Abstract
Here we report a novel finding of an antisense oligonucleotide (ASO) microwalk in which we examined the position-specific role of intronic residues downstream from the 5′ splice site (5′ ss) of SMN2 exon 7, skipping of which is associated with spinal muscular atrophy (SMA), a leading genetic cause of infant mortality. Our results revealed the inhibitory role of a cytosine residue at the 10th intronic position (10C), which is neither conserved nor associated with any known splicing motif. Significance of 10C emerged from the splicing pattern of SMN2 exon 7 in presence of a 14-mer ASO (L14) that sequestered two adjacent hnRNP A1 motifs downstream from 10C and yet promoted SMN2 exon 7 skipping. Another 14-mer ASO (F14) that sequestered both, 10C and adjacent hnRNP A1 motifs, led to a strong stimulation of SMN2 exon 7 inclusion. The inhibitory role of 10C was found to be tightly linked to its unpaired status and specific positioning immediately upstream of a RNA:RNA helix formed between the targeting ASO and its intronic target. Employing a heterologous context as well as changed contexts of SMN2 intron 7, we show that the inhibitory effect of unpaired 10C is dependent upon a long-distance interaction involving downstream intronic sequences. Our report furnishes one of the rare examples in which an ASO-based approach could be applied to unravel the critical role of an intronic position that may not belong to a linear motif and yet play significant role through long-distance interactions.
Keywords: SMN, alternative splicing, intron 7, exon 7, ISS-N1, GC-rich, antisense oligonucleotide, ASO, SMA
INTRODUCTION
Alternative splicing is an essential process in the generation of protein diversity and has been a major contributory force to genome evolution (Xing and Lee 2007). Current estimates suggest that 95%–100% of human genes with two or more exons are alternatively spliced affecting all major aspects of cellular metabolism (Nilsen and Graveley 2010). Splicing is catalyzed by a spliceosome that represents one of the most complex macromolecular machines known (Nilsen 2003; Matlin and Moore 2007). Control of alternative splicing rests on nonspliceosomal factors that bind to pre-mRNA sequences called exonic splicing enhancers (ESEs) or intronic splicing enhancers (ISEs) and exonic splicing silencers (ESSs) or intronic splicing (ISSs) (Lin and Fu 2007; Martinez-Contreras et al. 2007; David and Manley 2008). Enhancer and silencer motifs promote or suppress splice-site (ss) selection, respectively. Methods to identify splicing motifs are continuing to evolve (Chasin 2007; Singh 2007a; Hertel 2008; Wang and Burge 2008; Yu et al. 2008). An additional regulatory role is provided by RNA structures that enforce accessibility to splicing elements, as well as bring two distantly located cis-elements in close proximity (Graveley 2005; Buratti et al. 2007; Singh et al. 2007; Shepard and Hertel 2008; Warf et al. 2009). Unraveling the mechanism by which splicing factors, RNA regulatory sequences, and structural motifs coordinate to regulate alternative splicing is an area of growing interest for evolving strategies to cure many human diseases associated with defective splicing (Garcia-Blanco 2006; Cooper et al. 2009; Tazi et al. 2009; Ward and Cooper 2010).
Despite a tremendous progress in our understanding of pre-mRNA splicing, validation of splicing motifs in the context of an endogenous gene has remained a daunting task. Antisense oligonucleotide (ASO)-based approaches offer an extraordinary potential to address this issue with additional implication to therapy of human diseases (Hua et al. 2008; Bauman et al. 2009). Refinements of ASO-based strategies have capability to provide information regarding the accessibility and the role of certain regions within RNA sequence. For example, ASO scanning arrays have been employed to probe alternative folding of RNA sequences (Ooms et al. 2004). The most exciting future application of an ASO-based approach includes identification of the critical role of an individual residue in pre-mRNA splicing using endogenous context. Such studies would address the role of the nonlinear motifs with implications to high-order interactions and structural rearrangements.
Humans have two nearly identical copies of the Survival Motor Neuron (SMN) gene: SMN1 and SMN2 (Lefebvre et al. 1995). The two SMN genes code for identical proteins; however, SMN2 predominantly generates a short transcript due to skipping of exon 7, producing a truncated SMN that is highly unstable (Vitte et al. 2007). The inability of SMN2 to compensate for the loss of SMN1 results in spinal muscular atrophy (SMA), a debilitating disease of children and infants (Wirth et al. 2006). Since SMN2 is almost universally present in SMA patients, correction of SMN2 exon 7 splicing holds the promise for cure. Due to anticipated target specificity, a large number of studies have focused on ASO-based approaches to restore SMN2 exon 7 inclusion (Bauman et al. 2009). Among these studies, our earlier reported intronic splicing silencer N1 (ISS-N1) has emerged as a leading target in a systematic antisense microwalk (Singh et al. 2006; Hua et al. 2008). Consistently, a recent report has independently confirmed the high therapeutic potential of ISS-N1 by demonstrating that blocking ISS-N1 with an ASO substantially elevated the SMN levels in brain of SMA mice (Williams et al. 2009).
ISS-N1 is a mixed composition sequence spanning from the 10th to 24th positions of intron 7 (Fig. 1A; Singh et al. 2006). Due to its strategic location and strong negative impact, ISS-N1 has been termed as the master checkpoint (Buratti et al. 2006). The 15-nucleotide (nt)-long ISS-N1 harbors two putative hnRNP A1 motifs that have been proposed to be responsible for its inhibitory impact (Fig. 1A; Hua et al. 2008). Using an ultra-refined antisense microwalk, we have recently shown that ISS-N1 overlaps with an 8-nt-long GC-rich sequence spanning from the seventh to 14th position of intron 7 (Fig. 1A; Singh et al. 2009). We have also shown that sequestering of this GC-rich sequence with an 8-mer ASO fully restores SMN2 exon 7 inclusion in SMA patient cells (Singh et al. 2009). Interestingly, the stimulatory effect of the 8-mer ASO comes without full sequestration of any of the validated hnRNP A1 binding sites. Thus, our results open a possibility of the role of an additional negative element and/or structural motif associated with the GC-rich sequence in the vicinity of the 5′ ss. We have earlier reported the inhibitory role of a RNA structure (terminal stem–loop 2 or TSL2) sequestering the 5′ ss of SMN2 exon 7 (Singh et al. 2007). It remains to be seen if a factor interacting with TSL2 makes a secondary contact with the downstream GC-rich sequence or vice versa.
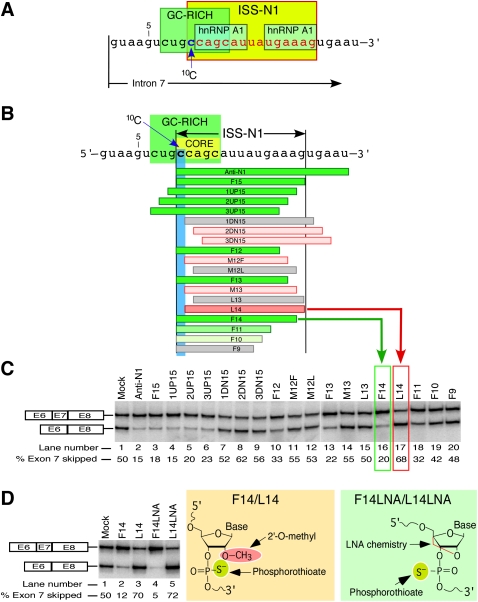
FIGURE 1.
An ultra-refined antisense microwalk reveals the significance of a cytosine residue at the 10th position (10C) of SMN2 intron 7. (A) Diagrammatic representation of cis-elements at the 5′ end of SMN2 intron 7. ISS-N1 is highlighted in yellow with all but the first nucleotide shown in red. Two hnRNP A1 motifs are indicated as described by Hua et al. (2008). GC-rich sequence is highlighted in green (Singh et al. 2009). Numbering of nucleotides starts from the first position of SMN2 intron 7. The location of 10C is marked by an arrow. (B) Diagrammatic representation of ASOs and their annealing positions relative to ISS-N1 region. Numbering of nucleotides starts from the first position of intron 7. Boundaries of ISS-N1 are demarcated. The first ISS-N1 residue, 10C, is highlighted in blue. The GC-rich sequence (Singh et al. 2009) is highlighted in green. The core sequence of the antisense target is highlighted in yellow. ASOs are shown as horizontal bars. Green bars represent ASOs that promote SMN2 exon 7 inclusion (exon 7 skipping is ≤45%). The intensity of green color reflects the strength of ASO stimulatory effect. Gray bars represent ASOs that have no effect on SMN2 exon 7 inclusion. Pink bars represent ASOs that promoted SMN2 exon 7 skipping (exon 7 skipping is ≥55%). To emphasize that L14 caused the most dramatic increase in exon 7 skipping, it is shown as a dark pink bar. (C) Splicing pattern of endogenous SMN2 after treatment with ASOs shown in panel B. SMA type I patient fibroblasts (GM03813) were treated with 20 nM of different ASOs and the total RNA for in vivo splicing assay was isolated 24 h post-transfection. The upper band corresponds to exon 7–included product; the lower band corresponds to exon 7–skipped product. The percentage of exon skipping was calculated from the total value of exon-included and exon-skipped products. The values represent mean of three independent experiments. The standard deviations were <5% of mean. The effect of F14 and L14 are highlighted with a green and red box, respectively. (D) Splicing pattern of endogenous SMN2 after treatment with 50 nM of indicated ASOs. The chemistry of the ASOs used is shown on the right. In vivo splicing assays were performed and analyzed as described in panel C. The values represent mean of three independent experiments. The standard deviations were <5% of mean.
Given the large number of ASOs tested (against ISS-N1), ISS-N1 stands out among other splicing-correcting antisense targets reported thus far. Nevertheless, not all size combinations of ASOs targeting ISS-N1 and its neighboring sequences have been examined. Consequently, the published results do not address the role of specific residues that fall beyond the traditional definition of motifs associated with the known splicing factors. Here we take advantage of an additional antisense microwalk to reveal the critical role of a cytosine residue at the 10th position of human SMN2 intron 7 in conferring the nature of the antisense response. We refer this residue as 10C hereafter. 10C represents the first residue of ISS-N1 but does not belong to any of the known motifs, including hnRNP A1 (Fig. 1A). Our study revealed a rare finding in which two identical size ASOs whose targets differed by a single nucleotide (one ASO sequestered 10C, whereas other did not) produced an opposite effect on SMN2 exon 7 splicing. We confirmed that unsequestered 10C plays a strong negative role when combined with an ASO targeting a 14-nt sequence immediately downstream from 10C. We also show that the inhibitory effect of unsequestered 10C is dependent upon a long-distance interaction involving downstream intronic sequences. Our findings underscore the potential of an ASO-based approach in unraveling the paramount significance of a single intronic nucleotide due to its location relative to other splicing cis-elements within an entire intron.
RESULTS
Identification of a master position within the core of the antisense target
Substantial evidence and independent validations confirm that ISS-N1 is an ideal target for an ASO-mediated correction of aberrant splicing associated with a major genetic disease (Singh et al. 2006, 2009; Hua et al. 2008; Williams et al. 2009). Based on a stimulatory effect of ASOs targeting ISS-N1 and the GC-rich sequence, we have established that the first five residues of ISS-N1 constitute the core of antisense target (Singh et al. 2009). 10C occupies the first position of this core and is strategically located in the middle of the GC-rich sequence (Fig. 1). The significance of 10C emerges from the fact that it may participate in high-order interaction because it does not fall within the confines of the recently described hnRNP A1 motifs proposed to be a cause for the negative effect associated with ISS-N1 (Hua et al. 2008; Singh et al. 2009).
To further examine the role of 10C as an integral part of the antisense target, we performed an additional ultra-refined antisense microwalk. We used ASOs of varying sizes that blocked different portions of ISS-N1 with or without sequestering 10C. Of note, since ISS-N1 is a 15-nt-long element, we confined our comparison of antisense effect to 15-mer or shorter ASOs (Anti-N1 being the only exception). Annealing positions of ASOs are diagrammatically shown in Figure 1B. Sequences of all ASOs used in this study are given in Table 1. For the sake of simplicity we used an antisense nomenclature comprised of a letter(s) followed by a number that represents the size of an ASO. F series ASOs possess the identical 3′ ends with the first position of ISS-N1 being complementary to the 3′-terminal nucleotides of ASOs. L series ASOs possess the identical 5′ ends with the last position of ISS-N1 being complementary to the 5′-terminal nucleotides of ASOs. M series ASOs block sequences in the middle of ISS-N1. Other ASOs containing UP and DN letters sequester residues upstream of and downstream from ISS-N1, respectively. Here, we performed antisense screening at a relatively low ASO concentration of 20 nM. Our earlier reported ASOs that sequestered 10C served as positive controls (Fig. 1B, green bars).
TABLE 1.
Sequences of ASOs used
As shown in Figure 1C, ASOs that did not sequester 10C failed to produce any stimulatory response on SMN2 exon 7 inclusion and in some instances even caused an increase in SMN2 exon 7 skipping. The most striking example was L14, particularly when the effect of this ASO was compared with the effect of F14. Note that both ASOs are 14-nt long, the GC content of their targets is the same, and their annealing positions differ by only 1 nt, 10C. Yet F14 and L14 produce opposite effects on exon 7 splicing: F14 that sequestered 10C efficiently restored SMN2 exon 7 inclusion, while L14 that did not block 10C increased exon 7 skipping (Fig. 1C). Antagonistic effects of F14 and L14 were verified by three different batches of ASOs synthesized at different times. Also, we performed experiments at various concentrations ranging from 1 nM to 1 μM. Stimulatory effect of F14 increased with the increasing concentrations of F14, whereas the inhibitory effect of L14 increased with the increasing concentrations of L14 (data not shown). Of note, L14 sequestered both hnRNP A1 motifs within ISS-N1 and yet promoted SMN2 exon 7 skipping. Other short ASOs that did not sequester 10C had less pronounced negative effects. For example, L13 and M13 that are only 1 nt shorter than L14 showed substantially reduced if any negative effects on SMN2 exon 7 splicing. Similar results were observed with M12F and M12L that annealed to the 12-nt-long sequences in the middle of ISS-N1. We also tested 15-mer ASOs that did not sequester 10C but targeted sequences immediately downstream from 10C. These 15-mer ASOs produced inhibitory response albeit at higher concentrations (data not shown).
Next, we wished to verify that the antagonistic effects of F14 and L14 on splicing of SMN2 exon 7 were not specific to the chemistry of ASOs. Note that in our ultrarefined microwalk described above, the ASOs had uniform phosphorothioate backbone and 2′-O-methyl modifications (Fig. 1D). For comparison, we chose F14 and L14 with locked nucleic acid (LNA) chemistry in which an extra-bridge that connects 2′-oxygen and 4′-carbon of ribose sugar is added (Fig. 1D). LNAs display unprecedented hybridization affinity toward complementary single-stranded RNA and have been widely used in a variety of applications (Veedu and Wengel 2009). In LNAs used in our experiments, we also replaced uracyl residues with thymidine residues. Such replacement has a potential to improve base pairing with adenosine residues. To increase the intracellular stability of LNAs, we incorporated uniform phosphorothioate backbone. LNAs corresponding to F14 and L14 were named as F14LNA and L14LNA, respectively. As shown in Figure 1D, F14LNA and L14LNA retained their characteristic antagonistic effects. These results validated the fact that sequestering of 10C is essential for producing the stimulatory response irrespective of the chemistry of ASO. In addition, our results showed that inhibitory effect of unsequestered 10C does not depend on the chemistry of the duplex formed between an ASO and its ISS-N1 target.
Antisense effect is specific to base paring with target
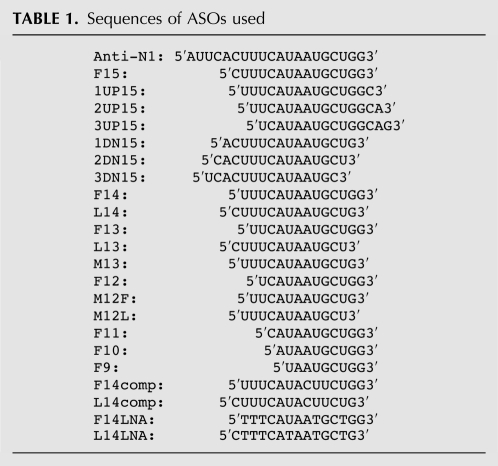
Having discovered that two antisense targets differing by a single nucleotide could produce antagonistic effect upon annealing to their cognate ASOs, we next examined the efficacy and specificity of these ASOs using SMN2 minigene system. For this, HeLa cells were cotransfected with the minigene (0.1 μg) and an ASO of interest (50 nM), and the effect on splicing was assessed by RT-PCR. To avoid the off-target effect, we deliberately chose a lower range of minigene and ASO concentrations that we have determined to be optimum for modulation of SMN2 exon 7 splicing (Singh et al. 2009). As shown in Figure 2A, the antisense effect on splicing of SMN2 minigene was consistent with the results for the endogenous SMN2 in SMA patient fibroblasts: F14 increased exon 7 inclusion, while L14 increased exon 7 skipping. These results validate the necessity of sequestration of 10C for the stimulatory response of ASOs targeting ISS-N1. Our results also suggest that sequences upstream of exon 6 do not modulate the response of ASOs targeting ISS-N1. When F14 and L14 were mutated giving rise to ASOs we called F14comp and L14comp, their effect on SMN2 exon 7 splicing was obviated (Fig. 2A). These results suggest that the positive effect of F14 and negative effect L14 on splicing of exon 7 are dependent on ASO base pairing with their respective target sites.
FIGURE 2.
Effect of ASOs are specific to their targets. (A) Diagrammatic representation of the target area in intron 7 of SMN2 minigene. ISS-N1 is highlighted in light gray. Numbering of nucleotides starts from the first position of intron 7. The location of 10C is marked by an arrow. The sequences of four ASOs and their annealing positions in ISS-N1 region are shown. Mutations are indicated in white letters and highlighted in black. Effect of ASOs on splicing of SMN2 minigene is shown on the right. HeLa cells were cotransfected with 50 nM of a given ASO and 0.1 μg of SMN2 minigene. Splicing was determined 24 h after transfection. The percentage of exon 7 skipping was calculated as described in Figure 1C. (B) Diagrammatic representation of the target area in intron 7 of SMN2/I7-08 minigene. ISS-N1 is highlighted in gray. Numbering of nucleotides starts from the first position of intron 7. Location of 10C is marked by an arrow. Sequences of four ASOs and their annealing positions are shown. Mutations in ISS-N1 area as well as in the ASOs are indicated in white letters and highlighted in black. Effect of the ASOs on splicing of SMN2/I7-08 minigene is shown on the right. In vivo splicing assays were performed and analyzed as described in panel A.
To further validate that the opposite effects of F14 and L14 are not due to interactions with nonspecific targets, we performed a key experiment in which ASOs were cotransfected with our earlier described mutant minigene, SMN2/I7-08. This minigene has C-to-A and U-to-G substitutions at the fifth and the seventh positions of ISS-N1, respectively. We have shown that these substitutions have capability to weaken the RNA:RNA duplex formed between Anti-N1 and ISS-N1 (Singh et al. 2006). Consistently, both F14 and L14 lost their respective antisense effects when cotransfected with SMN2/I7-08 (Fig. 2B), confirming their target specificity. At the same time, F14comp and L14comp, which showed perfect complementarity with the mutated ISS-N1 within SMN2/I7-08, produced the respective antisense effect similar to the outcome of F14 and L14 sequestration of the wild-type ISS-N1 (Fig. 2B). These results also underscore that the inhibitory effect of untargeted 10C is not linked to the sequence of duplex formed between an ASO and its target within ISS-N1 region.
Site-specific mutations confirm the negative impact of unpaired 10C
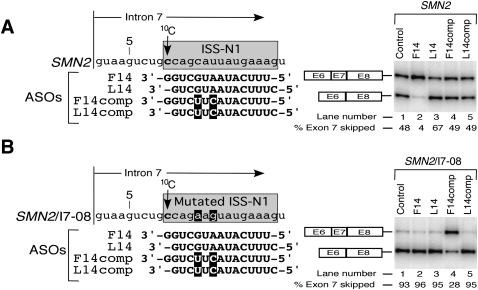
We next wished to investigate a possible mechanism behind the negative effect of L14 on SMN2 exon 7 splicing. We began addressing this issue by employing a SMN2 mutant minigene (SMN2Δ64) in which 10C was deleted bringing the target of L14 1 nt closer to the 5′ ss. As shown in Figure 3A, the deletion of 10C somewhat increased exon 7 skipping compared with the wild-type SMN2, indicating that the negative effect of ISS-N1 could be retained without 10C. Interestingly, when cotransfected with SMN2Δ64, L14 showed a strong stimulatory response on exon 7 inclusion (Fig. 3A). This result validated the inhibitory nature of unsequestered 10C in the context of the L14:target duplex (L14-duplex) and underscored a rare finding that deletion of a single intronic position upstream of a target sequence could reverse the impact of antisense response. Interestingly, F14 maintained its stimulatory effect in SMN2Δ64 despite a 1-nt overhang at the 3′ end due to the deletion of 10C. However, improvement of exon 7 inclusion in the presence of F14 was milder than the one produced by L14, probably due to the fact that the RNA:RNA duplex formed by L14 and its target was longer by 1 nt.
FIGURE 3.
Sequestration of 10C decides the outcome of antisense response. Numbering of nucleotides starts from the first intronic position. ISS-N1 sequence is highlighted in gray. The first C residue in ISS-N1 is marked as 10C. (A) Diagrammatic representation of the 5′ portion of intron 7 of SMN2 minigene and its mutant, SMN2/Δ64. The location of 10C and its deletion are indicated. Effect of ASOs on splicing of wild-type and mutated SMN2 minigene is shown on the right. Cotransfections and analyses were done as in Figure 2A. (B) Diagrammatic representation of wild-type and mutated ISS-N1 targeted by different ASOs. Sequences of ASOs and their base pairing with the corresponding target are shown. Note that wild-type and mutated ISS-N1 element are located within intron 7 of SMN2 and SMN2/64A minigene, respectively. Arrows mark 10C and 10A positions. The 3′-overhang of ASOs are highlighted. Effect of ASOs on splicing of SMN2 minigene and its mutant, SMN2/64A, is shown in the bottom panel. Cotransfections and splicing analyses were done as in Figure 2A.
To further confirm that the inhibitory effect of L14 is due to untargeted 10C in the context of L14-duplex, we used 5′1U-F15. This ASO is identical to L14 except it has an additional uracyl residue added at its 3′ end. Note that this residue does not base pair with 10C but is capable of sterically preventing the accessibility of 10C (Fig. 3B). As shown in Figure 3B, 5′1U-F15 completely overcame the inhibitory response of L14. However, it was unable to produce a stimulatory response since it did not effectively sequester 10C.
To test whether the inhibitory effect of L14-duplex is specifically linked to the type of residue at the 10th intronic position, we used SMN2/64A minigene in which 10C residue was replaced with an adenosine residue (10A). This mutation did not change the splicing pattern of SMN2 (Fig. 3B), allowing more accurate comparison of the antisense response with and without the sequestration of the 10th intronic nucleotide. Surprisingly, L14 produced a strong stimulatory effect in the context of 10A, confirming that the inhibitory effect of L14-duplex in the wild-type context is solely linked to an untargeted cytosine residue at the 10th intronic position. Consistent with the neutral role of the unpaired 10A, ASOs such as F15, F14, and 1DN15 also produced stimulatory effect on exon 7 inclusion in SMN2/64A minigene. The effects of these ASOs were similar to 5′1U-F15 that sequestered the 10A (Fig. 3B).
Effect of the immediate context on the antisense response
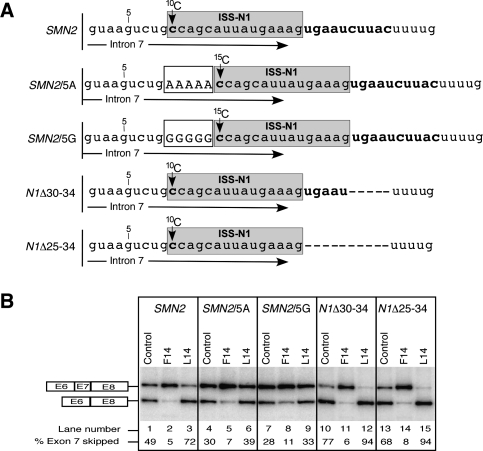
In order to examine the role of the immediate context of target sequence on the antisense response, we used our previously described SMN2 minigene mutants (Singh et al. 2006). In SMN2/5A and SMN2/5G five adenosine and guanosine residues were inserted before ISS-N1, respectively. Such arrangement will place 10C away from the 5′ ss and alter the nature of a presumptive long-distance and/or short-distance interactions that are specific to a nucleotide located at a particular position. When transfected in HeLa cells, SMN2/5A and SMN2/5G showed somewhat less skipping of exon 7 compared with the wild-type SMN2 minigene (Fig. 4B). Upon treatment with F14, both SMN2/5A and SMN2/5G showed increased exon 7 inclusion, suggesting that the effect of F14 is exclusively based on the blocking of a linear cis-element comprised of two putative hnRNP A1 binding sites. Our results also confirmed that the effect of F14 is not sensitive to the precise location of target site with respect to the 5′ ss. On the other hand, L14 produced a decreased negative effect, suggesting that a combination of unsequestered 15C and L14-duplex is less inhibitory than a combination of unsequestered 10C and L14-duplex. Overall our results support that relative positioning of an antisense target with respect to the 5′ ss may be important for determining the impact of an unsequestered residue preceding an RNA:RNA duplex formed between the target and an ASO.
FIGURE 4.
Effect of the local context on antisense response of ISS-N1-targeting ASOs. (A) Diagrammatic representation of the target area in intron 7 of SMN2 minigene and its four mutants. ISS-N1 is highlighted in gray. The first C residue in ISS-N1 is marked as 10C or 15C, depending on its position relative to the beginning of intron 7. Of note, numbering of intronic residues starts with the first position of intron 7. Five-nt-long insertions immediately upstream of ISS-N1 in SMN2/5A or SMN2/5G minigenes are boxed and shown in capital letters. (B) In vivo splicing pattern of wild-type and mutant SMN2 minigenes shown in panel A. The minigenes were cotransfected with an ASO of interest. Cotransfections and splicing analysis were done similarly as in Figure 2A.
To further understand the impact of the local context on the antisense response, we used N1Δ30-34 and N1Δ25-34 minigenes in which five and 10 residues were deleted immediately downstream from ISS-N1, respectively (Fig. 4A). These deletions are likely to break any secondary structure overlapping ISS-N1. When transfected in HeLa cells, transcripts derived from both of the above mutants displayed increased exon 7 skipping (Fig. 4B). In both of these mutants, F14 and L14 produced strong stimulatory and inhibitory effect, respectively. Since these mutants retain the relative positioning of ISS-N1 with respect to the 5′ ss, the antisense response was very similar to the one in the wild-type context. These results also suggest that the inhibitory effect of the combination of an untargeted 10C and the downstream L14-duplex might not be affected by the sequences immediately downstream from ISS-N1. However, our results do not rule out the role of further downstream intronic sequences that may interact with the untargeted 10C during the catalytic core formation.
Effect of the heterologous context on the antisense response
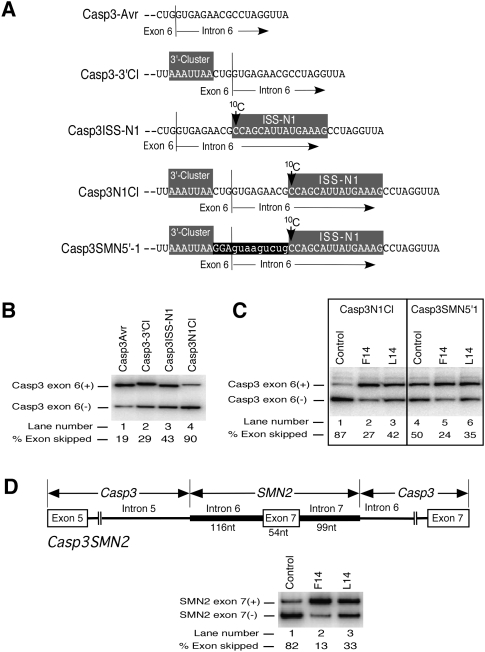
Studies in a heterologous system provide valuable information regarding portability of a cis-element in question. Linear cis-elements are easily portable in a heterologous context, whereas complex cis-elements involving long-distance interactions are generally not portable. We have previously shown that ISS-N1 is a portable cis-element because its insertion at the identical location within intron 6 of Caspase 3 (Casp3) minigene promotes skipping of Casp3 exon 6 (Singh et al. 2006). Indeed, when transfected in HeLa cells, Casp3ISS-N1 minigene showed 43% of Casp3 exon 6 skipping compared with 19% observed in Casp3Avr minigene that lacks ISS-N1 (Fig. 5A,B). 3′-Cluster is another negative element located upstream of the 5′ ss of SMN2 exon 7 (Singh et al. 2004a). Interestingly, insertion of this element at the similar location relative to the 5′ ss (3 nt upstream of the exon/intron junction) (Fig. 5A) in Casp3 exon 6 (Casp3-3′Cl minigene) caused moderate increase in skipping of this exon (Fig. 5B). When both 3′-Cluster and ISS-N1 were inserted in Casp3 minigene (Casp3N1Cl mutant) (Fig. 5A), skipping of Casp3 exon 6 increased to 90% (Fig. 5B). Note that in Casp3N1Cl mutant we tried to recreate “SMN2” arrangement of ISS-N1 and 3′-Cluster relative to each other and the 5′ ss. For this purpose ISS-N1 and 3′-Cluster were inserted 9 nt and 3 nt downstream from and upstream of the 5′ ss of Casp3 exon 6, respectively (Fig. 5A). Since Casp3N1Cl showed the highest amount of exon skipping (Fig. 5B), we decided to use this minigene to test the effect of ISS-N1 targeting by F14 and L14 on splicing of the “heterologous” exon. As shown in Figure 5C, when Casp3N1Cl was cotransfected with F14, inclusion of Casp3 exon 6 increased significantly. Surprisingly, L14 also produced a moderate stimulatory response despite the unsequestered 10C (Fig. 5C). We observed similar results using Casp3-SMN5′-1 minigene in which 12 residues between 3′-Cluster and ISS-N1 (last three exonic and first nine intronic residues) were replaced with SMN sequence (Fig. 5A). The replaced residues combined with the 3′-Cluster and ISS-N1 introduced the 5′ ss “environment” of SMN2 exon 7 in Casp3 minigene. Yet, in Casp3 context the combination of the unsequestered 10C and L14-duplex was unable to produce an inhibitory effect observed in SMN2. These results suggested a possible role of additional sequences in conferring the negative effect associated with unsequestered 10C.
FIGURE 5.
Effect of the heterologous context on antisense response of ISS-N1-targeting ASOs. (A) Diagrammatic representation of exon 6/intron 6 junction in Casp3 minigene variants. 3′-Cluster and ISS-N1, which are shaded in gray, were inserted either individually or together 3 nt upstream of and 9 nt downstream from the exon/intron junction, respectively. In Casp3-SMN5′-1 minigene, the entire region from the beginning of 3′-Cluster to the end of ISS-N1 corresponds to SMN sequence. The last 3 nt of SMN exon 7 and the first 9 nt of SMN intron 7 are highlighted in black. As a result of this insertion, Casp3 exon 6 now has the 5′ ss of SMN exon 7 followed by ISS-N1 element. The location of 10C is marked by an arrow. (B) Effect of 3′-Cluster and ISS-N1 insertions on Casp3 exon 6 splicing. HeLa cells were transfected with 0.8 μg of Casp3 minigene variant, and the total RNA for in vivo splicing assay was isolated 24 h post-transfection. Exon 6-included and exon 6-skipped spliced products are indicated. Percentage of exon skipping was calculated from the total value of exon-included and exon-skipped products similarly as in Figure 2. (C) In vivo splicing patterns of two Casp3 minigene variants in the presence of ASOs. Cotransfections and splicing analysis were done as in Figure 2A. (D) In vivo splicing patterns of Casp3SMN2 minigene in the presence of ASOs. Diagram explaining the composition of this hybrid minigene is shown on the top. Cotransfections and splicing analysis were done as in panel C.
Absence of negative effect of L14 in heterologous context could be due to the lack of cooperative interactions among cis-elements that define both, the 3′ and 5′ ss of SMN2 exon 7. To address this issue, we created a hybrid minigene (Casp3SMN2) in which we replaced the entire middle exon and flanking intronic sequences of Casp3 splicing cassette with 269 nt of SMN2 containing entire exon 7, 116 nt of upstream intron 6, and 99 nt of downstream intron 7 (Fig. 5D). Thus, Casp3SMN2 provided an opportunity to evaluate the effect of F14 and L14 in a “semi-heterologous” system in which large portions of wild-type context defining the 3′ and 5′ ss of SMN2 exon 7 were present. At the same time, Casp3SMN2 minigene retained the Casp3 context that defines the 5′ ss of upstream and 3′ ss of downstream exons. As shown in Figure 5D, when Casp3SMN2 was cotransfected with either F14 or L14, both ASOs promoted exon 7 inclusion. These results indicated that the inhibitory effect of unsequestered 10C is linked to a long-distance interaction that could not be formed in the context of Casp3SMN2. Our results also suggested that the splicing factors directly interacting with exon 7, and the flanking intronic sequences are not involved in producing inhibitory effect associated with unsequestered 10C upstream of L14-duplex.
Inhibitory effect of 10C is linked to an intra-intronic long-distance interaction
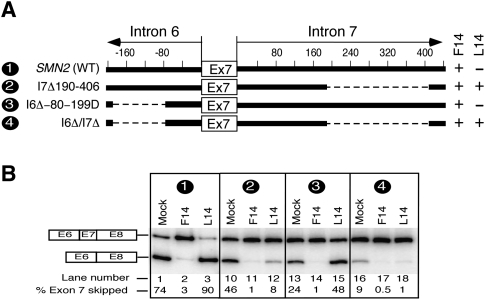
Based on the corroborative evidence of experiments described in Figures 3–5, the most plausible mechanism of inhibitory effect of unsequestered 10C would be involvement of a long-distance interaction(s). To explore such possibility, we generated SMN2 minigene mutants with large deletions within intron 6 and intron 7 farther away from the 3′ and 5′ ss of exon 7. These mutants were then used to test the effect of F14 and L14 on exon 7 splicing. As shown in Figure 6, when the 3′ portion of intron 7 was deleted (I7Δ190-406), the negative effect of L14 on exon 7 splicing was abrogated and it became almost as effective in restoring the inclusion of exon 7 as F14. However, in SMN2 mutant with a deletion in intron 6 (I6Δ-80-199D), L14 still retained its inhibitory effect on exon 7 splicing (Fig. 6). Interestingly, when I6Δ-80-199D deletion was combined with I7Δ190-406 deletion (mutant I6Δ/I7Δ), L14 again lost its inhibitory effect despite the unsequestered 10C (Fig. 6B). These results are in line with the effect of F14 and L14 on exon 7 splicing in the hybrid Casp3SMN2 minigene that lacks similar regions of intron 6 and intron 7 (Fig. 5D). The observation that L14 lost its ability to increase exon 7 skipping when the region between positions 188 and 407 of intron 7 was missing provided the first indication of a potential long-distance interaction between untargeted 10C and a sequence upstream of the 3′ ss of exon 8. In other words, our results suggest that the inhibitory effect of untargeted 10C upstream of L14-duplex is intimately linked to sequences upstream of the 3′ ss of exon 8.
FIGURE 6.
Effect of deletions within intron 6 and intron 7 on antisense response of ISS-N1-targeting ASOs. (A) Diagrammatic representation of the deleted regions in intron 6 and 7 of SMN2 minigene. Deletions are represented by dotted lines. Positive numbers indicate nucleotide positions within intron 7 and start from the first position of this intron. Negative numbers indicate nucleotide positions within intron 6 and start from the last position of this intron. Names of mutants are given on the left; numbers in names reflect positions of the first and the last deleted nucleotides. A deletion mutant producing a stimulatory effect in presence of an ASO is shown as plus, whereas a mutant producing a negative effect in presence of an ASO is shown as minus. (B) In vivo splicing pattern of wild-type and mutant SMN2 minigenes shown in panel A. Minigenes were cotransfected with an ASO of interest. Cotransfections and splicing analyses were done similarly as in Figure 2A.
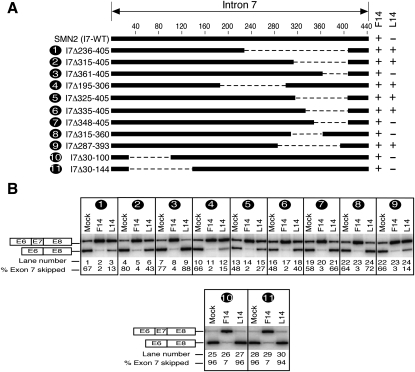
Next we wanted to determine if a specific sequence upstream of the 3′ ss of exon 8 was involved in long-distance interactions with untargeted 10C. Since the deletion in the 3′ portion of intron 7 that abolished the negative impact of untargeted 10C in the context of L14-duplex was rather large (from the 190th and 406th positions), we generated several SMN2 mutants with smaller overlapping deletions in this area and cotransfected them with F14 and L14. Based on the results shown in Figure 7, F14 treatment effectively restored inclusion of SMN2 exon 7 in all mutants, while the response to L14 appeared to depend upon size and location of deletion within intron 7. For example, several sets of overlapping deletions in the 3′ region of intron 7 resulted into the loss of inhibitory effect and/or gain of stimulatory effect on exon 7 inclusion when the deletion size was 72 nt or more (Fig. 7). In particular, when mutants SMN2-I7Δ195-306 and SMN2-I7Δ287-393 with deletions of 112 and 107 nt, respectively, were cotransfected with L14, exon 7 inclusion increased about 2.5 times compared with “mock” cotransfection (Fig. 7B). At the same time, L14 produced no stimulatory effect on mutant with equally large (115-nt-long) deletion in the 5′ portion of intron 7 (SMN2-I7Δ30-144) (Fig. 7B). Since ISS-N1 harbors the only target site for L14 within entire SMN2, the loss of inhibitory effect of L14 in deletion mutants could be directly linked to the loss of a long-distance interaction associated with the deleted sequences. Based on our results we conclude that long-distance interaction is area specific and not sequence specific. Our results of SMN2 intron 7 deletions do not support a straightforward mechanism of loss of L14 inhibitory effect as a consequence of the reduced size of intron 7. Further supporting this argument, a much larger downstream intron (∼1.6 kb) in the heterologous context of Casp3SMN2 fusion minigene did not recapitulate the inhibitory effect of an untargeted 10C upstream of L14-duplex (Fig. 5D).
FIGURE 7.
Effect of deletions within intron 7 on antisense response of ISS-N1-targeting ASOs. (A) Diagrammatic representation of the deleted regions within intron 7 of SMN2 minigene. Deletions are represented by dotted lines. Numbers indicate nucleotide positions and start from the first position of intron 7. Names of mutants are given on the left; numbers in the names reflect positions of the first and the last deleted nucleotides. A deletion mutant producing a stimulatory effect in presence of an ASO is shown as plus, whereas a mutant producing a negative effect in presence of an ASO is shown as minus. (B) In vivo splicing pattern of mutant SMN2 minigenes shown in panel A. Minigenes were cotransfected with an ASO of interest. Cotransfections and splicing analysis were done similarly as in Figure 2A.
Antagonistic effect of ASOs is not linked to the differential displacement of hnRNP A1
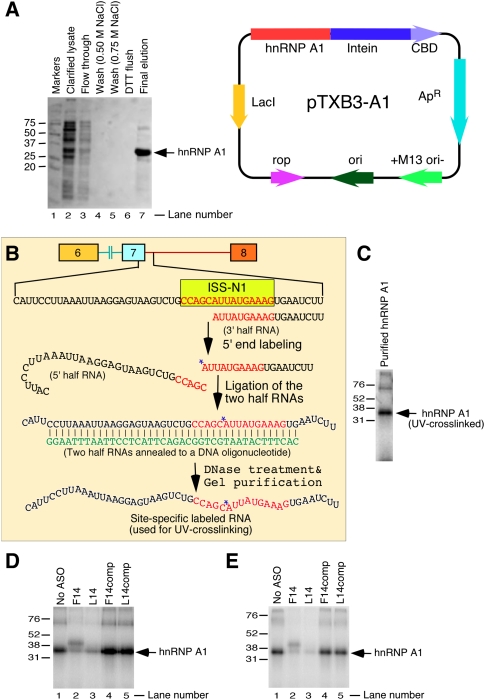
hnRNP A1 motifs located within ISS-N1 have been linked to skipping of SMN2 exon 7 (Hua et al. 2008). Since F14 and L14 produce an opposite effect on exon 7 splicing, we wished to test whether these ASOs display any disparity in their ability to prevent binding of hnRNP A1 to ISS-N1. For this purpose, we performed in vitro experiments using a purified recombinant hnRNP A1 protein. The purification of hnRNP A1 was done using an IMPACT (Intein Mediated Purification with an Affinity Chitin-binding Tag) system that allows a single-column purification of an Escherichia coli expressed protein without vector-derived amino acids or affinity tags. As shown in the Figure 8A, we were able to obtain a nearly homogenous (>90% pure) hnRNP A1 preparation.
FIGURE 8.
Site-specific UV-crosslinking of purified recombinant hnRNP A1 with ISS-N1. (A) Purification of recombinant hnRNP A1 protein expressed from pTXB3-A1 in E. coli ER2566 strain. Protein was purified by binding to a chitin affinity column, followed by DTT-induced self-cleavage of the M. xenopi GyrA intein. Sample aliquots were collected at different purification steps and used for SDS-polyacrylamide gel electrophoresis. (Lane 1) Molecular weight markers; (lane 2) clarified lysate; (lane 3) flow through from the column; (lanes 4,5) wash with increasing concentration of NaCl; (lane 6) DTT flush; and (lane 7) pooled hnRNP A1-containing fractions eluted after overnight DTT-induced intein self-cleavage at 4°C. Band corresponding to hnRNP A1 is indicated by an arrow. Right panel shows the diagrammatic representation of pTXB3-A1 construct. This construct contains hnRNP A1 ORF with a C terminus fused in frame to the M. xenopi GyrA intein/chitin binding domain (CBD). (B) Diagrammatic representation of steps for site-specific 32P-labeling of RNA probe. The sequence of probe and its relative location within SMN are given. ISS-N1 is highlighted in yellow and its sequence shown in red letters. Position of the 32P-radioisotope incorporation is indicated by a star. The sequence of the bridging DNA oligonucleotide is shown in green. (C) Autoradiogram showing hnRNP A1-crosslinked product. Site-specifically 32P-labeled RNA probe was UV-crosslinked with purified recombinant hnRNP A1 followed by RNase digestion and fractionation on 13% SDS-polyacrylamide gel. (D) UV-crosslinking in the presence of different ASOs under “denaturing” condition. Here, RNA probe was first refolded in the presence of a corresponding ASO to ensure ASO annealing. After that, the purified hnRNP A1 was added to the reaction mixture followed by UV-crosslinking. Analysis of hnRNP A1-crosslinked products was done as in panel C. (E) UV-crosslinking in the presence of different ASOs under “native” condition. Here the RNA probe was denatured and refolded prior to an ASO and hnRNP A1 addition to the reaction mixture. UV-crosslinking reaction and analysis of the products were performed as in panel C.
We then used site-specific UV-crosslinking to probe the interaction of the purified hnRNP A1 with ISS-N1. The 50-nt-long RNA probe used for UV-crosslinking contained the last 18 residues of exon 7 and the first 32 residues of intron 7 of SMN2, including the ISS-N1 region (Fig. 8B). To capture a direct interaction of hnRNP A1 with ISS-N1, a single radioactive 32P-moeity was introduced between the fifth and the sixth residues of ISS-N1. These residues fall within the first hnRNP A1 motif of ISS-N1 (Fig. 1A). The conditions of UV-crosslinking experiments were optimized to obtain the sufficient amount of hnRNP A1-crosslinked product following RNase digestion. The results of site-specific UV-crosslinking confirmed that purified hnRNP A1 binds at the site containing radioactive moiety (Fig. 8C). Having established that hnRNP A1 can be site-specifically crosslinked to ISS-N1, we sought to investigate whether a sequestration of ISS-N1 with F14 or L14 is able to prevent hnRNP A1 binding. For this purpose, the site-specifically labeled RNA probe was denatured and refolded in the presence of either F14 or L14 prior to addition of hnRNP A1 and UV crosslinking. As controls we used the mutant ASOs, F14comp and L14comp. As described earlier, these ASOs produce no antisense effect due to the mismatch mutations (Fig. 2). As shown in Figure 8D, F14 and L14 were equally efficient in preventing the binding of hnRNP A1 to ISS-N1 as indicated by the substantial decrease in the amount of hnRNP A1-crosslinked product compared with “No ASO” and F14comp/L14comp controls.
In the next experiment, the site-specifically labeled RNA probe was refolded prior to addition of the ASOs and hnRNP A1. Since the ASOs were added to the reaction mixture after the RNA probe was refolded, this experiment validated the accessibility of the target under the native conditions. Also it tested the affinity of hnRNP A1 for its “native” target in the presence of ASOs. Here again we used F14comp and L14comp as control ASOs. As shown in Figure 8E, again F14 and L14 were equally efficient in preventing hnRNP A1 binding, whereas control ASOs had no effect. Our results also confirmed that sequestration of 10C was not required for the displacement of hnRNP A1.
DISCUSSION
An untargeted cytosine decides the outcome of the target-specific antisense response
ISS-N1 has emerged as one of the best-studied intronic antisense targets for splicing correction in a major human disease. The 15-nt-long ISS-N1 has the distinction of harboring two putative hnRNP A1 motifs covering its last 14 residues (Hua et al. 2008). The first five residues of ISS-N1 together with the three upstream residues constitute an 8-nt-long GC-rich motif (Fig. 1A). Sequestration of either GC-rich motif or first 14 nt of ISS-N1 has the capability to fully restore SMN2 exon 7 inclusion in SMA patient cells at low nanomolar concentrations (Singh et al. 2006, 2009). Based on these findings, the first five residues of ISS-N1 constitute the core of the above two antisense targets (Singh et al. 2009). Although, the GC-rich sequence is the shortest known target for effective splicing correction in a patient cell line, the mechanism by which sequestration of this sequence restores SMN2 exon 7 inclusion is not known. Upon annealing to its target, an ASO has a capability to break the local context by introducing a double helical structure that affects the orientation of residues upstream of and downstream from the helix. Hence, an ASO-based approach provides a unique opportunity to test the significance of certain residues at specific positions even though these positions are not directly targeted by an ASO. Here we report a rare finding in which an untargeted cytosine residue at the 10th intronic position (10C) decides the outcome of the antisense response of ASOs that target sequences immediately downstream from 10C. A critical role of 10C was found to stem from its unique location within the GC-rich motif at a precise distance from the 5′ ss of exon 7.
The significance of unsequestered 10C in conferring the outcome of ISS-N1-targeting ASOs was best captured by F14 and L14. Targets of F14 and L14 differ by a single nucleotide. While F14 sequestered 10C and produced an expected strong stimulatory response on SMN2 exon 7 inclusion, L14 that did not target 10C triggered SMN2 exon 7 skipping. The negative effect of L14 was highly surprising, since L14 fully sequestered both of the hnRNP A1 motifs. The opposite effects of F14 and L14 were observed in the context of both, the endogenous gene and the minigene containing genomic sequences from SMN2 exon 6 through exon 8. This confirmed that a specific promoter sequence and/or any region upstream of exon 6 do not drive the antisense effect.
To validate the target specificity of F14 and L14, we took advantage of our meticulously designed mutant minigenes. In the absence of any off-target effect, it is expected that the lack of annealing of ASOs with the mutated target would eliminate the antisense response. Indeed, validating the target specificity, F14 and L14 lost their ability to affect exon 7 splicing in the mutated SMN2/I7-08 minigene. In a counter-experiment, mutant F14 and L14 that reinstated the base pairing with the mutated ISS-N1 fully restored the original antisense effects. These results confirmed that the antisense effects were driven by duplexes formed between ASOs and their respective intended targets. However, in case of L14, the inhibitory effect was linked to an untargeted 10C since the deletion of 10C reversed the effect of L14 on SMN2 exon 7 splicing (Fig. 3A). Further support for the inhibitory role of untargeted 10C came from SMN2/64A mutant in which 10C was replaced by 10A. Since this C-to-A change did not affect SMN2 exon 7 splicing pattern, 10A provided an ideal substitution to probe the role of an untargeted residue immediately upstream of L14-duplex. L14 was able to fully restore exon 7 inclusion in SMN2/64A. This surprising result constitutes one of the rare findings in which a target-specific antisense response was reversed by an otherwise neutral single nucleotide substitution at the untargeted position. Our results also confirmed that the specific location of untargeted 10C upstream of L14-duplex with respect to the 5′ ss is responsible for the inhibitory effect of L14. Supporting this argument, moving the target sequence away from the 5′ ss significantly decreased the inhibitory response of L14 (Fig. 4).
Unique antisense response is modulated by a context-specific long-distance interaction
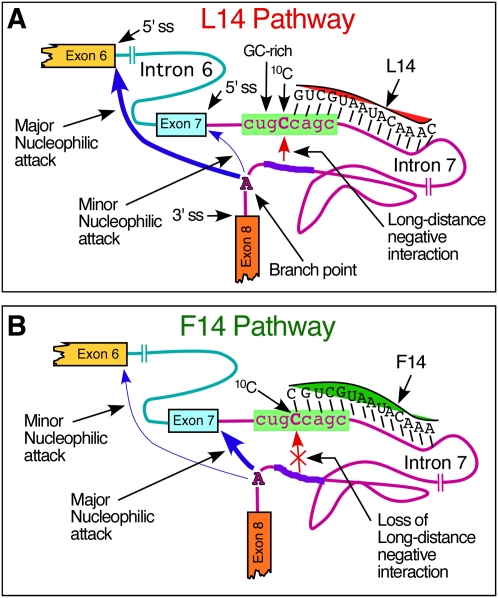
We used Casp3 minigene containing ISS-N1 to compare the impact of F14 and L14 in the context of a heterologous system. We have earlier shown that insertion of ISS-N1 downstream from the 5′ ss of Casp3 exon 6 promotes skipping of this exon. As expected, F14 fully restored Casp3 exon 6 inclusion. However, L14 also produced a noticeable stimulatory response, clearly suggesting that the inhibitory effect of unsequestered 10C upstream of L14 duplex is not dependent upon a linear cis-element (Fig. 5). These results provided the first evidence implicating the role of a long-distance interaction that is generally hard (if not impossible) to predict by available algorithms. Our subsequent experiments with a series of deletion mutations in the second half of intron 7 furnished the strong proof in support of such interaction (Figs. 6, 7). Our results suggested that an unsequestered 10C in presence of L14-duplex interacts with intron 7 sequences upstream of the 3′ ss of exon 8. It is conceivable that the annealing of L14 changes the local structure so that 10C becomes particularly “accessible” for interactions due to for example flipping. However, further experiments are required to confirm this possibility. One can hypothesize that a long-distance interaction with accessible 10C might interfere with a catalytic core formation at the 5′ ss of exon 7. Consequently, the competing 5′ ss of exon 6 becomes the favorable substrate for the transesterification reaction leading to exon 7 skipping (Fig. 9). On the other hand, 10C sequestered in F14-duplex would be no longer available for interactions, leading to usage of the 5′ ss of exon 7 (Fig. 9).
FIGURE 9.
Model of SMN2 exon 7 splicing modulation by 10C in the presence of L14 and F14. (A) L14 pathway: L14 leaves 10C unsequestered and accessible for a long-distance interaction with intronic sequences upstream of the branch point. This arrangement interferes with the catalytic core formation at the 5′ ss of exon 7. Consequently, the competing 5′ ss of exon 6 becomes the favorable substrate for the transesterification reaction leading to exon 7 skipping. (B) F14 pathway: F14 sequesters 10C and prevents a long-distance interaction with intronic sequences upstream of the branch point. This arrangement favors catalytic core formation at the 5′ ss of exon 7. Consequently, SMN2 exon 7 inclusion is promoted.
Recent updates reaffirm that spliceosome complexes are massive, dynamic ribonucleoprotein assemblies that undergo extensive remodeling and exchange of components as spliceosomes are constructed, activated, and recycled (Smith et al. 2008; Wahl et al. 2009; Newman and Nagai 2010). Prp8 is the largest spliceosomal protein that plays a significant role in formation of catalytic core of spliceosome by establishing contacts with the 5′ ss, 3′ ss, and branch point (Grainger and Beggs 2005). It is possible that specific orientation of untargeted 10C prevents recruitment of Prp8 and/or its interacting partners that include RNA helicases with unwinding activity of RNA:RNA duplexes. Independent reports suggest that the structural rearrangement within spliceosome must release branch point–binding complexes for the first transesterification reaction to take place (Golas et al. 2005; Lardelli et al. 2010). Hence, it is probable that the specific orientation of the untargeted 10C affects release of the branch point-binding complexes. Role of specific orientation of the untargeted 10C is somewhat supported by the observation that 5′1U-F15 ASO that has a 3′ uridine overhang eliminates the inhibitory effect associated with L14-duplex (Fig. 3B). Since the overhang is positioned opposite to 10C, one may hypothesize that either a noncanonical base pairing with 10C partially alters the orientation of 10C or the overhang sterically shields 10C, making it inaccessible. As a consequence, the inhibitory effect of untargeted 10C is reduced. Irrespective of the possible mechanism, our results clearly indicate that the inhibitory effect of unsequestered 10C is not linked to differential recruitment of hnRNP A1 to ISS-N1 region since L14 and F14 were equally efficient in displacing this inhibitory factor from ISS-N1 (Fig. 8). The discovery of the prominent role of 10C in this study combined with our recent report regarding splicing modulation by an 8-mer ASO targeting GC-rich sequence, we are tempted to suggest that effect of these ASOs are realized at least in part due to direct remodeling of catalytic core and not merely due to displacement of an inhibitory factor.
Despite the fact that human U2 introns have no preference for a particular residue at the 10th intronic position (Burge et al. 1999), a majority (five out of eight) of human SMN introns contain C residue at the 10th intronic position. Four of these 10Cs are also conserved between human and mice SMN introns. In regard to 10C of ISS-N1, the entire GC-rich sequence is not conserved between human and mouse. It remains to be seen if evolutionary preference for 10Cs in most human SMN introns is merely a coincidence or a part of a yet to be identified regulatory network. Although, not all 10C-containing SMN introns are associated with skipping of exons, it cannot be ruled out that the specific residues at the 10th intronic position may augment/delay the process of catalytic core formation and intron removal. Our finding that the stimulatory effect in presence of the untargeted 10C was not at par with the sequestered 10C in all contexts supports this hypothesis.
The number of reported regulatory elements in the vicinity of the 5′ ss of SMN2 exon 7 continues to grow (Singh 2007b; Hua et al. 2008; Gladman and Chandler 2009). Majority of these cis-elements are absent in mouse Smn and seem to be specific to humans. Linear cis-elements and RNA secondary structures define most of these regulatory elements. Deletions and substitution mutations have been able to validate the role of all of the SMN cis-elements described so far. In this study, use of an ASO-based approach was able to establish the significance of a single cytosine residue at the 10th intronic position that falls within a unique GC-rich sequence. Since several of 10C deletion/substitution mutations did not change the splicing pattern of SMN2 exon 7 and yet reversed the antisense response, it became obvious that 10C is an integral part of the regulatory network involving long-distance and/or secondary interactions. Role of such interactions have been implicated in several systems (Bartel et al. 1991; Matsuura et al. 2001; Singh et al. 2004c). Future experiments would address the detailed mechanistic aspects that define the very unique 5′ ss of a critical exon, skipping of which is associated with a major human disease.
MATERIALS AND METHODS
Minigenes and expression vectors
Minigene splicing cassettes pSMN2ΔI6, SMN2/I7-8, SMN2/5A, SMN2/5G, N1Δ30-34, N1Δ25-34, Casp3Avr, Casp3ISS-N1, and SMN2/64A were described earlier (Singh et al. 2004b, 2006, 2009). Mutations, deletions, and insertions within minigenes were introduced by PCR using Phusion High-Fidelity DNA polymerase (New England Biolabs). Minigene splicing cassettes Casp3-3′Cl and Casp3ISS-N13′Cl were generated by inserting TTAAATTAA sequence in Casp3Avr and Casp3ISS-N1, respectively, using the AvrII restriction site. The exact locations of TTAAATTAA sequence are shown in Figure 5A. In the Casp3-SMN5′-1 minigene, the entire SMN sequence from the 43rd position of exon 7 to 24th position of intron 7 was inserted in Casp3Avr minigene using two-step high fidelity PCR as described earlier (Singh et al. 2007). The same PCR approach was used to generate Casp3SMN2 hybrid minigene, in which the last 116 residues of intron 5 and the entire exon 6 of Casp3 were substituted with the last 116 nt of intron 6, the entire exon 7, and the first 99 nt of intron 7 of SMN2 (Fig. 5D). All minigene constructs were verified by sequencing. All oligonucleotides for cloning and sequencing were obtained from Integrated DNA Technologies.
To generate a bacterial expression vector for human hnRNP A1 recombinant protein, hnRNP A1 coding sequence was inserted in frame with the downstream Mycobacterium xenopi GyrA intein/chitin binding domain in pTXB3 plasmid (New England Biolabs). hnRNP A1 sequence was amplified from an hnRNP A1 expression vector provided by Dr. Benoit Chabot (Université de Sherbrooke) (LaBranche et al. 1998). For cloning purposes, position 51 in hnRNP A1 coding sequence was mutated by a two-step PCR amplification. The 5′ portion of hnRNP A1 was amplified with a pair of primers 5′-RNPA-imp2 (GTGGTGGTACCATGGCCTCTAAGTCAGAGTCTCCTAAAGAGCCCGAACAG) and 3′-RNPA-U mut (CCCTCCAATaAAGAGCTTCCTCAGCTGTTCGGGCTC), where NcoI restriction site is underlined and an A-to-T mutation at position 51 is indicated by a lowercase letter. This mutation is translationally silent but destroys a SapI restriction site in a wild-type hnRNP A1. As a part of our cloning strategy, a GCC codon was added at the beginning of hnRNP A1 sequence in 5′-RNPA-imp2 primer (indicated in italics). This codon helps to create an NcoI restriction site, which itself [site] contains an ATG sequence used for translation initiation. To amplify the 3′ portion of hnRNP A1 sequence, we used a pair of primers 5′-RNPA-U mut (AGGAAGCTCTTtATTGGAGGGTTGAGCTTTGAAAC) and 3′-RNPA-imp (CTGGTGGTTGCTCTTCCGCAAAATCTTCTGCCACTGCCATAGCTAC) with an A-to-T mutation at position 51 indicated by a lowercase letter and the SapI restriction site underlined. The 3′-RNPA-imp primer deletes the termination codon of hnRNP A1 and introduces the SapI restriction site followed by the first codon of M. xenopi GyrA intein fused to the last codon of hnRNP A1. This primer design guarantees that after intein-mediated cleavage of the fusion protein (see Protein Purification Procedure), the recombinant hnRNP A1 will have no additional amino acids at its C terminus. The 5′ and 3′ portions of hnRNP A1 were gel-purified and ligated in the second step PCR reaction using the primers 5′-RNPA-imp2 and 3′-RNPA-imp. The PCR product corresponding to the full-length hnRNP A1 was gel-purified, digested with NcoI and SapI and cloned between the corresponding sites of pTXB3 vector. The resulting hnRNP A1 bacterial expression vector, pTXB3-A1, was confirmed by sequencing. As a result of the cloning strategy, the hnRNP A1 protein expressed from pTXB3-A1 contains one extra amino acid appended to the N-terminus.
Cell culture
The E. coli strain ER2566 (New England Biolabs) was used for expression of the recombinant hnRNP A1 protein. The strain was grown in LB medium supplemented with 100 μg/mL of ampicillin. HeLa cells obtained from the American Type Culture Collection were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Primary fibroblast cell line from SMA type I patient (repository no. GM03813) was obtained from Coriell Cell Repositories. These cells were maintained in MEM supplemented with 2 mM GlutaMAX-I and 15% FBS. All tissue culture media and supplements were purchased from Invitrogen.
Antisense oligonucleotides
RNA ASOs were synthesized by Dharmacon Inc. or by TriLink Biotechnologies. ASOs incorporated 2′-O-methyl modification and phosphorothioate backbone (2OMePS) as described earlier (Singh et al. 2006). LNAs containing uniform phosphorothioate backbone were synthesized by Exiqon.
Transfections and in vivo splicing assays
Transient transfections of cells with plasmid DNA and/or with ASOs were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's recommendations. Briefly, cells were plated 24 h prior to transfection so that their density on the day of transfection was ∼80%. Oligonucleotide concentration varied and is specified in the legends of Figures 1 and 2. Depending on the experiment, the amount of the plasmid used for transfection was either 0.1 or 0.8 μg. In a given experiment, the total amount of ASO was maintained constant by adding the control oligonucleotide (5′-UUGCCUUUCU-3′). The transfection efficiency of an ASO was measured in a parallel experiment with the fluorescent labeled control ASO along with the experimental ASO. Total RNA was isolated at the indicated time points using Trizol reagent (Invitrogen). To generate cDNA, reverse-transcription was carried out using SuperScript III Reverse Transcriptase (Invitrogen) and Oligo (dT) primer (Invitrogen). Minigene-specific spliced products were identified using Taq DNA polymerase (Invitrogen) and the following pairs of primers: P1 and P2 for SMN2 minigene (Singh et al. 2004b), P1 and P56 for Casp3Avr-based minigenes (Singh et al. 2006), and N-24 and P2 for endogenous SMN (Singh et al. 2006). PCR reactions were performed in the presence of a trace amount of [a-32P] dATP (3000 Ci/mmol, Perkin-Elmer). Analysis and quantifications of spliced products were performed using a FPL-5000 Image Reader and Multi Gauge software (Fuji Photo Film Inc.). Results were confirmed by at least three independent experiments. Standard deviation was less than 5% of the mean.
Purification of hnRNP A1 protein
E. coli ER2566 was transformed with pTXB3-A1 expression plasmid and plated on LB-agar plates containing ampicillin. A single freshly grown colony from the LB plate was inoculated into 5 mL of LB medium with ampicillin and grown overnight at 37°C. The entire 5 mL of the overnight culture was then used to inoculate 500 mL of fresh LB medium with ampicillin, and bacterial growth continued until the OD600 of the culture reached 0.8. At this moment, the culture was shifted to 30°C, and the protein expression was induced with 0.2 mM IPTG for 5 h. All subsequent steps were carried out at 4°C. The cells were harvested by centrifugation at 6000 rpm for 10 min (Sorvall Legend RT+ centrifuge, Thermo Scientific), and the cell pellet was resuspended in 10 mL of ice-cold column buffer (CB; 20 mM Na-HEPES at pH 8.5, 1 mM EDTA, 500 mM NaCl) supplemented with Protease Inhibitor Cocktail (Roche). Cells were lysed by sonication on ice at 5 W for five 10-sec bursts, with 60-sec intervals (Microson Ultra sonic cell disruptor, Misonix Inc.) followed by centrifugation at 12,000 rpm for 15 min to remove cell debris. For affinity chromatography, the lysate was diluted to 50 mL with ice-cold CB and slowly loaded on a chitin (New England Biolabs) column (Bio-Rad; 10 mL plastic column with a 5 mL bed volume) equilibrated at 4°C with 50 mL of CB. The column was then washed with 100 mL of CB, followed by another wash with 50 mL of CB, in which NaCl concentration was increased from 500 to 700 mM. The hnRNP A1 protein was released by inducing an on-column intein self-cleavage in the presence of DTT. To induce intein self-cleavage, the column was quickly flushed with 15 mL of CB containing 50 mM DTT. The column flow was stopped, and the column was left overnight at 4°C. The freed hnRNP A1 protein was eluted from the column with CB. Elution fractions of 1 mL were collected, and the hnRNP A1 protein was recovered in the first nine fractions. The pooled fractions were desalted using NAP columns (GE Healthcare) and stored in 50% glycerol at −80°C. The eluted protein was analyzed by electrophoresis in 10% SDS-polyacrylamide gels, stained with Coomassie blue. Protein concentration was determined against BSA standards run on a SDS-polyacrylamide gel.
Generation of site-specifically labeled RNA probe
All synthetic RNA oligonucleotides used to generate a site-specifically 32P-labeled probe were obtained from Dharmacon. The 3′ portion of the probe, 5DN-18 (AUUAUGAAAGUGAAUCUU), was 5′-end-labeled using [γ-32P] ATP (3000 Ci/mmole, Perkin-Elmer) and T4 polynucleotide kinase (New England Biolabs) followed by phenol:chloroform extraction and ethanol precipitation. The 5′-end-labeled 5DN-18 fragment was then ligated to the 5′ portion of the probe, TSL2-U1-32 (CAUUCCUUAAAUUAAGGAGUAAGUCUGCCAGC) using T4 DNA ligase (New England Biolabs) and a bridging DNA oligonucleotide (GGAATTTAATTCCTCATTCAGACGGTCGTAATACTTTCAC). The bridging oligonucleotide was complementary to 28- and 12-nt-long segments on the 5′ and 3′ portions of the probe, respectively (Fig. 8B). Briefly, two RNA fragments were hybridized to the bridging DNA by mixing 50 pmol of 5DN-18, 50 pmol TSL2-U1-32, and 100 pmol of bridging DNA and 2 μL 10× DNA ligation buffer (New England Biolabs) in a 16 μL reaction mixture. The mixture was heated for 2 min at 75°C and shifted to 37°C. At this point 20 units of Superase (Ambion) was added to the reaction mixture, and the incubation proceeded for 3 h. The hybridized substrates were then ligated by adding 20 nmol of ATP and 400 units of T4 DNA ligase (New England Biolabs). The ligation was carried out for 4 h at 37°C. The ligated RNA product was gel purified in a denaturing 16% polyacrylamide gel containing 8 M urea. The RNA product was eluted overnight at 37°C using the “crush and soak” method (Singh et al. 2006) and precipitated with ethanol.
UV-crosslinking
For UV-crosslinking under native condition, the site-specifically 32P-labeled RNA probe was first denatured for 3 min at 90°C and refolded for 1 h at 37°C. Fifty nanograms of the refolded probe was then used in a 50 μL crosslinking reaction containing 20 mM Tris-Cl (pH 7.6), 200 mM KCl, and 2 mM MgCl2. When needed, 0.6 μM of an ASO of interest was used in crosslinking reaction. Initial experiments were done in native condition in which RNA probe was first refolded by heating for 3 min at 90°C followed by slow cooling to room temperature. Next, refolded RNA probe and the ASO were incubated for 1 h at 37°C before 2.5 μg of hnRNP A1 was added; the reaction was then moved to ambient temperature, and the incubation continued for another 10 min. The UV-crosslinking of RNA–protein complexes was carried out on ice at a distance of 0.5 cm for 15 min using a hand-held UV-transilluminator (254 nm, UVG 54, UVP). After crosslinking, the RNA was digested with 1 unit of RNase T1 (USB) and 1 μg RNase A (TEKnova) for 30 min at 37°C. The crosslinked products were resolved by electrophoresis on a 13% SDS-polyacrylamide gel, which was then dried and analyzed using a FPL-5000 Image Reader (Fuji Photo Film Inc.).
For UV-crosslinking under denaturing condition, 50 ng of the RNA probe was added to a 50 μL crosslinking reaction containing 20 mM Tris-HCl (pH 7.6), 200 mM KCl, and 2 mM MgCl2 with or without 0.6 μM ASO. The reaction mixture was heated for 3 min to 90°C and moved to 37°C for an overnight incubation. This refolding of the RNA and an ASO together ensures that the latter will anneal to its target sequence. Following the overnight incubation, 2.5 μg of hnRNP A1 was added to the reaction, and it was shifted to an ambient temperature for 10 min. UV-crosslinking and detection of RNA-hnRNP A1 crosslinked products were done similarly as described for “native” conditions.
ACKNOWLEDGMENTS
We thank Drs. Benoit Chabot and Peter Mouritzen for providing hnRNP A1 expression vector and for providing help with design of LNAs, respectively. This work was supported by a grant from United States National Institutes of Health (R01NS055925) to R.N.S. R.N.S. acknowledges support from a Salsbury Endowment at Iowa State University. N.N.S. is supported in part by the Center for Integrated Animal Genomics at Iowa State University. R.N.S. and N.N.S. are also supported in part by the Iowa Center for Advanced Neurotoxicology.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2154310.
REFERENCES
- Bartel DP, Zapp ML, Green MR, Szostak JW 1991. HIV-1 Rev regulation involves recognition of non-Watson–Crick base pairs in viral RNA. Cell 67: 529–536 [DOI] [PubMed] [Google Scholar]
- Bauman J, Jearawiriyapaisarn N, Kole R 2009. Therapeutic potential of splice-switching oligonucleotides. Oligonucleotides 19: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle M, Baralle FE 2006. Defective splicing, disease and therapy: Searching for master checkpoints in exon definition. Nucleic Acids Res 34: 3494–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Dhir A, Lewandowska MA, Baralle FE 2007. RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion events. Nucleic Acids Res 35: 4369–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world (ed. Gesteland RF et al. ), 2nd ed, pp. 525–560 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Chasin LA 2007. Searching for splicing motifs. Adv Exp Med Biol 623: 85–106 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL 2008. The search for alternative splicing regulators: New approaches offer a path to a splicing code. Genes Dev 22: 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco MA 2006. Alternative splicing: Therapeutic target and tool. Prog Mol Subcell Biol 44: 47–64 [DOI] [PubMed] [Google Scholar]
- Gladman JT, Chandler DS 2009. Intron 7 conserved sequence elements regulate the splicing of the SMN genes. Hum Genet 126: 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Lührmann R, Stark H 2005. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol Cell 17: 869–883 [DOI] [PubMed] [Google Scholar]
- Graveley BR 2005. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123: 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ 2008. Combinatorial control of exon recognition. J Biol Chem 283: 1211–1215 [DOI] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet 19: 199–202 [DOI] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR 3rd, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80: 1–5 [DOI] [PubMed] [Google Scholar]
- Lin S, Fu XD 2007. SR proteins and related factors in alternative splicing. Adv Exp Med Biol 623: 107–122 [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B 2007. hnRNP proteins and splicing control. Adv Exp Med Biol 623: 123–147 [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Moore MJ 2007. Spliceosome assembly and composition. Adv Exp Med Biol 623: 14–35 [DOI] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20: 7259–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AJ, Nagai K 2010. Structural studies of the spliceosome: Blind men and an elephant. Curr Opin Struct Biol 20: 82–89 [DOI] [PubMed] [Google Scholar]
- Nilsen TW 2003. The spliceosome: The most complex macromolecular machine in the cell. Bioessays 25: 1147–1149 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Verhoef K, Southern E, Huthoff H, Berkhout B 2004. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res 32: 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2008. Conserved RNA secondary structures promote alternative splicing. RNA 14: 1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN 2007a. Unfolding the mystery of alternative splicing through a unique method of in vivo selection. Front Biosci 12: 3263–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN 2007b. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol 4: 7–10 [DOI] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ, Singh RN 2004a. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit Rev Eukaryot Gene Expr 14: 271–285 [DOI] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ, Singh RN 2004b. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun 315: 381–388 [DOI] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ, Singh RN 2004c. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA 10: 1291–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ, Singh RN 2006. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol 26: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Singh RN, Androphy EJ 2007. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res 35: 371–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN 2009. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol 6: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. ‘Nought may endure but mutability’: Spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S 2009. Alternative splicing and disease. Biochim Biophys Acta 1792: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veedu RN, Wengel J 2009. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol 6: 321–323 [DOI] [PubMed] [Google Scholar]
- Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J 2007. Refined characterization of the expression and stability of the SMN gene products. Am J Pathol 171: 1269–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA 2010. The pathology of splicing. J Pathol 220: 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Diegel JV, von Hippel PH, Berglund JA 2009. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci 106: 9203–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ 2009. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci 29: 7633–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Hahnen E 2006. Spinal muscular atrophy and therapeutic prospects. Prog Mol Subcell Biol 44: 109–132 [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee C 2007. Relating alternative splicing to proteome complexity and genome evolution. Adv Exp Med Biol 623: 36–49 [DOI] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Lührmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]