Abstract
BACKGROUND
Current laboratory methods for monitoring response to therapy for tuberculosis (TB) rely on mycobacterial culture. Their clinical usefulness is therefore limited by the slow growth rate of Mycobacterium tuberculosis. Rapid methods to reliably quantify response to anti-TB drug efficacy are desirable.
METHODS
We developed two real-time PCR assays using hydrolysis probes to target DNA of the IS6110 insertion element and mRNA of antigen 85B, respectively, extracted directly from sodium hydroxide-N-acetyl-L-cysteine decontaminated and concentrated sputum specimens. We prospectively compared these assays to sputum mycobacterial culture in patients receiving anti-TB therapy.
RESULTS
Sixty-five patients with newly diagnosed tuberculosis and receiving a standardized first-line anti-TB drug regimen were evaluated at week 2, and months 1, 2 and 4 after initiating therapy. Both DNA PCR (98.5% positive) and mRNA RT-PCR (95.4% positive) were better than standard Ziehl-Neelsen staining techniques (83.1%) for detecting M. tuberculosis in culture-positive sputum specimens. Overall agreement between culture and mRNA RT-PCR among all 286 sputum specimens was 87.1%, and compared to culture the mRNA RT-PCR diagnostic sensitivity and specificity were 85.2% and 88.6%, respectively. For monitoring efficacy of therapy, mRNA RT-PCR results paralleled culture results at the follow-up time points.
CONCLUSIONS
The continued presence of viable M. tuberculosis by culture and antigen 85B mRNA by RT-PCR correlated clinically with anti-TB drug resistance, whereas the DNA PCR assay showed a high false positive rate. This mRNA RT-PCR assay may allow rapid monitoring of response to anti-TB therapy.
Keywords: Mycobacterium tuberculosis, mRNA, culture
Introduction
Mycobacterium tuberculosis grows slowly and requires several weeks to detect in clinical specimens using standard culture techniques. This may delay the microbiologic diagnosis of TB as compared to most other bacterial infections. The slow growth of M. tuberculosis also delays the availability of drug susceptibility assay results, which may be necessary to guide therapy. Conversion of sputum culture from positive to negative within the initial weeks or months of therapy correlates with the sterilizing activity (i.e. killing of M. tuberculosis in tissues) of the anti-TB drug regimen, and is considered the best predictor of treatment success (1). Effective treatment regimens rapidly decrease the number of viable M. tuberculosis organisms in sputum, with the number of cultivable bacilli typically reduced by approximately 10-fold within the first 1 to 2 weeks (2). However, because of the slow growth rate of M. tuberculosis, results of early on-treatment cultures, if such cultures were obtained, would not be available in a timely manner.
Anti-TB therapy regimen selection is largely empiric. Patients are generally prescribed a standard first line anti-TB regimen comprised of four antimicrobial agents. Treatment may be modified weeks or months later as results of antimicrobial susceptibility tests become available. A rapid, raliable method that reflects effective anti-TB drug activity is extremely desirable. In recent years tests based on nucleic acid amplification techniques have been developed for the direct detection of M. tuberculosis in clinical specimens (3–9). Molecular techniques have the potential to improve clinical care through a dramatic reduction in the time required for detection and may provide substantial savings in the overall cost of care of a patient (4, 10). Because the half-life of bacterial mRNA is extremely short compared to rRNA or genomic DNA, assays that target mycobacterial mRNA better reflect mycobacterial viability (11). The ability of mRNA-based assays to distinguish viable from nonviable organisms suggests that such assays should also be useful in monitoring the efficacy of anti-TB therapy (12), and others have described their use in this context (12–16).
In 2006, 142 TB cases per 100.000 population were reported in the Republic of Georgia, among the highest rates in the WHO European region. Although case detection rate for new smear-positive TB cases has increased in recent years, the treatment success rate remains low at only 73% of new smear-positive cases in the 2005 cohort (17). A major challenge for TB control is the high rate of multidrug-resistant (MDR) TB. In 2006 MDR TB strains caused 6.8% of new cases of active TB, while 26.4% of cases in previously treated individuals were caused by MDR strains (18–20). Approximately 25% of all TB cases are managed in Tbilisi, the capital city.
We have developed two real-time PCR assays using hydrolysis probes to detect M. tuberculosis-specific DNA and mRNA directly from sputum specimens. In a prospective study we evaluated assay performance for monitoring response to anti-TB therapy compared to mycobacterial culture among patients referred to the National Center of Tuberculosis and Lung Diseases (NCTLD).
Materials and Methods
STUDY COHORT
The study cohort included new cases of pulmonary TB managed at the inpatient and outatient departments of the NCTLD. Participants were >15 years of age and resided in Tbilisi. In accordance with the WHO definition, a new case was defined as a patient who had never had treatment for TB or who had taken antituberculosis drugs for less than one month (21). All patients in this study were enrolled within the first or second day of treatment initiation. Each new case of TB was diagnosed and recruited initially from clinical signs, symptoms, and chest radiograph, and confirmed in all cases by the growth of M. tuberculosis from sputum culture. Patients were enrolled into the study from January 2006 through June 2007. The study was approved by the Georgian NCTLD Ethics Committee, and all participants provided written inform consent prior to study entry. A questionnaire was used to assess clinical symptoms, history of contact with an active TB case, previous diagnosis of TB, and demographic information in accordance with current standard practices at NCTLD. Other clinical examinations and procedures, such as radiographic studies, blood hematology, smear/stain/culture examination, and drug susceptibility testing were conducted according to the National Guidelines. HIV status was also checked for all enrolled patients. Patients were treated with isoniazid (INH), rifampin (RIF), ethambutol (EMB) and either streptomycin (STM) or pyrazinamide (PZA), and evaluated every two weeks initially and monthly thereafter. Thirteen patients who were enrolled and started on therapy were subsequently found to have negative TB cultures and were excluded from study analysis.
SPECIMEN COLLECTION AND PROCESSING
Sputum specimens were collected pre-therapy, at week 2, and 1, 2, and 4 months after initiating anti-TB therapy. Standard Ziehl-Neelsen techniques using carbol fushin stain were used to detect acid-fast bacilli (AFB) in sputum smears. For mycobacterial culture, sputum specimens were decontaminated with sodium hydroxide-N-acetyl-L-cysteine (NaOH-NALC), neutralized with hydrochloric acid, and centrifuged at 3,000 ×g for 20 min. Treated specimens were incubated on Lowenstein-Jensen medium for 8 weeks at 37°C. Presumptive M. tuberculosis isolates were initially speciated based on rapidity of growth and ability to grow on selective media as previously described (20). For each 0.2 ml NaOH-NALC treated specimen, 0.9 mL of Lysis Buffer (bioMerieux Inc., Durham, NC) was added and the mixture stored and shipped at 4°C to Vanderbilt University Medical Center for nucleic acid extraction and PCR testing (see below). Potential amplicon carryover contamination was diminished by uracil N-glycosylase-based chemical modification (5, 22).
RECOMBINANT PLASMIDS AND CLINICAL ISOLATES
The IS6110- and 85B-specific fragments were generated by real-time PCR amplification (see below) and were subsequently cloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA). The DNA concentration of the recombinant plasmid standards was calibrated by spectrophotometry at 260nm (22). Each inserted plasmid was adjusted to 10,000 copies/μL and stored at −80°C. A panel of 12 M. tuberculosis complex strains, which were either ATCC strains or well characterized clinical isolates, was included in the study. Non-M. tuberculosis complex strains, including M. avium-intracelluare complex, M. kansasii, M. scrofulaceum, M. paratuberculosis, and M. marinum, were included in the study for analytical specificity determination (23).
PHENOTYPIC ANTI-MYCOBACTERIAL SUSCEPTIBILITY TESTING
Antimicrobial susceptibility testing for anti-mycobacterial drugs was performed using the absolute concentrations method on Lowenstein-Jensen agar slants (24). In brief, mycobacterial suspension was performed from the primary culture and the turbidity adjusted to 1 McFarland standard with sterile saline. A series of 10-fold dilutions were prepared and 0.2 ml inoculated onto the following first-line TB drug-containing media: STM (4 mg/L); RIF (40 mg/L); and EMB (2 mg/L). The INH (0.2 mg/L) containing media was inoculated with 0.2 ml of a 100-fold dilution of the suspension. All inoculated sets were incubated at 37°C for 28–42 days. Isolates resistant to both INH and RIF were defined as MDR-TB.
NUCLEIC ACID EXTRACTION
Total nucleic acids were extracted from NaOH-NALC decontaminated and concentrated sputum specimens using the NucliSens easyMAG system (bioMerieux Inc., Durham, NC). Briefly, 200 μL of the Lysis Buffer/NaOH-NALC-treated sputum specimen mixture was placed in the instrument using the default extraction protocol (25). Total nucleic acids were eluted in 55 μL of Elution Buffer (bioMerieux Inc.) and 5μL of each extract was used for nucleic acid amplification. The recombinant plasmids were serially diluted in triplicate by pooled negative sputum specimens prior to the nucleic acid extraction. Human β-actin gene was amplified as an internal amplification control. The primers and fluorophore hydrolysis probes for human β-actin gene and real-time PCR protocol were published previously (22).
REAL-TIME DNA PCR ASSAY
A real-time assay targeting the M. tuberculosis-specific IS6110 insertion element was used (26). Briefly, 5μl of extracted nucleicacid was added to 20 μL of reaction mixture containing 0.8 μM of each primer and 0.4 μM fluorophore hydrolysis probe (final concentration), and mixed with 25 μL of TaqMan Universal PCR Master Mix (Applied Biosystems). The thermocycling conditions were a 2 min degradation of the pre-amplified templates at 50 °C and then 40 cycles of denaturation at 95 °C for 15 s and annealing and extension at 58 °C for 60 s (22). Primers (MTB-IS6110-791F: 5′-TAA CCG GCT GTG GGT AGC A-3′; MTB-IS6110-864R: 5′-CGG TGA CAA AGG CCA CGT A-3′) and the fluorophore hydrolysis probe (MTB-IS6110-830MGB: 5′-CTG GGC AGG GTT C-3′) were modified from those published previously (5, 27). Probes were dual labeled with the reporter dye FAM (6-carboxyfluorescein) at the 5′ end and the quencher MGB (minor groove binder) at the 3′ end. The assay was performed using an ABI PRISM 7700 Sequence Detection System (Applied Biosystems).
REAL-TIME mRNA RT-PCR ASSAY
A real-time assay targeting the M. tuberculosis-specific antigen 85B gene was used (28). In brief, 25 μl reaction mixture containing 5 μl extracted total nucleic acids, 0.5 μM each primer, and 0.2μM hydrolysis probe was mixed with 25 μl TaqMan One-Step RT-PCR 2× Master Mix (Applied Biosystems). Reaction conditions were designed as follows: RT at 48 °C for 30 min, initial denaturation at 95°C for 10 min, and 40 cycles of denaturation (95 °C for 15 s) and annealing/extension (60 °C for 1 min) (29). Primers (MTB-85B-693F: 5′-CGA CCC TAC GCA GCA GAT C-3′; MTB-85B-758R: 5′-TTC CCG CAA TAA ACC CAT AGC-3′) and the fluorophore hydrolysis probe (MTB-85B-719MGB: 5′-TGG TCG CAA ACA ACA C-3′) were modified from those published previously (14). Probes were dual labeled with the reporter dye FAM at the 5′ end and the quencher MGB at the 3′ end. The assay was performed using an ABI PRISM 7700 Sequence Detection System.
STATISTICAL ANALYSIS
Qualitative results were obtained for each specimen tested and PCR efficiencies were calculated as guided (23). Specimens with quantification cycle (Cq) values between 38 and 40 were repeated and considered positive if the repeat Cq value was ≤ 38. Groups were compared by chi-squared, Fisher’s exact and McNemar tests using Epi Info software (version 3.4; Centers for Disease Controland Prevention, Atlanta, GA) or SAS (version 9.1; SAS Institute Inc., Cary, NC). Odd ratios (OR), 95% confidence limits (CI) and P values were calculated; p values ≤0.05 were considered statistically significant.
Results
Experiments were first performed to determine the assay limit of detection by testing the recombinant plasmid standards spiked with pooled negative sputum specimens. Plasmids covering 0–1,024 copies/reaction at 2-fold dilutions were included in the experiment and each dilution was performed in triplicate. The limits of detection of the two PCR assays for the IS6110- and 85B-specific fragments were the same at 4 copies per reaction, which was equivalent to 1,000 copies/ml of sputum. Standard curves are shown in Supplemental Figures 1 and 2. PCR efficiencies were 1.91 for IS6110- and 1.66 for 85B-specific targets respectively. Both PCR assays were specific for M. tuberculosis strains, and samples spiked with mycobacteria other than tuberculosis including M. avium-intracellulare, M. kansasii, M. scrofulaceum, M. paratuberculosis, and M. marinum were not detected by PCR.
During the 1.5 year study period, 65 individuals with culture-confirmed TB enrolled into the study. The study group included 36 males and 29 females, with a mean age of 36.5 ± 13.7 years. All enrolled patients were sero-negative for HIV, had newly diagnosed TB, and had either never previously taken TB drugs or had only started TB drugs within two days prior to recruitment into the study. Follow-up sputum specimens were available at week 2, and 1, 2 and 4 months of anti-TB therapy from 65, 63, 56 and 37 individuals, respectively.
Human β-actin gene was amplified in all nucleic acid extracts from NaOH-NALC-decontaminated and concentrated sputum and spiked plasmid specimens, indicating that no total inhibition occurred in the nucleic acid amplification reaction. For sputum specimens collected before treatment and at each follow-up visit, total nucleic acids were tested by both DNA PCR and mRNA RT-PCR assays with hydrolysis probes. Of the 65 sputum specimens collected at the recruitment visit, 54 were AFB smear-positive, whereas 64 and 62 were positive by DNA PCR and mRNA RT-PCR, respectively. The diagnostic sensitivities of DNA PCR (98.5%, OR=13.04, 95% CI=1.65–278.71, P=0.003) and mRNA RT-PCR (95.4%, OR=4.21, 95% CI=1.01–20.17, P=0.024) were significantly higher than with the AFB smear (83.1%) (Table 1). The agreement of testing results between AFB and DNA PCR or mRNA RT-PCR assay was significantly different (P=0.006 or P=0.039 respectively, McNemar’s test) and DNA PCR and mRNA RT-PCR were comparable (P=0.5, McNemar’s test).
Table 1.
Performance of AFB, DNA PCR and mRNA RT-PCR for detecting Mycobacterium tuberculosis in culture-positive sputum specimens collected at the time of recruitment.
| Tests | Number tested | Number positive (%) |
|---|---|---|
| AFB (carbol fushin stain) | 65 | 54 (83.1) |
| DNA PCR | 65 | 64 (98.5) |
| mRNA RT-PCR | 65 | 62 (95.4) |
When all 286 sputum specimens collected at the time of recruitment and during on-treatment follow-up were compared, DNA PCR assay showed higher diagnostic sensitivity (93.8% vs. 85.2%, OR=2.61, 95% CI=1.03–6.81, P=0.025) and lower diagnostic specificity (51.3% vs.88.6%, OR=7.39, 95% CI=3.99–13.82, P<0.0001) than mRNA RT-PCR, using results of mycobacterial culture as the standard (Table 2). Overall agreement was 87.1% between culture and RT-PCR, which was significantly better than between culture and DNA PCR (87.1% vs.70.3%, OR=2.85, 95% CI=1.82–4.47, P<0.0001) (Table 2).
Table 2.
Performance DNA and mRNA real-time assays with hydrolysis probes for detecting M. tuberculosis-specific DNA and mRNA in sputum specimens in comparison to mycobacterial culturea
| Real-time assay | C+P+ (True positives) | C+P− (False negatives) | C−P+ (False positives) | C−P− (True negatives) | Diagnostic sensitivity, % | Diagnostic specificity, % | Agreement with culture, % |
|---|---|---|---|---|---|---|---|
| DNA PCR | 120 | 8 | 77 | 81 | 93.8 | 51.3 | 70.3 |
| mRNA RT-PCR | 109 | 19 | 18 | 140 | 85.2 | 88.6 | 87.1 |
Abbreviations: C, culture; P, DNA or mRNA real-time PCR assay; +, positive; −, negative.
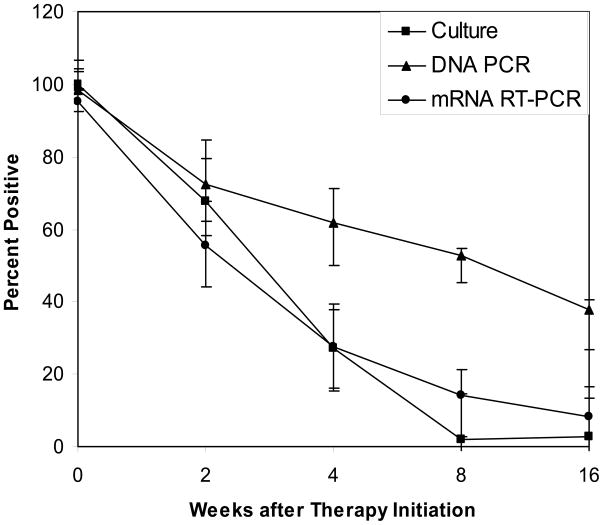
Conversion of assay results from positive to negative during therapy was used to indicate effective anti-TB therapy. At two weeks of therapy, culture and mRNA RT-PCR were negative in 21 (32.3%) and 29 (44.6%) of cases, respectively. After one, two and four months of therapy, 46 (63.0%), 55 (98.2%) and 36 (97.3%) were negative by culture. In comparison, 45 (72.3%), 48 (86.7%), and 34 (91.9%) were negative by mRNA RT-PCR. The mRNA RT-PCR assay agreed with culture at most follow-up time points in 36 (55.4%) of the cases, and converted to negative earlier than culture in 13 (20.0%), and later than culture in 13 (20.0%) (Figure 1). Compared to culture, while these differences were not statistically significant (p>0.05), mRNA RT-PCR appeared to be more diagnostically sensitive for monitoring treatment response at two-week follow up, but less sensitive at later weeks. A significantly high false positive rate was noticed in the late follow-up stage when the DNA PCR assay was used for therapy monitoring (Figure 1).
Fig. 1.
The negative conversion of mycobacterial culture, DNA PCR and RT-PCR during anti-TB therapy. Positive percentages along with lower and upper bars were presented.
Relationships between time to conversion to negative during anti-TB therapy and antimicrobial susceptibility patterns at treatment initiation were explored. Among 59 non-MDR TB cases, 47 (79.7%) of cultures turned negative within one month on therapy while among 6 MDR TB cases, 5 (83.4%) did not turn negative until two months therapy or after (OR=19.58, 95% CI=1.88–488.24, Fisher exact P=0.0037) (Table 3). The trend was similar when an RT-PCR assay was used to follow conversion on anti-TB therapy (OR=15.00, 95% CI=1.46–370.50, Fisher exact P=0.0086), indicating that those with non-MDR tended to convert to negative more rapidly than those caused by MDR isolates as determined by both culture and mRNA assays. There was not a significant difference in time to conversion to negative between culture and RT-PCR (Fisher exact P>0.5) (Table 3).
Table 3.
Relationship between conversion time to negative and drug resistance patterns
| Assay | Drug resistance | Number of cases | Number converted to negative (%) | |
|---|---|---|---|---|
| ≤1 month | ≥ 2 months | |||
| Culture | MDR | 6 | 1 (16.7) | 5 (83.4) |
| Non-MDR | 59 | 47 (79.7) | 12 (20.3) | |
| RT-PCR | MDR | 6 | 1 (16.7) | 5 (83.4) |
| Non-MDR | 56* | 42 (75.0) | 14 (25.0) | |
Three cases were excluded as culture-positive but mRNA RT-PCR-negative at the time of recruitment.
Discussion
In this prospective study we used M. tuberculosis-specific DNA PCR and mRNA RT-PCR assays to monitor response to anti-TB therapy. Conversion of assay results from positive to negative during follow-up was considered an indicator of effective therapy. Results with mRNA RT-PCR assay agreed with culture results at every time point after the initiation of therapy in 55.4% of cases, and predicted conversion earlier in 20.0% and later in 20.0% of cases. Validation of the performance of molecular assays by comparison to a conventional culture may be associated with artifactually decreased diagnostic specificity. Nevertheless, the mRNA RT-PCR assay described here may provide a rapid real-time tool for monitoring anti-TB therapeutic efficacy.
The emergence and spread of MDR and extensively drug-resistant (XDR) strains of M. tuberculosis pose a serious threat to current anti-TB therapy regimens (30). Current therapy involves the initial administration of four first-line drugs. The initial regimen is subsequently modified as necessary based on results of antimicrobial resistance assays that may not become available for several months. Current laboratory methods for monitoring the efficacy of TB therapy rely on mycobacterial culture, and the slow growth of M. tuberculosis causes delays in determining results. Our data indicated that patients with single drug resistance tended to convert to negative more rapidly than those caused by MDR isolates as determined by both culture and mRNA assays, which makes the mRNA assay attractive especially in a population with higher MDR prevalence. DNA target-based molecular methods have not yet met the need for a more rapid test (10). While these assays can shorten the time to verify the presence of M. tuberculosis in a clinical specimen, they are not specific enough to be used as an index of “test of cure” because DNA may persist long after bacteria have been killed (27).
Several groups of investigators have reported efforts to use mRNA target-based assays to rapidly monitor treatment efficacy (12–16). Concentrations of M. tuberculosis mRNA decline after initiation of therapy, as do viable M. tuberculosis colony counts, with 90% of patients becoming negative for both markers within 2 months of treatment (13, 14). The rapid disappearance of M. tuberculosis mRNA from sputum suggests that it is a good index of microbial viability, and a useful marker for assessing response to therapy. In our study, we used a large quantity of high concentration guanidine isothiocyanate in Lysis Buffer to stabilize M. tuberculosis mRNA in sputum specimens as soon as they were decontaminated and concentrated by NaOH-NALC. Our evaluation of 65 new tuberculosis cases showed that mRNA RT-PCR agreed well with culture at most follow-up time points. In addition, at the first 2-week follow-up time point, mRNA turned negative in 13 (20.0%) cases while culture remains positive. Similar to culture, time to mRNA conversion to negative correlated with anti-TB drug susceptibilities. Cases involving strains with only INH-resistance responded to anti-TB therapy more quickly than MDR-TB cases. The mRNA RT-PCR assay can be used as a rapid and real-time tool for assessing the clinical response to anti-TB therapy, replacing more time-consuming phenotypic antimicrobial susceptibility assays.
Our RT-PCR protocol was designed to amplify and detect mRNA only from concentrated and decontaminated sputum specimens. Because the easyMAG extraction system recovers total nucleic acids, we could not avoid residual background amplification of the antigen 85B gene DNA. This may explain a slightly delayed TB conversion at later follow-up time-points as compared to culture. Techniques that can either specifically recover mRNA or destroy residual DNA during extraction may enhance the performance of mRNA RT-PCR (31, 32). Another potential way to enhance performance is based on emerging evidence that certain host cytokine responses are associated with effective therapy (33–35). The combination of assays to measure M. tuberculosis transcript and changes in the host response may ultimately prove even better for monitoring the response to anti-TB therapy (36–38).
Supplementary Material
Acknowledgments
The authors thank patients, laboratorians, nurses and physicians at the National Center of Tuberculosis and Lung Diseases, Republic of Georgia for their excellent assistance, Yuwei Zhu and William Dupont for their statistical assistance and Timothy Sterling, Charles Stratton and David Haas for critically reviewing the manuscript. This study was funded in part by grant GEB2-2605-TB-04 from the U.S. Civilian Research and Development Foundation.
Footnotes
This study was presented in part at the 108th General Meeting of the American Society for Microbiology, Boston, Massachusetts, 1–5 June, 2008
References
- 1.Mitchison DA. Assessment of new sterilizing drugs for treating pulmonary tuberculosis by culture at 2 months. Am Rev Resp Dis. 1993;147:1062–3. doi: 10.1164/ajrccm/147.4.1062. [DOI] [PubMed] [Google Scholar]
- 2.Jindani A, Aber VR, Edwards EA, Mitchison DA. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Resp Dis. 1980;121:939–49. doi: 10.1164/arrd.1980.121.6.939. [DOI] [PubMed] [Google Scholar]
- 3.Brisson-Noel A, Gicquel B, Lecossier D, Levy-Frebault V, Nassif X, Hance AJ. Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet. 1989;2:1069–71. doi: 10.1016/s0140-6736(89)91082-9. [DOI] [PubMed] [Google Scholar]
- 4.Kaul KL. Molecular detection of Mycobacterium tuberculosis: impact on patient care. Clin Chem. 2001;47:1553–8. [PubMed] [Google Scholar]
- 5.Tang YW, Meng S, Li H, Stratton CW, Koyamatsu T, Zheng X. PCR Enhances acid-fast bacillus stain-based rapid detection of Mycobacterium tuberculosis. J Clin Microbiol. 2004;42:1849–50. doi: 10.1128/JCM.42.4.1849-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thwaites GE, Caws M, Chau TT, Dung NT, Campbell JI, Phu NH, et al. Comparison of conventional bacteriology with nucleic acid amplification (amplified mycobacterium direct test) for diagnosis of tuberculous meningitis before and after inception of antituberculosis chemotherapy. J Clin Microbiol. 2004;42:996–1002. doi: 10.1128/JCM.42.3.996-1002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shankar P, Manjunath N, Mohan KK, Prasad K, Behari M, Shriniwas, Ahuja GK. Rapid diagnosis of tuberculous meningitis by polymerase chain reaction. Lancet. 1991;337:5–7. doi: 10.1016/0140-6736(91)93328-7. [DOI] [PubMed] [Google Scholar]
- 8.Drosten C, Panning M, Kramme S. Detection of Mycobacterium tuberculosis by real-time PCR using pan-mycobacterial primers and a pair of fluorescence resonance energy transfer probes specific for the M. tuberculosis complex. Clin Chem. 2003;49:1659–61. doi: 10.1373/49.10.1659. [DOI] [PubMed] [Google Scholar]
- 9.Walker GT, Nadeau JG, Linn CP, Devlin RF, Dandliker WB. Strand displacement amplification (SDA) and transient-state fluorescence polarization detection of Mycobacterium tuberculosis DNA. Clin Chem. 1996;42:9–13. [PubMed] [Google Scholar]
- 10.Tang YW, Procop GW, Persing DH. Molecular diagnostics of infectious diseases. Clin Chem. 1997;43:2021–38. [PubMed] [Google Scholar]
- 11.Fenhalls G, Stevens L, Moses L, Bezuidenhout J, Betts JC, Helden Pv P, et al. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect Immun. 2002;70:6330–8. doi: 10.1128/IAI.70.11.6330-6338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aellen S, Que YA, Guignard B, Haenni M, Moreillon P. Detection of live and antibiotic-killed bacteria by quantitative real-time PCR of specific fragments of rRNA. Antimicrob Agents Chemother. 2006;50:1913–20. doi: 10.1128/AAC.00869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardin LE, Perkins MD, Wolski K, Haun S, Teixeira L, Chen Y, et al. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Resp Crit Care Med. 1999;160:203–10. doi: 10.1164/ajrccm.160.1.9811006. [DOI] [PubMed] [Google Scholar]
- 14.Jou NT, Yoshimori RB, Mason GR, Louie JS, Liebling MR. Single-tube, nested, reverse transcriptase PCR for detection of viable Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:1161–5. doi: 10.1128/jcm.35.5.1161-1165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eltringham IJ, Drobniewski FA, Mangan JA, Butcher PD, Wilson SM. Evaluation of reverse transcription-PCR and a bacteriophage-based assay for rapid phenotypic detection of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:3524–7. doi: 10.1128/jcm.37.11.3524-3527.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Mangan JA, Dhillon J, Sole KM, Mitchison DA, Butcher PD, Coates AR. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol. 2000;182:6358–65. doi: 10.1128/jb.182.22.6358-6365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO. Global tuberculosis control: surveillance, planning, financing. Geneva: World Health Organization Publication; 2008. [Google Scholar]
- 18.Mdivani N, Zangaladze E, Volkova N, Kourbatova E, Jibuti T, Shubladze N, et al. High prevalence of multidrug-resistant tuberculosis in Georgia. Int J Infect Dis. 2008 doi: 10.1016/j.ijid.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstock DM, Hahn O, Wittkamp M, Sepkowitz KA, Khechinashvili G, Blumberg HM. Risk for tuberculosis infection among internally displaced persons in the Republic of Georgia. Intl J Tubercu Lung Dis. 2001;5:164–9. [PubMed] [Google Scholar]
- 20.Gegia M, Mdivani N, Mendes RE, Li H, Akhalaia M, Han J, et al. Prevalence of and molecular basis for tuberculosis drug resistance in the Republic of Georgia: validation of a QIAplex system for detection of drug resistance-related mutations. Antimicrob Agents Chemother. 2008;52:725–9. doi: 10.1128/AAC.01124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. Treatment of tuberculosis: Guidelines for national porgrammes. 3. Geneva: World Health Organization Publication; 2003. [Google Scholar]
- 22.Li H, Dummer JS, Estes WR, Meng S, Wright PF, Tang YW. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J Clin Microbiol. 2003;41:187–91. doi: 10.1128/JCM.41.1.187-191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 24.Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from Northwestern Russia: implications for genotypic resistance testing. J Clin Microbiol. 2002;40:3810–3. doi: 10.1128/JCM.40.10.3810-3813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang YW, Sefers SE, Li H, Kohn DJ, Procop GW. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J Clin Microbiol. 2005;43:4830–3. doi: 10.1128/JCM.43.9.4830-4833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cave MD, Eisenach KD, McDermott PF, Bates JH, Crawford JT. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 27.Desjardin LE, Chen Y, Perkins MD, Teixeira L, Cave MD, Eisenach KD. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J Clin Microbiol. 1998;36:1964–8. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Wit L, de la Cuvellerie A, Ooms J, Content J. Nucleotide sequence of the 32 kDa-protein gene (antigen 85 A) of Mycobacterium bovis BCG. Nucleic Acids Res. 1990;18:3995. doi: 10.1093/nar/18.13.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, McCormac MA, Estes RW, Sefers SE, Dare RK, Chappell JD, et al. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol. 2007;45:2105–9. doi: 10.1128/JCM.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dye C. Doomsday postponed? Preventing and reversing epidemics of drug-resistant tuberculosis. Nat Rev Microbiol. 2009;7:81–7. doi: 10.1038/nrmicro2048. [DOI] [PubMed] [Google Scholar]
- 31.Desjardin LE, Perkins MD, Teixeira L, Cave MD, Eisenach KD. Alkaline decontamination of sputum specimens adversely affects stability of mycobacterial mRNA. J Clin Microbiol. 1996;34:2435–9. doi: 10.1128/jcm.34.10.2435-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honore-Bouakline S, Vincensini JP, Giacuzzo V, Lagrange PH, Herrmann JL. Rapid diagnosis of extrapulmonary tuberculosis by PCR: impact of sample preparation and DNA extraction. J Clin Microbiol. 2003;41:2323–9. doi: 10.1128/JCM.41.6.2323-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mistry R, Cliff JM, Clayton CL, Beyers N, Mohamed YS, Wilson PA, et al. Gene-expression patterns in whole blood identify subjects at risk for recurrent tuberculosis. J Infect Dis. 2007;195:357–65. doi: 10.1086/510397. [DOI] [PubMed] [Google Scholar]
- 34.Siawaya JF, Bapela NB, Ronacher K, Beyers N, van Helden P, Walzl G. Differential expression of interleukin-4 (IL-4) and IL-4 delta 2 mRNA, but not transforming growth factor beta (TGF-beta), TGF-beta RII, Foxp3, gamma interferon, T-bet, or GATA-3 mRNA, in patients with fast and slow responses to antituberculosis treatment. Clin Vaccine Immunol. 2008;15:1165–70. doi: 10.1128/CVI.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribeiro-Rodrigues R, Resende Co T, Johnson JL, Ribeiro F, Palaci M, Sa RT, et al. Sputum cytokine levels in patients with pulmonary tuberculosis as early markers of mycobacterial clearance. Clin Diagn Lab Immunol. 2002;9:818–23. doi: 10.1128/CDLI.9.4.818-823.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietze R, Teixeira L, Rocha LM, Palaci M, Johnson JL, Wells C, et al. Safety and bactericidal activity of rifalazil in patients with pulmonary tuberculosis. Antimicrob Agents Chemother. 2001;45:1972–6. doi: 10.1128/AAC.45.7.1972-1976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien RJ. Studies of the early bactericidal activity of new drugs for tuberculosis: a help or a hindrance to antituberculosis drug development? Am J Respir Crit Care Med. 2002;166:3–4. doi: 10.1164/rccm.2205007. [DOI] [PubMed] [Google Scholar]
- 38.Sirgel FA, Donald PR, Odhiambo J, Githui W, Umapathy KC, Paramasivan CN, et al. A multicentre study of the early bactericidal activity of anti-tuberculosis drugs. J Antimicrob Chemother. 2000;45:859–70. doi: 10.1093/jac/45.6.859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.