the energy from ATP hydrolysis to dissociate annealed strands. This can happen either by acting as a wedge to force the strands apart, or by twisting the duplex DNA to force it to separate. Passive helicases, on the other hand, take advantage of thermal fluctuations in the DNA between annealed and unannealed states, using ATP hydrolysis to preserve single-stranded DNA from reannealing. If RecQ4 and Mcm2-7 work as an active-passive pair, one helicase actively unwinds the DNA, while the other trails behind and passively maintain the separation (Fig. 5A). Mcm2-7, which is capable of passing double-stranded DNA through the center of its hexameric ring, could act to actively destabilize duplex DNA (reviewed in Remus et al., 2009 and Bochman and Schwacha, 2009). RecQ4 would then advance to passively maintain the separation. The converse pairing (active RecQ4, passive Mcm2-7) is difficult to conceive with Mcm2-7 remaining bound to double-stranded DNA, because this puts it ahead of the replication fork.
Figure 5.
The coordination of RecQ4's helicase domain with Mcm2-7 in replication. In the following models RecQ4 is represented as a monomer, as indicated by initial biochemical experiments (Capp et al., 2009). The rest of the replicative complex is assumed, but not drawn. (A) Active Mcm2-7, passive RecQ4: Mcm2-7 actively destabilizes duplex DNA (1), causing it to transiently come apart. RecQ4 then enters the new single-stranded region (2), and prevents reannealing. (B) Active RecQ4, Mcm2-7 as double-stranded DNA pump: DNA is pushed by Mcm2-7 towards RecQ4 (1), but is not immediately unwound. This allows RecQ4 to actively unwind double-stranded DNA (2). (C) Active RecQ4, active Mcm2-7 (the Ploughshare model): Mcm2-7 pushes double-stranded DNA into RecQ4, which is simultaneously moving forward, unwinding the DNA (both 1 and 2). (D) RecQ4 as a specialty helicase: Mcm2-7 acts as the primary replicative helicase; RecQ4 is at the replication fork because of the licensing function of the Sld2 domain, but does not act as a helicase. Occasionally a replication block is encountered which Mcm2-7 is unable to unwind. This blocks the replicative complex and fork, but not Mcm2-7, which is around the double-stranded DNA (1). The replicative complex reconfigures to allow activity of the RecQ4 helicase domain (2). RecQ4 unwinds the blockage, which is repaired or bypassed (3). The replicative complex reconfigures again (4), and Mcm2-7 continues as the primary helicase.
It has been proposed that Mcm2-7 acts as a double-stranded DNA pump (Bochman and Schwacha, 2009). This suggests another means of cooperating with RecQ4 at the replication fork (Fig. 5B). Mcm2-7 may move ahead of the replication fork, pushing double-stranded DNA towards RecQ4, which then unwinds it. In this scenario RecQ4 would fulfill the role of active helicase. Mcm2-7, on the other hand, would act as a DNA translocase. In this capacity, it would serve as a motor for the replicative complex, and to clear off DNA-bound proteins ahead of the fork.
RecQ4 and Mcm2-7 may also cooperate as active helicases (Fig. 5C), in a variation of the Ploughshare model (Bochman and Schwacha, 2009). The Ploughshare model proposes that Mcm2-7 unwinds DNA by pushing it towards a rigid wedge, either on Mcm2-7 itself, or on an associated protein. It is possible that rather than a simple wedge, another helicase (RecQ4) is involved. In this situation, Mcm2-7 would push the DNA directly into RecQ4, which would actively unwind it. This coordination between the two helicases would lessen the burden borne by either individually.
It is also possible that Mcm2-7 is the primary replicative helicase, and RecQ4 does not actually function as a helicase at the replication fork. When the replicative complex encounters structures that Mcm2-7 is unable to unwind, such as certain forms of DNA damage, or topologically constrained DNA structures like Holliday junctions, RecQ4 may be required (Fig. 5D). In such cases, Mcm2-7 would pass the blockage through its central pore, but the replicative complex would be paused until RecQ4 unwound it. This would imply a role for RecQ4 consistent with that of the other RecQ helicases, which also act on topologically constrained DNA structures. Any role in DNA repair would also primarily involve unwinding such topological abnormalities.
Much remains to be discovered about RecQ4. A number of experiments clearly demonstrate both that the enzyme acts during replication, and that it acts as a helicase. It is not immediately obvious how this is so, but there may be an advantage to combining the non-enzymatic replication licensing function of Sld2 with the enzymatic activity of a RecQ helicase. Exactly how these two functions are coordinated will likely be a key element in DNA replication or repair, and will certainly be an area of active investigation in the future.
Acknowledgements
We are grateful to Stefanie Hartman Chen and Jo Anna Wiersma for assistance in preparing this manuscript. We would also like to thank Randolph Capp for aiding in figure preparation.
Footnotes
Declarations of Interest This work is funded by NIH Grant GM29006.
References
- Araki H. Regulatory mechanism of the initiation step of DNA replication by CDK in budding yeast. Biochim. Biophys. Acta. 2009;1804:520–523. doi: 10.1016/j.bbapap.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. The Mcm Complex: Unwinding the Mechanism of a Replicative Helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burks LM, Yin J, Plon SE. Nuclear import and retention domains in the amino terminus of RECQL4. Gene. 2007;391:26–38. doi: 10.1016/j.gene.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Cabral REC, Queille S, Bodemer C, Prost Y, Neto JBC, Sarasin A, Daya-Grosjean L. Identification of new RECQL4 mutations in Caucasian Rothmund-Thomson patients and analysis of sensitivity to a wide range of genotoxic agents. Mutat. Res. 2008;643:41–47. doi: 10.1016/j.mrfmmm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Capp C, Wu J, Hsieh T. Drosophila RecQ4 Has a 3'-5' DNA Helicase Activity That Is Essential for Viability. J. Biol. Chem. 2009;284:30845–30852. doi: 10.1074/jbc.M109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy T, Shevelev I, Pena-Diaz J, Huhn D, Kuenzle S, Mak R, Miah MF, Hess D, Fey M, Hottiger MO, et al. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 2009;122:1258–1267. doi: 10.1242/jcs.037747. [DOI] [PubMed] [Google Scholar]
- Foe VE, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 149–300. [Google Scholar]
- Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, Fukumura R, Nakamura M, Takahashi H, Noda Y, Kito S, et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- Ichikawa K, Noda T, Furuichi Y. Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nippon Yakurigaku Zasshi. 2002;119:219–226. doi: 10.1254/fpj.119.219. [DOI] [PubMed] [Google Scholar]
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Liu H, Zhang Y, Otta SK, Plon SE, Wang LL. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum. Genet. 2008;123:643–653. doi: 10.1007/s00439-008-0518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair. 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell. Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhata T, Araki R, Fukumura R, Kuroiwa A, Matsuda Y, Tatsumi K, Abe M. Cloning, genomic structure and chromosomal localization of the gene encoding mouse DNA helicase RecQ helicase protein-like 4. Gene. 2000;261:251–258. doi: 10.1016/s0378-1119(00)00498-4. [DOI] [PubMed] [Google Scholar]
- Park SJ, Lee YJ, Beck BD, Lee SH. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted Loading of Mcm2-7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Diffley JFX. Eukaryotic DNA replication control: Lock and load, then fire. Curr. Opin. Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Schurman SH, Hedayati M, Wang ZM, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, III, et al. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell Cycle Regulation of DNA Replication. Annu. Rev. Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Otsuki M, Ishii Y, Tada S, Enomoto T. RecQ family helicases in genome stability: lessons from gene disruption studies in DT40 cells. Cell Cycle. 2008;7:2472–2478. doi: 10.4161/cc.7.16.6462. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Shimamoto A, Koshiji M, Pedeux R, Rusin M, Spillare EA, Shen JC, Huang LE, Lindor NM, Furuichi Y, et al. Tumor suppressor p53 represses transcription of RECQ4 helicase. Oncogene. 2005;24:1738–1748. doi: 10.1038/sj.onc.1208380. [DOI] [PubMed] [Google Scholar]
- Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- Siitonen HA, Sotkasiira J, Biervliet M, Benmansour A, Capri Y, Cormier-Daire V, Crandall B, Hannula-Jouppi K, Hennekam R, Herzog D, et al. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kohno T, Ishimi Y. DNA Helicase Activity in Purified Human RECQL4 Protein. J. Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tak YS, Araki H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Division. 2007a;2 doi: 10.1186/1747-1028-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007b;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Falaschi A, Vindigni A. The human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol. 2010 doi: 10.1128/MCB.01290-09. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennos EM, Collins M, James WD. Rothmund-Thomson syndrome: Review of the world literature. J. Am. Acad. Dermatol. 1992;27:750–762. doi: 10.1016/0190-9622(92)70249-f. [DOI] [PubMed] [Google Scholar]
- Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp. Cell Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Wu J, Capp C, Feng L, Hsieh TS. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev. Biol. 2008;323:130–142. doi: 10.1016/j.ydbio.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009a;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lei Z, Huang H, Dui W, Liang X, Ma J, Jiao R. dRecQ4 Is Required for DNA Synthesis and Essential for Cell Proliferation in Drosophila. PLoS One. 2009b;4 doi: 10.1371/journal.pone.0006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Kwon YT, Varshavsky A, Wang W. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]