Abstract
AIM: To investigate whether dicoumarol, a potent inhibitor of NAD(P)H quinone oxidoreductase-1 (NQO1), potentiates gemcitabine to induce cytotoxicity in cholangiocarcinoma cells (CCA) and the role of reactive oxygen generation in sensitizing the cells.
METHODS: Four human cell lines with different NQO1 activity were used; the human CCA cell lines, KKU-100, KKU-OCA17, KKU-M214, and Chang liver cells. NQO1 activity and mRNA expression were determined. The cells were pretreated with dicoumarol at relevant concentrations before treatment with gemcitabine. Cytotoxicity was determined by staining with fluorescent dyes. Oxidant formation was examined by assay of cellular glutathione levels and reactive oxygen species production by using dihydrofluorescein diacetate. Measurement of mitochondrial transmembrane potential was performed by using JC-1 fluorescent probe. Western blotting analysis was performed to determine levels of survival related proteins.
RESULTS: Dicoumarol markedly enhanced the cytotoxicity of gemcitabine in KKU-100 and KKU-OCA17, the high NQO1 activity and mRNA expressing cells, but not in the other cells with low NQO1 activity. Dicoumarol induced a marked decrease in cellular redox of glutathione in KKU-100 cells, in contrast to KKU-M214 cells. Dicoumarol at concentrations that inhibited NQO1 activity did not alter mitochondrial transmembrane potential and production of reactive oxygen species. Gemcitabine alone induced activation of NF-κB and Bcl-XL protein expression. However, gemcitabine and dicoumarol combination induced increased p53 and decreased Bcl-XL levels in KKU-100, but not in KKU-M214 cells.
CONCLUSION: NQO1 may be important in sensitizing cells to anticancer drugs and inhibition of NQO1 may be a strategy for the treatment of CCA.
Keywords: NAD(P)H quinone oxidoreductase-1, Dicoumarol, Cholangiocarcinoma, Chemotherapy, Oxidative stress
INTRODUCTION
NAD(P)H quinone oxidoreductase-1 (NQO1 or DT-diaphorase) is a ubiquitous flavoprotein localized widely in body tissues. It is an obligated two-electron reductase that reduces quinones to hydroquinones, thus bypassing the toxic semiquinone intermediates, and these resultant hydroquinones are thus ready for further conjugation and excretion[1]. Several functions of NQO1 have been proposed including xenobiotic detoxification, superoxide scavenging, maintenance of endogenous antioxidant, modulation of p53 and proteasomal degradation[2-4]. It is conceivable that NQO1 functions primarily to protect normal cells from oxidant stress and electrophilic attack. A number of experimental models and epidemiological studies support the concept that intake of dietary phytochemicals confers a cancer chemoprevention effect and these chemicals have been shown to induce increased expression of phase II drug detoxifying enzymes including NQO1[5,6]. The cytoprotective role of NQO1 is supported by reports that disruption of the NQO1 gene or genetic polymorphism increase the risk of chemical-induced toxicity and carcinogenesis[7,8]. The expressions of NQO1 and antioxidant enzymes are recognized as an adaptive response to chemical stress[6,9]. On the other hand, analysis of several solid tumors found an over-expression of the NQO1 gene in cancers of the liver, thyroid, breast, colon, lung, and pancreas[10,11]. Under these circumstances, NQO1 probably functions to protect cancer cells by eliminating oxidant species and making cells resistant to anticancer drugs that induce oxidative injury[12].
Inhibition of NQO1 activity by dicoumarol has been shown to suppress urogenital and pancreatic cancer cell growth and potentiate cytotoxicity of cisplatin and doxorubicin[13,14]. The inhibition of NQO1 with dicoumarol was suggested to stimulate formation of superoxide, oxidative stress and subsequent suppression of pancreatic cancer cell growth and induction of apoptosis[15,16]. However, dicoumarol has been shown to induce formation of reactive oxygen species (ROS) independently from NQO1 activity by inhibition of the mitochondrial electron transport chain[17]. Therefore, the question as to whether inhibition of NQO1 renders cancer cells more sensitive to chemotherapeutic agents is still not clear. More study is necessary to define the role of NQO1 in cancer cells. Sensitizing cancer cells to be more susceptible to radiotherapy or chemotherapy may be an important strategy to overcome resistance in cancer chemotherapy.
Cholangiocarcinomas (CCA) are rare types of liver cancers arising from the biliary duct system. Surgical resection with a histologically free margin is the only chance for cure[18]. Unfortunately, only few patients are eligible for surgery. Furthermore, current chemotherapy and radiotherapy regimens do not substantially improve survival in CCA patients[18,19]. Gemcitabine has been the most important nucleoside analog which has wide spectrum activity against various solid and hematological tumors[20]. It shows some efficacy and is well tolerated in CCA patients[21]. It is essential to gain insight into molecular mechanisms by which cancer cells operate to survive and evade the attack by chemotherapeutic agents.
Because NQO1 appears to be a potential target for exploitation in cancer chemotherapy, we investigated whether dicoumarol, a potent inhibitor of NQO1, sensitized CCA cells to respond to the cytotoxicity of gemcitabine. Furthermore, we examined whether dicoumarol- and gemcitabine-induced cell killing was associated with ROS generation, mitochondrial dysfunction and apoptotic protein expression.
MATERIALS AND METHODS
Human cell line cultures
Three human CCA cell lines established in our institute, KKU-100, KKU-OCA17 and KKU-M214, were derived from human intrahepatic CCA tissues with the histological types of poorly differentiated, well differentiated and moderately differentiated adenocarcinoma, respectively[22,23]. Chang liver cells were also used in the study. CCA cells and Chang liver cells were routinely cultured as previously described[24] in Ham’s F12 media, supplemented with 4 mmol/L L-glutamine, 12.5 mmol/L N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES), at pH 7.3, 100 U/mL penicillin 100 μg/mL streptomycin sulfate and 10% fetal calf serum. The media was renewed every 3 d, trypsinized with 0.25% trypsin-EDTA and subcultured in the same media.
NQO1 activity assay
NQO1 assay was performed essentially according to a previously published method[25]. Cells were seeded onto 96-well cultured plates overnight. Cells were then lysed with 50 μL of 0.8% digitonin in 2 mmol/L EDTA at room temperature for 10 min. The assay was performed using menadione and MTT [3-(4,5-dmethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole] in the substrate coupling reaction and measured as rate-kinetics in a micro-plate reader at a wavelength of 620 nm. The initial velocity of reaction was calculated as NQO1 activity using the extinction coefficient of formazan of MTT of 11 300 L/mol per cm and correction factor for the light path. In the determination of the enzymatic property of NQO1 in each cell line, cells were incubated with varying concentrations of dicoumarol for 10 min before carrying out assays as above.
Cytotoxicity assay
CCA cells (KKU-100, KKU-OCA17, and KKU-M214) and Chang liver cells were seeded onto 96-well cultured plates at a density of 5 × 103 cells/well (KKU-M214 and Chang cells) or 7.5 × 103 cells/well (KKU-100 and KKU-OCA17) for an overnight, then media was renewed with fresh media containing test compound and further incubated for the indicated times. The cytotoxicity was determined by fluorescence microscopy[26]. In brief, cells were washed once with phosphate-buffered saline (PBS) and the following added: 4 μL mixture of acridine orange, ethidium bromide (each 1 μg/mL) and trace amount of hemoglobin. The cells were examined using a Nikon Eclipse TS100 inverted microscope with excitation and emission filters of 480 and 535 nm, respectively. The microphotographs were taken at predetermined three areas per well in triplicate wells per concentration with a Nikon Coolpix digital camera. The numbers of viable and non-viable cells were counted. The viable cells were colored green with intact nuclei. The non-viable cells included necrotic and apoptotic cells which showed bright orange staining and green fluorescence, with appearance of cell shrinkage and condensation and fragmentation of the nuclei, respectively. The antiproliferation value was calculated as = (number of viable cells in control wells - viable cells in treatment wells)/(number of viable cells in control wells) × 100.
Determinations of glutathione and glutathione disulfide
Total glutathione assay was performed essentially according to Tietze methods[27]. Glutathione disulfide (GSSG) was assayed by the previously described method[28] using 1-methyl-2-vinylpyridinium triflate (M2VP) as a glutathione scavenger. Cell cultures were trypsinized and washed three times with cold PBS buffer and centrifuged at 1500 g, 4°C for 10 min and resuspended in PBS buffer. Cell suspension of 100 μL was reacted with M2VP (33 mmol/L in DI) or without M2VP. The solution was mixed gently and stored frozen at -20°C until analysis. An aliquot of cell suspension was saved for protein determination by Bradford’s dye binding assay.
Measurement of mitochondrial transmembrane potential
To measure the change in mitochondrial transmembrane potential (∆Ψm), the lipophilic cation fluorescent dye JC-1 (5,5’,6,6’-tetrachloro-1,1’,3,3’-tetraethyl-benzimidazolylcarbocyanine iodide) was used. After treatment with dicoumarol, gemcitabine or the combination at defined period of times, cultured cells were loaded with JC-1 mitochondrial membrane potential assay kit (Clayman Chemical, Ann Arbor, Michigan) by incubation for 30 min at 37°C. After that, cultured cells were rinsed, incubated in JC-1 assay buffer and mitochondrial transmembrane potential was analyzed by a fluorescent plate reader. In healthy mitochondria, JC-1 forms J-aggregates which display strong fluorescent intensity with excitation and emission wavelength at 560 and 595 nm, respectively. In depolarized mitochondria, JC-1 exists as J monomers which show strong fluorescence with excitation and emission wavelength at 485 and 535 nm, respectively. The shift down in ratio of fluorescent intensity of JC-1 aggregates to fluorescent intensity of monomers is used as an indicator of depolarization of ∆Ψm.
Determination of formation of reactive oxygen species
Reactive oxygen species (ROS) levels generated from cultured cells were determined by incubating the cells in Hank’s buffer supplemented with 15 mmol/L HEPES containing 1.2 μg/mL dihydrofluorescein diacetate (H2-DHFDA) (Sigma-Aldrich, St. Louis, MO) for 30 min at 37°C. H2-DHFDA was taken up into the cells, hydrolyzed and oxidized to the fluorescent product DHF by ROS. The fluorescent signal was determined by a fluorescent plate reader with a setting of the excitation and emission wavelengths at 485 and 520 nm, respectively.
RNA isolation and reverse transcription-polymerase chain reaction
Total RNA was extracted from liver tissues and the four cell lines using Trizol®LS reagent following the manufacturer’s instructions. Total RNA (3 μg) was reverse-transcribed in 20 μL containing 0.5 μg of oligo(dT)15 primer, 20 U of RNasin® ribonuclease inhibitor and 200 U of ImProm-II™ reverse transcriptase in 10 × PCR buffer, 3 mmol/L MgCl2, and 1 mmol/L dNTPs. The first-strand cDNA was synthesized at conditions of 42°C for 60 min. The reverse transcription products served as a template for real-time PCR. PCR amplification was performed using specific primers for the NQO1 and the internal control using FDFT1. The PCR primer sequences were as follows: NQO1; forward primers: 5'GGCAGAAGAGCACTGATCGTA3', NQO1; reverse primers: 5'TGATGGGATTGAAGTTCATGGC3', GenBank accession number BC007659.2, FDFT1; forward primers: 5'TTTAACTTCTGTGCTATTCCAC3', FDFT1; reverse primers: 5'TCTCCAGTCTGAACATAGTC3', GenBank accession number NM_004462.3. The real-time fluorescence PCR, based on SYBR Green, was carried out in a final volume of 20 μL containing 1 × SYBR Green PCR Master Mix (DyNAmo™ Flash SYBR® Green qPCR Kit), 0.5 μmol/L of each NQO1 or FDFT1 primer. Thermal cycling was performed for each gene in duplicate on cDNA samples in 96-well reaction plates using the ABI 7500 Sequence Detection system (Applied Biosystems). A negative control was included in the experimental runs. The negative control was set up by substituting the template with deionized H2O and this routinely had a high Ct value which represented the lower detection limit. Real-time PCR was conducted with the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s and 72°C for 45 s. To verify the purity of the products, a melting curve analysis was produced after each run. Upon completion of 40 PCR amplification cycles, there was a dissociation step of ramping temperature from 60°C to 95°C steadily for 20 min, while the fluorescence signal was continually monitored, for melting curve analysis. The relative expression ratio (R) of target genes is calculated based on efficiency (E) and Ct deviation and expressed in comparison to a reference gene. The corresponding real time PCR efficiencies were calculated according to the equation E = 10[-1/slope]. All data were analyzed using Sequence Detector Software Version 1.4 (Applied Biosystems).
Western blotting analysis of whole cell and nuclear protein extracts
The whole cell lysates and nuclear protein were prepared according to a previous report[29]. Treated KKU-100 and KKU-M214 cells were washed with PBS, collected and lysed at 4°C with cell lysis buffer [20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L Na2EDTA, 1 mmol/L EGTA, 1% Triton, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 μg/mL leupeptin, 1 mmol/L dithiothreitol, 0.1 mmol/L phenylmethylsulfonyl fluoride (PMSF)] with vigorous shaking. Following centrifugation at 10 000 g for 15 min, supernatant was collected and stored at -80°C until use. Nuclear protein was prepared by lysis of cultured cells with hypotonic buffer A [10 mmol/L HEPES-KOH pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L DTT, 0.2 mmol/L PMSF, 0.1 mmol/L EGTA], incubated in an ice bath for 15 min and then 1% NP-40 was added, cells were centrifuged at 12 000 g, 4°C for 15 min, the nuclear pellet was resuspended in ice-cold buffer B (20 mmol/L HEPES-KOH pH 7.9, 25% glycerol, 1.5 mmol/L MgCl2, 420 mmol/L NaCl, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, 0.2 mmol/L PMSF, and 1 mmol/L EGTA), followed by incubation at 0°C for 45 min. After vortex mixing, the suspension was centrifuged at 12 000 g, 4°C for 30 min. The supernatant containing nuclear proteins was stored at -80°C for the NF-κB Western immunoblot analysis.
The protein samples were mixed with 5 × loading dye buffer, heated at 95°C for 5 min and proteins were separated by electrophoresis in 10% SDS-polyacrylamide gel. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes at 50 V for 2 h. The PVDF membranes were blocked for 1 h at room temperature with 5% (w/v) skimmed milk powder in Tris buffer saline (TBS) with 0.1% Tween-20. PVDF membrane was incubated overnight at 4°C with primary antibodies of rabbit polyclonal IgG NF-κB p65 subunits (dilution 1:500) (sc-109: Santa Cruz Biotechnology), mouse monoclonal IgG Bcl-XL (1:1000) (sc-8392), rabbit polyclonal IgG Bax (1:2000) (sc-493), rabbit polyclonal IgG cyclin D1 (1:1500) (sc-718), and mouse monoclonal IgG1 β-actin (1:2500) (sc-8432) diluted with TBS. The primary antibody was then removed and the blots were extensively washed with TBS/Tween-20. Blots were then incubated for 2 h at room temperature with the secondary antibody horseradish peroxidase goat anti-mouse IgG (sc-2005) and goat anti-rabbit IgG (sc-2004) at 1:5000 dilutions in TBS buffer. After removal of the secondary antibody and washes in TBS buffer, the blots were incubated in the ECL substrate solution (Supersignal® West Pico Chemiluminescent Substrate). Densities of the specific bands of NF-κB, Bcl-XL, Bax, cyclin D1, and β-actin were visualized and captured by Imagequant 350 (GE Healthcare).
Statistical analysis
Data are expressed as mean ± SE of duplicate assays from three independent experiments. An analysis of variance with repeated measurement was used to determine significant differences between each experimental group. The level of significance was set at P < 0.05.
RESULTS
Activity and expression of NQO1 in CCA cells
Three CCA cell lines with different histological backgrounds were employed in this study for assessing the status of NQO1. The CCA cells illustrated varying degrees of NQO1 activity and mRNA expression. KKU-100 and KKU-OCA17 cells had high NQO1 activity, whereas KKU-M214 showed low NQO1 activity. Moreover, Chang liver cells, which were derived from normal liver tissue, showed relatively low NQO1 activity and were comparable to KKU-M214 cells (Figure 1A). Consistently, NQO1 mRNA expression in KKU-100 cells showed higher levels than in the other cell lines (Figure 1B).
Figure 1.
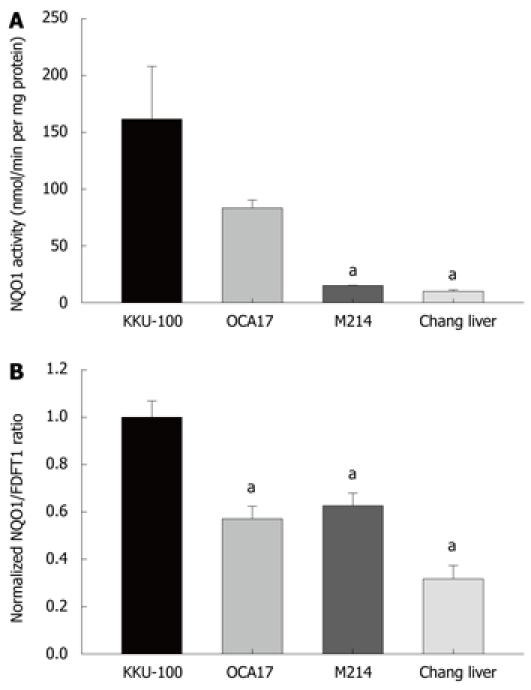
NQO1 activity and mRNA expression of cultured cells. A: NQO1 activity: Cholangiocarcinoma cells, KKU-100, KKU-OCA17, KKU-M214, and Chang liver cells were cultured in 96-well plates for assay of NQO1 activity by enzymatic methods; B: Expression of NQO1 mRNA: The cells were cultured in 6-well plates. Total RNA was extracted by Trizol reagent, converted to cDNA and analyzed by real-time PCR using FDFT1 as internal control. Bars represent mean ± SE, each from 3 experiments. aP < 0.05 vs control group.
Sensitivity of NQO1 to dicoumarol
Dicoumarol is a very potent inhibitor of NQO1 activity. All four cell lines were tested against dicoumarol. NQO1 in CCA and Chang cell lysates were assayed for kinetics of inhibition by dicoumarol. The inhibition of NQO1 activity was rapid, within 10 min, and was apparent with a comparable potency among the 4 cell lines (Figure 2) with IC50 of 0.15 ± 0.07, 0.15 ± 0.07, 0.24 ± 0.14 and 0.10 ± 0.22 μmol/L for KKU-100, KKU-OCA17, KKU-M214 and Chang cells, respectively. Since the enzyme activity in all cell lines was almost completely abolished at a dicoumarol concentration of 10 μmol/L, in subsequent experiments dicoumarol was used at the concentration of 10 μmol/L.
Figure 2.
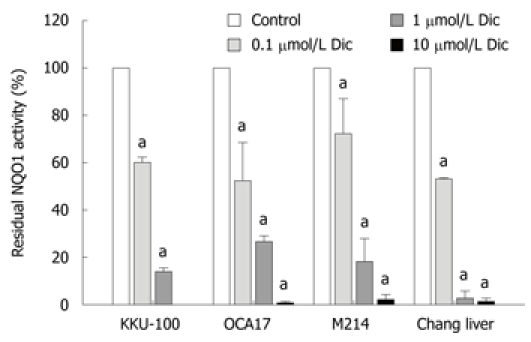
Concentration response of NQO1 inhibition by dicoumarol. KKU-100, KKU-OCA17, KKU-M214, and Chang liver cells were cultured in 96-well plates overnight. The NQO1 activity was assayed in the presence of the inhibitor dicoumarol at concentrations from 0.1-10 μmol/L. Bars represent mean ± SE, each from 3 experiments. aP < 0.05 vs control group.
Enhancement of cytotoxicity of gemcitabine by dicoumarol
To investigate whether inhibition of cellular NQO1 activity was associated with increased anticancer drug sensitivity, CCA cells were preincubated with dicoumarol at varied concentrations for 4 h followed by an addition of gemcitabine at predetermined concentrations. The concentrations of gemcitabine that caused cytotoxicity of about 20%-30% were used in the experiments. Those for KKU-100, KKU-OCA17, KKU-M214 and Chang cells were 1, 10, 1 nmol/L and 10 μmol/L, respectively. Treatment with dicoumarol alone at concentrations of 0.1-10 μmol/L caused modest cytotoxicity after incubation for 24 h. Dicoumarol at a concentration of 10 μmol/L induced ≤ 10% cytotoxicity in all cell types.
Combination of gemcitabine and dicoumarol produced a markedly enhanced cytotoxic effect, particularly in KKU-100 and KKU-OCA17 cells. The enhanced cytotoxic effect in both cell lines was conceivably more than a simple additive effect of the drug and dicoumarol (Figure 3). The cytotoxicity of gemcitabine in KKU-100 cells was enhanced from 34% to 68% and in KKU-OCA17 cells from 25% to 47% in the presence of dicoumarol. On the other hand, the drug combination produced only an additive cytotoxicity in KKU-M214 and Chang cells, i.e. increased from 23% to 36% and 23% to 40%, respectively. It is noted that KUU-100 and KKU-OCA17 cells are high NQO1 activity cells when compared with KKU-M214 and Chang cells.
Figure 3.
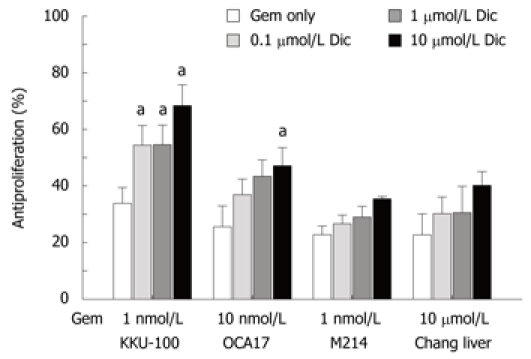
Potentiation of cytotoxicity of gemcitabine by dicoumarol. KKU-100, KKU-OCA17, KKU-M214, and Chang liver cells were cultured in 96-well plates overnight. Cultured cells were pretreated with dicoumarol (0.1-10 μmol/L) for 4 h before treatment with gemcitabine at 1 nmol/L, 10 nmol/L, 1 nmol/L and 10 μmol/L, for KKU-100, KKU-OCA17, KKU-M214, and Chang liver cells, respectively. Antiproliferation was analyzed by staining the cells with fluorescent dyes before examination under fluorescent microscope. Bars represent mean ± SE, each from 3 experiments. aP < 0.05 vs control group.
Dicoumarol-induced oxidative stress in CCA cells
The interactive effect of dicoumarol with gemcitabine in enhancing cytotoxicity was explored with regard to whether dicoumarol induced cellular stress, rendering the cells more susceptible to gemcitabine. In this study, KKU-100 and KKU-M214 cells were employed as representatives of high and low NQO1 activity cells, respectively. The basal total GSH levels in KKU-M214 were higher than that in KKU-100 cells and significantly increased by the treatment with dicoumarol (10 μmol/L) (Figure 4A). On the other hand, glutathione disulfide (GSSG) levels in KKU-100 were dramatically increased with the treatment of dicoumarol, whereas the levels in KKU-M214 were decreased (Figure 4B). This indicates that the two cell types have different oxidative responses after the treatment with dicoumarol.
Figure 4.
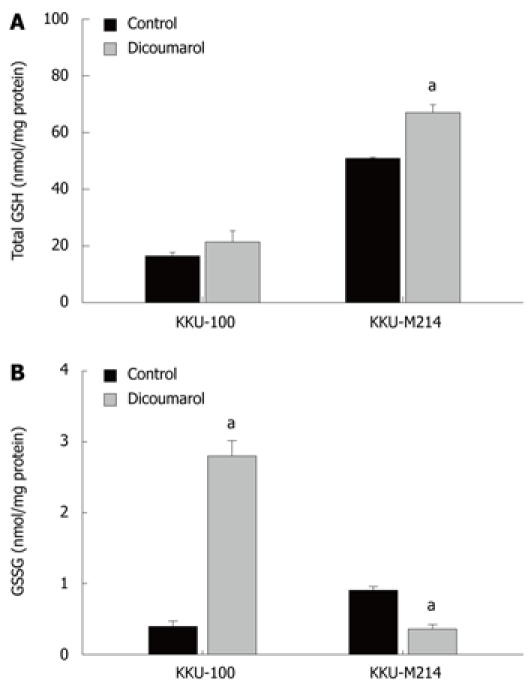
Glutathione redox status in cells treated with dicoumarol. KKU-100 and KKU-M214 cells were cultured and treated with dicoumarol (10 μmol/L) for 4 h, and cells were scraped for assays of (A) total glutathione and (B) glutathione disulfide. Bars represent mean ± SE, each from 3 experiments. aP < 0.05 vs control group.
Determination of the mitochondrial transmembrane potential and formation of reactive oxygen species
Disruption of ∆Ψm is recognized as one of the critical steps leading to apoptotic cell death. The integrity of the inner mitochondrial membrane can be assessed by monitoring of the potential gradient across the membrane using the fluorescent dye JC-1. There was no change in ∆Ψm in cells after treatment with dicoumarol, gemcitabine or the drug combination in both cell types, at an early time (6 h) as well as at 24 h (Figure 5A and B). In contrast, dicoumarol at excessive concentrations (50 and 150 μmol/L) showed a rapidly depolarized ∆Ψm, as shown by decrease in the ratio of fluorescent intensity of JC-1 aggregates/JC-1 monomers after 3 h incubation (Figure 5C). The formation of ROS by cultured cells was monitored by a fluorescent dye, DHFDA. It was apparent that there was no increase in ROS formation in cells treated with dicoumarol, gemcitabine or the combination in both cell lines (data not shown), even at a concentration and time where cell apoptosis was evident. High concentrations of dicoumarol also did not induce ROS production, even at those concentrations which caused cell death.
Figure 5.

Assay of mitochondrial transmembrane potential and reactive oxygen in CCA cells. The mitochondrial transmembrane potential was analyzed by using JC-1 fluorescent probe. Fluorescent readings of the J-aggregates and J monomers were used as measurement of mitochondrial transmembrane potential. KKU-100 and KKU-M214 cells were cultured in 96-well black plates. The cultured cells were pretreated with dicoumarol at 10 μmol/L for 4 h, then gemcitabine at 1 nmol/L was added and incubated at various times. A: Incubation for 6 h; B: Incubation for 24 h; C: Other cultured cells were treated with dicoumarol at 50 and 150 μmol/L for 3 h. Bars represent mean ± SE, each from triplicate assay. aP < 0.05 vs control group.
Alteration of survival response proteins induced by gemcitabine and dicoumarol
In order to understand how the combination of gemcitabine and dicoumarol enhanced cytotoxicity in high NQO1 activity cells, proteins related to cell survival were analyzed by Western immunoblotting. Protein p53 is a tumor suppressor protein that responds to various noxious stimuli. Dicoumarol or gemcitabine treatment induced increased p53 levels in KKU-100 cells and the level was further increased during the treatment with the drug combination. On the other hand, dicoumarol or gemcitabine or the combination did not alter p53 levels in KKU-M214 cells. Bcl-2 proteins consist of proapoptotic (Bax) and antiapoptotic proteins (Bcl-XL) where they regulate mitochondria outer membrane permeabilization. Bcl-XL levels were induced by gemcitabine in both cell types, however, the combination of gemcitabine with dicoumarol suppressed elevated Bcl-XL levels in KKU-100 cells but not in KKU-M214 cells. On the other hand, the changes in Bax levels were found with a similar pattern in both cell types, i.e. Bax levels were induced with gemcitabine, while drug combination induced no changes when compared with controls. Cyclin D1 levels were decreased upon the treatment with dicoumarol in both cell types, but not by gemcitabine. However, the drug combination apparently suppressed cyclin D1 levels when compared with gemcitabine alone.
The expression of the nuclear p65 subunit of NF-κB was slightly increased after the treatment with gemcitabine and showed little change induced by the co-treatment with dicoumarol in both cell lines. Figure 6 shown the Western analysis of proteins related to survival.
Figure 6.
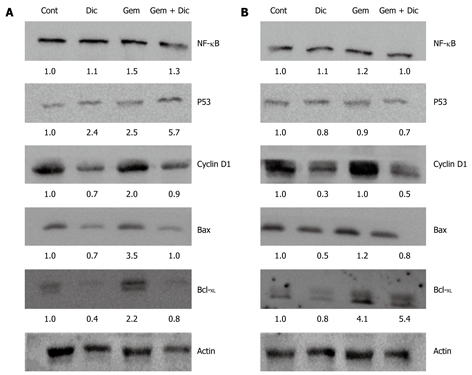
Western blotting analysis of proteins related to survival. KKU-100 and KKU-M214 cells were cultured overnight, pretreated with 10 μmol/L dicoumarol for 4 h before being treated with 1 nmol/L gemcitabine for 24 h. Cultured cells were collected for Western blotting analysis using β-actin as an internal control for equal protein loading. A: KKU-100 cells; B: KKU-M214 cells. Cont: Controls; Dic: Dicoumarol alone; Gem: Gemcitabine alone; Gem + Dic: Combination of gemcitabine and dicoumarol. Values were an average from two experiments of the target protein normalized with the internal control.
DISCUSSION
NQO1 is one of the attractive targets in development of chemotherapy, since NQO1 may function to protect normal cells, as well as tumor cells, particularly when it is highly expressed[12,30]. Suppression of NQO1 induced by proinflammatory cytokines or genetic defects may render normal cells susceptible to oxidant-mediated cell toxicity and carcinogenesis[7,8,22]. Inhibition of NQO1 activity with dicoumarol at relevant concentrations which produced modest cytotoxicity when administered alone potentiated gemcitabine-induced cytotoxicity in cells with high NQO1 activity. The combination of dicoumarol with gemcitabine enhanced p53 and decreased Bcl-XL expression, and this may be related to the mechanism of cell sensitization and killing.
NQO1 is well known to function as a drug metabolizing and antioxidant enzyme, protecting the cell from oxidative injury[3]. Previous studies suggested that NQO1 modulates p53 expression by interference with 20S proteasome-mediated degradation of p53[3,31]. P53 is a tumor suppressor gene that upon stimulation by DNA damage or oxidative stress, induces either growth arrest or apoptosis[32]. Our study showed that dicoumarol increased levels of p53 in KKU-100 cells, but these levels were unchanged in KKU-M214 cells. The increased p53 expression, together with decreased Bcl-XL protein expression, was associated with enhanced cytotoxicity of gemcitabine. However, dicoumarol has been previously reported to decrease p53 protein levels and this was associated with either inhibition or induction of apoptosis in myeloid leukemic cells or urogenital cancer cells, respectively[14,31]. The discrepancy in p53 levels induced by the treatment with dicoumarol is not readily comprehensible, although our study used much lower concentrations of dicoumarol. Moreover, treatment of gemcitabine apparently induced the activation of NF-κB and expression of Bcl-XL. This increase of antiapoptotic proteins may be a survival signal engaged in response to gemcitabine[33]. Interestingly, the combination with dicoumarol diminished the expression of Bcl-XL in KKU-100 cells concurrent with potentiation of the cytotoxicity of gemicitabine, while this did not happen in KKU-M214 cells.
Protein p53 and the Bcl-2 protein family regulate mitochondrial outer membrane permeabilization[34]. Induction of apoptotic cell death is often associated with disruption of the mitochondrial inner membrane which is reflected as dissipation of ∆Ψm and formation of mitochondrial permeability transition pore (MPTP) with subsequent cell death[35]. In the treatment with low concentrations of dicoumarol with or without gemcitabine, ∆Ψm appeared to be well maintained according to the assay of JC-1, despite the drug combination inducing marked CCA cell killing. This suggests no disruption of the mitochondrial inner membrane at the concentrations that potentiate gemcitabine cytotoxicity. Likewise, it was shown that prostaglandin A2 induced human promyelocytic leukemia cell death associated with cytochrome c release, with no change in ∆Ψm[36]. However, dicoumarol at the high concentrations caused disruption of ∆Ψm and rapid cell killing which is consistent with many previous reports[14,17].
Dicoumarol has been suggested to trigger cell killing via increased formation of ROS[15-17]. We did not observe an increased ROS formation when cells were treated with dicoumarol or dicoumarol with gemcitabine combination. Dicoumarol, even at high concentrations (150 μmol/L) that rapidly killed both types of CCA cells within 3 h of incubation (data not shown), had no significant effect on ROS formation. This suggests that superoxide formation may not be an important mediator of cell killing by dicoumarol[17]; it is probable that dicoumarol exerts its cell killing mechanism on CCA cells differently from other epithelial cells.
GSH, a tripeptide found abundantly in cells, functions as antioxidant and regulates activities of various redox-sensitive proteins[37]. Although dicoumarol did not increase formation of ROS, it induced oxidant stress in KKU-100 cells, but not in KKU-M214 cells, evidently by increasing and decreasing GSSG levels, respectively. The increased cellular oxidant may be implicated in several signal transduction pathways which eventually lead to enhanced susceptibility of KKU-100 cells. Nonetheless, dicoumarol has been reported to show several different pharmacological activities[15,17,38] and apoptotic cell killing may be dissociated with NQO1 inhibition[39], as some synthetic mechanism-based NQO1 inhibitors have shown lack of association between enzyme inhibition and cytotoxic effect. The discrepancy of the reports may be explicable partly on the grounds that the sensitizing effect of dicoumarol may be dependent on inherent activity of NQO1 in these cell types.
In conclusion, NQO1 may play roles in the sensitivity of CCA cells to gemcitabine. When using dicoumarol at relevant concentrations to inhibit NQO1 activity, dicoumarol enhances gemcitabine cytotoxicity in high NQO1 activity CCA cells. Furthermore, the mechanism of dicoumarol-induced cell killing may not be mediated via disruption of mitochondrial function and formation of ROS, but may be related to suppression of the pro-survival response to the chemotherapy. NQO1 is a potential target in the development of chemotherapy for tumors with high enzyme expression.
COMMENTS
Background
NAD(P)H quinone oxidoreductase-1 (NQO1) is a drug metabolizing and antioxidant enzyme which plays important roles in protection of cells against electrophiles and oxidants. Some cancers exhibit high activity of NQO1 which suggests a cytoprotective role in the cancer.
Research frontiers
Inhibition of NQO1 activity may abate the adaptive survival response of cancer cells induced by gemcitabine therapy and render cancer cells more susceptible to the drug.
Innovations and breakthroughs
This report shows that dicoumarol, a potent inhibitor of NQO1 at low concentrations, potentiates cytotoxicity of gemcitabine in cholangiocarcinoma cells which inherently have high NQO1 activity. Disruption of cellular glutathione redox may be associated with modulation of redox-sensitive signaling proteins such as p53 and render enhanced cells susceptible to gemcitabine.
Applications
Inhibition of NQO1 is suggested to be a potential strategy for development in cancer chemotherapy, particularly for cholangiocarcinoma which was used in this study.
Peer review
The methodology and the results are appropriate and support the findings in the literature.
Footnotes
Supported by Thailand Research Fund, National Science and Technology Development Agency, research funding from Khon Kaen University; the Royal Golden Jubilee Ph.D. Program (to Kongpetch S); the Office of the Commission on Higher Education (to Buranrat B)
Peer reviewer: Ismail Matalka, MD, FRCPath, Professor, Department of Pathology and Laboratory Medicine, King Abdullah University Hospital and School of Medicine, Jordan University of Science and Technology, Irbid, Jordan
S- Editor Wang JL L- Editor Logan S E- Editor Ma WH
References
- 1.Talalay P, Dinkova-Kostova AT. Role of nicotinamide quinone oxidoreductase 1 (NQO1) in protection against toxicity of electrophiles and reactive oxygen intermediates. Methods Enzymol. 2004;382:355–364. doi: 10.1016/S0076-6879(04)82019-6. [DOI] [PubMed] [Google Scholar]
- 2.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nioi P, Hayes JD. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res. 2004;555:149–171. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, Berliner LJ, Ross D. NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol. 2004;65:1238–1247. doi: 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- 5.Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Lin W, Keum YS, Liew C, et al. Pharmacogenomics of phenolic antioxidant butylated hydroxyanisole (BHA) in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Pharm Res. 2006;23:2621–2637. doi: 10.1007/s11095-006-9099-x. [DOI] [PubMed] [Google Scholar]
- 6.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 7.Saldivar SJ, Wang Y, Zhao H, Shao L, Lin J, Spitz MR, Wu X. An association between a NQO1 genetic polymorphism and risk of lung cancer. Mutat Res. 2005;582:71–78. doi: 10.1016/j.mrgentox.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Radjendirane V, Joseph P, Lee YH, Kimura S, Klein-Szanto AJ, Gonzalez FJ, Jaiswal AK. Disruption of the DT diaphorase (NQO1) gene in mice leads to increased menadione toxicity. J Biol Chem. 1998;273:7382–7389. doi: 10.1074/jbc.273.13.7382. [DOI] [PubMed] [Google Scholar]
- 9.Aleksunes LM, Goedken M, Manautou JE. Up-regulation of NAD(P)H quinone oxidoreductase 1 during human liver injury. World J Gastroenterol. 2006;12:1937–1940. doi: 10.3748/wjg.v12.i12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 11.Siegel D, Ross D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic Biol Med. 2000;29:246–253. doi: 10.1016/s0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 12.Hu L, Miao W, Loignon M, Kandouz M, Batist G. Putative chemopreventive molecules can increase Nrf2-regulated cell defense in some human cancer cell lines, resulting in resistance to common cytotoxic therapies. Cancer Chemother Pharmacol. 2009:Epub ahead of print. doi: 10.1007/s00280-009-1182-7. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe J, Nishiyama H, Matsui Y, Ito M, Kawanishi H, Kamoto T, Ogawa O. Dicoumarol potentiates cisplatin-induced apoptosis mediated by c-Jun N-terminal kinase in p53 wild-type urogenital cancer cell lines. Oncogene. 2006;25:2500–2508. doi: 10.1038/sj.onc.1209162. [DOI] [PubMed] [Google Scholar]
- 14.Matsui Y, Watanabe J, Ding S, Nishizawa K, Kajita Y, Ichioka K, Saito R, Kobayashi T, Ogawa O, Nishiyama H. Dicoumarol enhances doxorubicin-induced cytotoxicity in p53 wild-type urothelial cancer cells through p38 activation. BJU Int. 2010;105:558–564. doi: 10.1111/j.1464-410X.2009.08732.x. [DOI] [PubMed] [Google Scholar]
- 15.Cullen JJ, Hinkhouse MM, Grady M, Gaut AW, Liu J, Zhang YP, Weydert CJ, Domann FE, Oberley LW. Dicumarol inhibition of NADPH:quinone oxidoreductase induces growth inhibition of pancreatic cancer via a superoxide-mediated mechanism. Cancer Res. 2003;63:5513–5520. [PubMed] [Google Scholar]
- 16.Lewis A, Ough M, Li L, Hinkhouse MM, Ritchie JM, Spitz DR, Cullen JJ. Treatment of pancreatic cancer cells with dicumarol induces cytotoxicity and oxidative stress. Clin Cancer Res. 2004;10:4550–4558. doi: 10.1158/1078-0432.CCR-03-0667. [DOI] [PubMed] [Google Scholar]
- 17.Du J, Daniels DH, Asbury C, Venkataraman S, Liu J, Spitz DR, Oberley LW, Cullen JJ. Mitochondrial production of reactive oxygen species mediate dicumarol-induced cytotoxicity in cancer cells. J Biol Chem. 2006;281:37416–37426. doi: 10.1074/jbc.M605063200. [DOI] [PubMed] [Google Scholar]
- 18.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 19.Uttaravichien T, Bhudhisawasdi V, Pairojkul C, Pugkhem A. Intrahepatic cholangiocarcinoma in Thailand. J Hepatobiliary Pancreat Surg. 1999;6:128–135. doi: 10.1007/s005340050095. [DOI] [PubMed] [Google Scholar]
- 20.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 Suppl 5:v7–v12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 21.Valle JW, Wasan H, Johnson P, Jones E, Dixon L, Swindell R, Baka S, Maraveyas A, Corrie P, Falk S, et al. Gemcitabine alone or in combination with cisplatin in patients with advanced or metastatic cholangiocarcinomas or other biliary tract tumours: a multicentre randomised phase II study - The UK ABC-01 Study. Br J Cancer. 2009;101:621–627. doi: 10.1038/sj.bjc.6605211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prawan A, Buranrat B, Kukongviriyapan U, Sripa B, Kukongviriyapan V. Inflammatory cytokines suppress NAD(P)H:quinone oxidoreductase-1 and induce oxidative stress in cholangiocarcinoma cells. J Cancer Res Clin Oncol. 2009;135:515–522. doi: 10.1007/s00432-008-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sripa B, Leungwattanawanit S, Nitta T, Wongkham C, Bhudhisawasdi V, Puapairoj A, Sripa C, Miwa M. Establishment and characterization of an opisthorchiasis-associated cholangiocarcinoma cell line (KKU-100) World J Gastroenterol. 2005;11:3392–3397. doi: 10.3748/wjg.v11.i22.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buranrat B, Prawan A, Sripa B, Kukongviriyapan V. Inflammatory cytokines suppress arylamine N-acetyltransferase 1 in cholangiocarcinoma cells. World J Gastroenterol. 2007;13:6219–6225. doi: 10.3748/wjg.v13.i46.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 26.Ribble D, Goldstein NB, Norris DA, Shellman YG. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005;5:12. doi: 10.1186/1472-6750-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 28.Somparn N, Kukongviriyapan U, Tassaneeyakul W, Jetsrisuparb A, Kukongviriyapan V. Modification of CYP2E1 and CYP3A4 activities in haemoglobin E-beta thalassemia patients. Eur J Clin Pharmacol. 2007;63:43–50. doi: 10.1007/s00228-006-0224-x. [DOI] [PubMed] [Google Scholar]
- 29.Jang JH, Surh YJ. Bcl-2 attenuation of oxidative cell death is associated with up-regulation of gamma-glutamylcysteine ligase via constitutive NF-kappaB activation. J Biol Chem. 2004;279:38779–38786. doi: 10.1074/jbc.M406371200. [DOI] [PubMed] [Google Scholar]
- 30.Dinkova-Kostova AT, Talalay P. Persuasive evidence that quinone reductase type 1 (DT diaphorase) protects cells against the toxicity of electrophiles and reactive forms of oxygen. Free Radic Biol Med. 2000;29:231–240. doi: 10.1016/s0891-5849(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 31.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci USA. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouchet BP, de Fromentel CC, Puisieux A, Galmarini CM. p53 as a target for anti-cancer drug development. Crit Rev Oncol Hematol. 2006;58:190–207. doi: 10.1016/j.critrevonc.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Fahy BN, Schlieman MG, Virudachalam S, Bold RJ. Inhibition of AKT abrogates chemotherapy-induced NF-kappaB survival mechanisms: implications for therapy in pancreatic cancer. J Am Coll Surg. 2004;198:591–599. doi: 10.1016/j.jamcollsurg.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 35.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. 2006;13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 36.Lee SY, Ahn JH, Ko KW, Kim J, Jeong SW, Kim IK, Kim J, Kim HS. Prostaglandin A2 activates intrinsic apoptotic pathway by direct interaction with mitochondria in HL-60 cells. Prostaglandins Other Lipid Mediat. 2010;91:30–37. doi: 10.1016/j.prostaglandins.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 38.Madari H, Panda D, Wilson L, Jacobs RS. Dicoumarol: a unique microtubule stabilizing natural product that is synergistic with Taxol. Cancer Res. 2003;63:1214–1220. [PubMed] [Google Scholar]
- 39.Dehn DL, Siegel D, Zafar KS, Reigan P, Swann E, Moody CJ, Ross D. 5-Methoxy-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]indole-4,7-dione, a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1, exhibits activity against human pancreatic cancer in vitro and in vivo. Mol Cancer Ther. 2006;5:1702–1709. doi: 10.1158/1535-7163.MCT-06-0105. [DOI] [PubMed] [Google Scholar]


