Abstract
The ribosome is a massive ribonucleoprotein complex (∼2.4 MDa) that utilizes large-scale structural fluctuations to produce unidirectional protein synthesis. Accommodation is a key conformational change during transfer RNA (tRNA) selection that allows movement of tRNA into the ribosome. Here, we address the structure–function relationship that governs accommodation using all-atom molecular simulations and single-molecule fluorescence resonance energy transfer (smFRET). Simulations that employ an all-atom, structure-based (Gō-like) model illuminate the interplay between configurational entropy and effective enthalpy during the accommodation process. This delicate balance leads to spontaneous reversible accommodation attempts, which are corroborated by smFRET measurements. The dynamics about the endpoints of accommodation (the A/T and A/A conformations) obtained from structure-based simulations are validated by multiple 100–200 ns explicit-solvent simulations (3.2 million atoms for a cumulative 1.4 μs), and previous crystallographic analysis. We find that the configurational entropy of the 3′-CCA end of aminoacyl-tRNA resists accommodation, leading to a multistep accommodation process that encompasses a distribution of parallel pathways. The calculated mechanism is robust across simulation methods and protocols, suggesting that the structure of the accommodation corridor imposes stringent limitations on the accessible pathways. The identified mechanism and observed parallel pathways establish an atomistic framework for interpreting a large body of biochemical data and demonstrate that conformational changes during translation occur through a stochastic trial-and-error process, rather than in concerted lock-step motions.
Keywords: energy landscape, ribosome, accommodation, all-atom simulation, tRNA selection
INTRODUCTION
Advances in X-ray crystallography (Schluenzen et al. 2000; Yusupov et al. 2001; Blaha et al. 2009; Schmeing et al. 2009; Zhang et al. 2009) and cryogenic electron microscopy (Cryo-EM) (Agirrezabala et al. 2008; Schuette et al. 2009) methods have afforded highly insightful snapshots of ribosome structures, each representing a local energetic minimum along the translation process (Fig. 1A–C). Single-molecule fluorescence resonance energy transfer (smFRET) has been used to characterize the transitions between these states (Blanchard 2009; Aitken et al. 2010). In parallel, pioneering modeling efforts (Malhotra et al. 1990; Malhotra and Harvey 1994) and advances in high-performance computing have ushered in an era in which molecular modeling and simulations can elucidate the relationship between static ribosome structures (Roberts et al. 2008; Villa et al. 2009), structural fluctuations about these local minima (Tama et al. 2003; Wang et al. 2004; Trylska et al. 2005), and ribosome function (Sanbonmatsu et al. 2005; Petrone et al. 2008; Gumbart et al. 2009). Here, we integrate molecular dynamics (MD) simulations and smFRET to describe the structural and dynamical features of the ribosome that govern translation, in atomic detail.
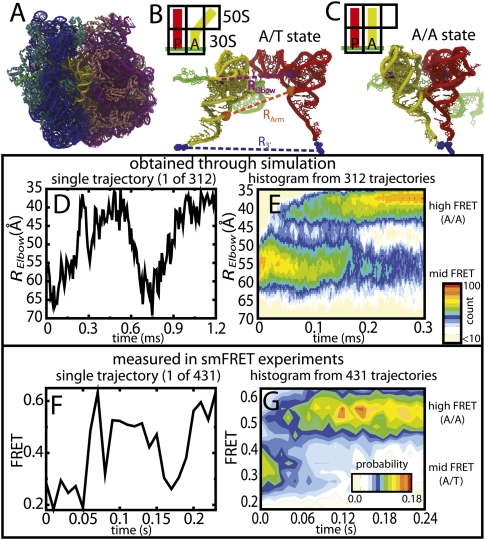
FIGURE 1.
Reversible fluctuations observed in simulations and smFRET. (A) Structure of the 70S ribosome with 16S rRNA (blue), 30S proteins (cyan), 23S rRNA (purple), 50S proteins (pink), mRNA (green), aa-tRNA (yellow), and p-tRNA (red) shown. aa-tRNA shown in the (B) A/T conformation and (C) A/A conformation. The purple dashed line, RElbow, is the distance between U47 of the aa-tRNA and U8 of the P-site tRNA (purple spheres), used experimentally, and computationally (distance between O3′ atoms), to measure aa-tRNA elbow accommodation. The orange dashed line, RArm, is the distance between the C3′ atom of G4 in the aa-tRNA and C5′ atom of G67 in P-site tRNA (orange spheres), which measures acceptor arm accommodation. The blue dashed line, R3′, is the distance between A-site and P-site amino acids (blue spheres), which measures 3′-CCA end accommodation into the peptidyltransferase center. (D) Time trace of RElbow for a single accommodation simulation. (E) P(RElbow, t), the unnormalized probability, was calculated from 312 independent accommodation transitions (12 million sampled structures). Values are colored off-white (low) to red (high), on a log scale, throughout all figures. Since FRET efficiencies are inversely related to RElbow , the Y-axes in D and E are inverted for easier comparison with F and G. (F) Single-molecule time trace monitored after stop-flow delivery of cognate EF-Tu·GTP·Phe-tRNAPhe(Cy5-acp3U47) to surface-immobilized ribosome complexes carrying fMet-tRNAOH in the P-site (Cy3-s4U8). (G) FRET population histogram of single FRET traces postsynchronized to a FRET value of 0.323. Population only includes traces where the tRNA reaches the A/A state (N = 431). Note: Reversible fluctuations were observed in many simulations and FRET traces and each trace has a unique profile. The uncanny similarities between the shapes of D and F are coincidental.
The fidelity mechanism underpinning transfer RNA (tRNA) selection on the ribosome is a critical determinant of the genetic code. In the case of cognate tRNA, this multistep process is rate limited by accommodation (Rodnina and Wintermeyer 2001), wherein conformational changes in the translating complex direct entry of the 3′-CCA end of aminoacyl-tRNA (aa-tRNA) into the peptidyltransferase center (PTC) located in the 50S subunit (Fig. 1B,C). While smFRET can provide time-resolved measurements of this biological process (Blanchard et al. 2004), technology allowing simultaneous measurement of multiple degrees of freedom is still in its infancy.
Although a fully atomistic picture of accommodation is available through simulation (Sanbonmatsu et al. 2005), acquiring statistics that are comparable to experiments represents a significant challenge due to the size of the ribosome, the relatively slow timescale of the process (τ ∼ 10–100 ms), and the large displacement (∼90 Å) associated with aa-tRNA entry into the PTC. While conventional MD simulations of small polymers (∼104 atoms, including solvent) have reached microsecond timescales (Freddolino et al. 2008; Garcia and Paschek 2008; Monticelli et al. 2008), and are rapidly approaching the biologically relevant millisecond regime (Klepeis et al. 2009), achieving similar timescales with the ribosome (>3 × 106 atoms) necessitates new strategies and alternative approaches.
One way to access longer timescale simulations of the ribosome is to utilize the energy landscape theory of biomolecular folding and function, which was originally developed in the context of protein folding (Bryngelson et al. 1995). It is now well acknowledged that the energy landscape for protein folding has sufficiently small energetic trapping, such that the geometrical features of transition states and folding intermediates can be determined from protein topology (Clementi et al. 2000). This has allowed the use of energetically smooth force fields to explore folding mechanisms. These force fields, which are defined by the native structure, are also less expensive than highly detailed empirically based force fields, facilitating longer timescale simulations and longer length scale simulations. Inspired by the many successes in protein and RNA folding and function (Hills and Brooks 2009; Pincus et al. 2009), we apply an all-atom structure-based (Gō-like) model (Whitford et al. 2009a,b) of the Thermus Thermophilus 70S ribosome (Fig. 1A) (the endpoints of each simulation were taken from Tung and Sanbonmatsu [2004] and Sanbonmatsu et al. [2005]) to simulate the process of aa-tRNA accommodation.
By employing an all-atom structure-based force field, we are exploring the possibility that the ribosome serves as a structural filter, where the incoming aa-tRNA is accepted once it successfully navigates the steric barriers imposed by the accommodation corridor. Since steric contributions will be similar across simulation protocols, if steric signatures exist, one should be able to identify them with a simplified set of energetic interactions. The electrostatic composition is an indispensable contributor to ribosome structural stability. Since the crystal structure is defined as the lowest energy conformation in this model, these stabilizing electrostatic contributions are implicitly included. Due to ionic screening we assume that these interactions may be approximated by shorter-range interactions. In the structure-based model, attractive atom–atom interactions range ∼5–7 Å (where the energetic minima are at <4 Å). We further assume the potential energy decreases as a function of the accommodation reaction, where local energetic roughness is small and can therefore be described by an effective diffusion coefficient (Bryngelson and Wolynes 1989). We also performed explicit-solvent explicit-electrostatics simulations (Supplemental Fig. S1) for hundreds of nanoseconds each (Supplemental Table S1) to ensure the overall dynamics of the A/T and A/A conformations are consistent between these disparate simulation techniques.
In this study, we employed four different approaches to study the accommodation process: unrestrained structure-based simulations, explicit-solvent simulations, targeted molecular dynamics simulations, and smFRET. These methods provide independent measures of the dynamics, which enables cross-validation of the results. The identification of accommodation pathways, steric barriers, and transient intermediates with associated transition states was achieved by simulating 312 independent, and unrestrained, full accommodation transitions (which totaled roughly 200 ms of sampling) (see Materials and Methods). Shorter-timescale targeted molecular dynamics (TMD) simulations (704 accommodation events) confirmed the presence of the predicted steric barriers. Conventional explicit-solvent simulations were used (3.2 million atoms for a total of 1.4 μs of simulated time) to validate the short timescale motions (tens, to hundreds, of nanoseconds) and the overall scale and distribution of the structural fluctuations in the structure-based simulations. All structure-based and explicit-solvent simulations were of the full 70S ribosome with aa-tRNAPhe and P-site tRNAPhe. High-spatial and -time resolution smFRET imaging of aa-tRNA entering the ribosome corroborates the existence of reversible large-scale rearrangements in the aa-tRNA elbow during accommodation, which serves as partial validation of the simulated dynamics. Together, these data provide an atomistic framework for interpreting experimental measurements and building a theoretical understanding of tRNA dynamics in the ribosome.
RESULTS
Structure-based dynamics consistent with crystallographic measurements, explicit-solvent simulations, and normal mode calculations
The structural fluctuations of the 16S and 23S rRNA observed in the structure-based simulations have the same magnitude and distribution as those described by crystallographic anisotropic B-factors and explicit-solvent simulations. The average spatial root-mean-squared displacement (RMSD) of the 16S and 23S rRNA is similar for the crystal structure (1.7 Å) (Korostelev et al. 2008),4 structure-based simulations (1.7 Å), and explicit-solvent simulations (1.9 Å). The three distributions are also well correlated (Supplemental Fig. S2). The crystal structure of Korostelev et al. (2008) was used for comparison since translation–libration–screw (TLS) refinement (Winn et al. 2001) was used to determine anisotropic B-factors, which can provide a more realistic description of the dynamics than isotropic B-factors. The largest variation in the RMSD obtained from these methods is found for the L1 and L11 stalks in the 50S subunit and the beak and spur of the 30S subunit. While both simulation methods predict larger structural fluctuations in the stalks than are seen in the crystal structures, smFRET measurements corroborate spontaneous large-scale rearrangements of the L1 stalk in solution (Blanchard 2009; Aitken et al. 2010). These predicted stalk motions are also consistent with normal mode calculations that indicate the lowest frequency modes (i.e., the modes with the smallest potential energy gradient) correspond to L1 and L11 stalk motions (Tama et al. 2003).
Reversible aa-tRNA motions observed in simulations and single-molecule FRET experiments
Each all-atom simulation, which employed a structure-based Hamiltonian (Whitford et al. 2009a,b), was initiated in the A/T conformation and was terminated when the endpoint of accommodation was reached (i.e., Fig. 1C, A/A state). In the A/T conformation, aa-tRNA adopts a kinked, and therefore strained, configuration (relative to the A/A state). The aa-tRNA might even be considered a loaded, highly nonlinear, spring (Frank et al. 2005). In this model, the strain energy associated with the A/T conformation is solely due to contact distances and dihedral angles being displaced from the values found in the A/A conformation. With this time-independent potential energy function the aa-tRNA explores the free-energy landscape and spontaneously reaches the fully accommodated (A/A) conformation (Supplemental Fig. S3A,B). Since previous biochemical data suggest EF-Tu dissociation is faster than accommodation (Rodnina and Wintermeyer 2001), EF-Tu was not included in the simulation, though future work will explore the interplay between accommodation and EF-Tu.
Both structure-based simulations and smFRET experiments reveal that reversible fluctuations in distance occur between the elbow regions of A- and P-site tRNAs, prior to achieving the final A/A state (Fig. 1D,F). Comparable statistics for the accommodation process were obtained from computational and experimental approaches (312 versus 431 independent events) (Fig. 1E,G), which highlights the potential for direct theoretical-experimental comparison via these methods. Fluctuations between high-FRET (with the elbow accommodated and the 3′-CCA end not accommodated) (Fig. 2E) and mid-FRET (Fig. 1B, A/T) configurations, observed in both simulation and smFRET experiments, show that such motions are intrinsic to the tRNA–ribosome–mRNA complex.
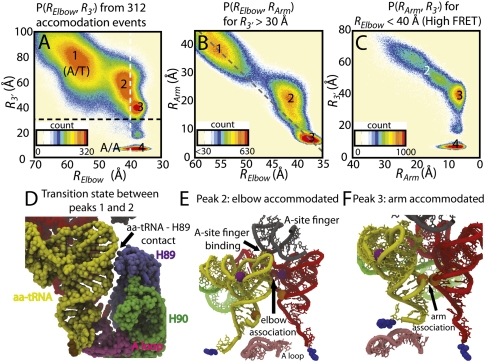
FIGURE 2.
Probability distributions reveal sequential elbow, arm, 3′-CCA end accommodation. (A) P(RElbow, R3′) shows four highly populated conformations: (1) A/T, (2) elbow accommodated, (3) acceptor arm accommodated, and (4) A/A states. Peaks are numbered consistently between plots, i.e., the same structures compose peak X (= 1,2,3,4) in each figure. (B) P(RElbow, RArm) prior to 3′-CCA entry into the PTC (R3′ > 30 Å, above the black dotted line in A). The bend in the probability distribution, centered about peak 2, indicates sequential elbow-arm accommodation. Simultaneous elbow and arm accommodation events would fall on the gray dashed line, which connects peaks 1 and 3. (C) P(RArm, R3′) for elbow-accommodated structures (RElbow < 30 Å, right of white dotted line in A). RArm and R3′ are correlated, but the arm reaches the A/A conformation, while the 3′-CCA end is ∼40 Å from the PTC (peak 3). (D) Representative snapshot of the transition state associated with elbow accommodation (between peaks 1 and 2 in A), where the tRNA closely approaches H89. (E) Snapshot of the elbow-associated state (peak 2) colored as in Figure 1 with the A-site finger (gray) and A loop (pink) also shown and rotated 90° from D. (F) Representative structure of arm-accommodated state (peak 3).
Since our simulated timescales are only estimated (see the supplemental material), one does not expect exact agreement with experimental timescales. Nonetheless, if one assumes elbow accommodation is a two-state process, where a single free-energy barrier separates the A/T and elbow-accommodated states, differences in timescale of this magnitude may be attributed to a difference in the free-energy barrier of ∼2–7 kT.5 A variety of features could lead to changes in barrier height of this magnitude. For example, in these simulations there is a coordinated displacement of H89, away from the A-site codon, during elbow accommodation (Fig. 3). Since this displacement is purely due to steric interactions (i.e., two atoms may not occupy the same space), if H89 movement were attenuated or amplified, the free-energy barriers should be altered, in both simulations and experiments. While it is beyond the scope of this study to predict which environmental conditions will have the largest effect on the conformation of H89, it is plausible that both ion concentrations and factor binding may be involved. Consistent with the predicted movement of H89, mutational analysis shows H89 is involved in the accommodation process and may act as an allosteric “finger” (Petrov et al. 2008) that regulates tRNA movement through the accommodation corridor.
FIGURE 3.
Displacement of H89 during aa-tRNA elbow accommodation. (A) Structure of A/T conformation with aa-tRNA (yellow), p-tRNA (red), mRNA (green), H89 (purple), and stem–loop of H89 (violet) shown. The distance between the stem–loop of H89 and the A-site codon (H89-codon), is indicated by the pink bar. The green bar indicates RElbow (as defined in Fig. 1B). (B) Probability distribution of H89-codon versus RElbow, calculated from 312 unrestrained structure-based simulations. Only structures in which the arm is not accommodated (RArm > 12 Å) are included in the distribution. The gray line is the average H89-codon distance as a function of RElbow. Note: X-axis is inverted for visual ease. As the aa-tRNA elbow accommodates (i.e., RElbow decreases) H89-codon increases when the aa-tRNA contacts the stem–loop of H89 (RElbow ∼ 49 Å) and then reduces after the aa-tRNA elbow has accommodated.
A multistate mechanism of accommodation
In the structure-based simulations, the underlying potential energy surface is downhill in the A/T to A/A direction. Since the potential energy profile in this model is devoid of energetic traps, thermodynamic barriers are the result of sharp decreases in configurational entropy. The structure of the accommodation corridor introduces configurational restrictions, which lead to entropic barriers that the aa-tRNA must navigate in order to reach the A/A conformation. As the aa-tRNA approaches an entropic barrier, it must search locally for a configuration that will lead to a decrease in energy. As discussed below, the asymmetric structure of aa-tRNA also contributes to such barriers, which reinforces the notion that aa-tRNA actively participates in the selection process (Cochella and Green 2005).
Entropic bottlenecks temporarily halt progress along the accommodation reaction coordinates, which leads to peaks in the probability distributions (Fig. 2A–C).6 Figure 2A–C, suggest that three entropic barriers correspond to the transitions states between the (1) A/T ensemble (Fig. 1B); (2) elbow accommodated state (Fig. 2E); (3) elbow and acceptor arm accommodated state (Fig. 2F); and (4) fully accommodated (A/A) state (Fig. 1C). The first entropic barrier corresponds to an isolated movement of the aa-tRNA elbow (as it associates with the A-site finger). Interactions between the tRNA acceptor arm (residues proximal to A64) and H89 (primarily C2471 and G2472) of the 23S rRNA (Fig. 2D) are formed in the transition state ensemble (defined as configurations where 44 Å < Relbow < 48 Å). Accordingly, elbow accommodation (i.e., movement of the elbow from the A/T ensemble to the A/A position) results in a temporary, reversible displacement of H89 (Fig. 3). Formation of the elbow-accommodated state (Fig. 2E) facilitates subsequent accommodation steps and is accompanied by promiscuous movements of the 3′-CCA end of aa-tRNA, where all possible orientations of the 3′-CCA end (i.e., all values of the pseudodihedral angle Φ2) are sampled (Fig. 4B; Supplemental Video 1). The next entropic barrier is encountered during movement of the aa-tRNA acceptor arm into an accommodated position (Fig. 2F, acceptor arm accommodation). Due to close interactions with the A loop, H89 and H90, the mobility of the 3′-CCA end (as measured by Φ2, Fig. 4B) is reduced (i.e., a decrease in entropy). From this state, accommodation is completed upon insertion of the 3′-CCA end into the PTC (fully accommodated state). These data suggest that the accommodation process itself is multistep in nature and that flexibility in the 3′-CCA end of the aa-tRNA is an integral component of the tRNA selection machinery.
FIGURE 4.
Entropy from 3′-CCA flexibility resists accommodation. (A) 3′-CCA rotation coordinate, Φ2 (Sanbonmatsu et al. 2005), formed by the O3′ atoms of residues 69, 71, 73, and O atom of Phe. (B) Probability as a function of Φ2 and R3′ indicates that in the A/T conformation (R3′ ∼ 80–90 Å) the flexible 3′-CCA end samples all angles with relatively high probabilities. During acceptor arm accommodation (R3′ = 40–60 Å), the restricted mobility of the 3′-CCA end indicates an entropic barrier. As the 3′-CCA end enters the PTC, the configurational entropy decreases as the 3′-CCA end proceeds via a (+) or (−) pathway (pink and black lines). Note that since Φ2 is a dihedral angle, (B) is periodic along the Y-axis with a modulus of 360°. Representative snapshots of 3′-CCA entry (C) along the major groove of H89, (D) by passing over the A loop, or (E) by passing between H89 and the A loop.
Flexible 3′-CCA end of aa-tRNA resists accommodation and accesses multiple pathways
In addition to the entropic barriers introduced by the ribosome structure, the flexible 3′-CCA end of the aa-tRNA also leads to a net loss in entropy upon accommodation (Fig. 4B). This predicted loss in configurational entropy is consistent with high-resolution crystallographic data that find the 3′-CCA end exhibits “relatively high disorder” (Shi and Moore 2000) in isolated tRNA and has low mobility when in complex with the 70S ribosome (Voorhees et al. 2009). This loss of entropy is physically intuitive: There are no external structural constraints on the single-stranded 3′-CCA end of aa-tRNA immediately after dissociation from EF-Tu, while enzymatic function of the ribosome requires specific interactions to be formed with the fully accommodated aa-tRNA molecule. Previous analysis of crystal structures led to the proposal of rotary motion of the 3′-CCA end during translocation (Agmon et al. 2005, 2006), which is similar to the motions observed here. A recent crystal structure of the ribosome complexed with EF-Tu and aa-tRNA (Schmeing et al. 2009) in the A/T conformation revealed large distortions in the 3′-CCA end, which also emphasizes its flexible nature. In addition to crystallographic evidence, explicit-solvent simulations corroborate the broadly distributed 3′-CCA configurations in the A/T ensemble and a narrow distribution of structures in the A/A state (Fig. 5A). Since the explicit-solvent simulations employ an empirically calibrated force field (for a recent review on empirical force fields, see Adcock and McCammon 2006), agreement partially validates the dynamics predicted by the structure-based simulations.
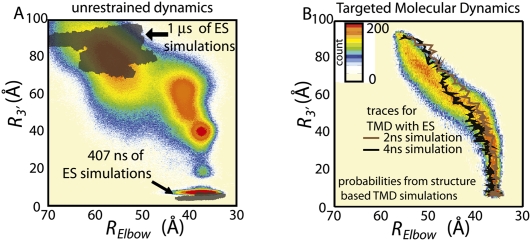
FIGURE 5.
Comparison of structure-based simulations and explicit-solvent (ES) simulations. (A) Color contour plot is the probability distribution obtained from structure-based simulations. Transparent gray shadows delimit the region of the phase space sampled in 1.4 μs of explicit-solvent simulations (a slight shift of the A/A ensemble, to lower R3′, is due to attraction between the aa-tRNA and p-tRNA amino acids). (B) Probability distribution from 704 TMD simulations with a structure-based force field. Two trajectories from explicit-solvent TMD (Sanbonmatsu et al. 2005) are shown as black and brown lines.
The flexibility of the single-stranded 3′-CCA end of tRNA allows it to navigate around the steric barriers surrounding the PTC via multiple pathways (Fig. 4B). During entry (R3′ ∼ 35–10 Å), the 3′-CCA end can take a (+) route where it passes between the 5′ end of the tRNA and H89 (Fig. 4C). Alternately, it may enter in the (−) direction where the 3′-CCA end moves between the aa-tRNA acceptor arm and the A loop (Fig. 4D), or take more extended structures and pass between the A loop and H89 (Fig. 4E; Supplemental Video 2). Comparison of recently obtained high-resolution crystal structures of T. thermophilus ribosomes reveals conformational variability in the PTC (Voorhees et al. 2009), which will be inextricably linked to the likelihood of observing each pathway. Since these results suggest that the dynamics of the incoming aa-tRNA may be influenced by the conformation of the ribosome, the inherent flexibility of the ribosome is also likely to be important during tRNA selection.
Structure of the accommodation corridor leads to a robust mechanism
As discussed above, in the structure-based simulations, the calculated mechanism of accommodation is a result of the steric interactions between the aa-tRNA and the ribosome. To verify that the mechanism is due to these structural restrictions, and not the model's energetics, we compared the predicted accommodation dynamics for a variety of parametric settings. Further, we compared the dynamics to those obtained using targeted molecular dynamics (TMD). We find that overall, the calculated accommodation pathways are similar, though there are some notable exceptions.
The overall elbow, then arm, then 3′-CCA accommodation mechanism is robust to changes in the energetics and temperature (Supplemental Fig. S4). We first reduced the strength of contacts that are unique to the A/A conformation (see Materials and Methods), effectively reducing the driving force of accommodation. While the relative populations of each state varied between parameter sets, the overall accommodation mechanism was consistent. Though, since these simulations are also structure-based, one might argue that the energy profile is still too similar to unambiguously partition the contributions of structure and energetics.
To further investigate which features of the mechanism are robust, we turn to TMD simulations (see Materials and Methods). In TMD simulations a time-dependent constraint induces a transition over a predetermined period of time. Accordingly, since TMD algorithms do not allow the system to fully equilibrate at each point along the transition, the energetic details of the force field become less relevant, such that the stochastic character of the transitions cannot be observed. Therefore, common mechanistic features are likely due to the molecular structure, and not the model's energetics. Comparison of the structure-based simulations without and with TMD (cf. Figs. 2A and 5B) reveals a conserved mechanism, albeit with a narrower distribution of pathways for the latter. We additionally compared the pathways from TMD simulations with an explicit-solvent force field (each 2–4 ns in duration) and a structure-based force field (each 8 μs in duration). Both explicit-solvent (Sanbonmatsu et al. 2005) and structure-based TMD pathways (Fig. 5B; Supplemental Fig. S5) fall within the region of phase space sampled in unrestrained simulations (Fig. 2). While for all simulations 3′-CCA end entry into the PTC is the final step of accommodation, the largest mechanistic difference observed between methods is the precise timing of elbow and arm accommodation. Specifically, in the TMD trajectories the elbow and arm accommodation was more correlated, on average. Since the work done by the TMD algorithm alters the relative timing of the elbow and arm, the balance between their motions may also be sensitive to the environmental conditions. In contrast, even with TMD, CCA entry is facilitated by successful arm accommodation.
DISCUSSION
This study represents the first example in which all-atom simulations of ribosomal conformational transitions have been compared with single-molecule fluorescence experiments. In the context of tRNA accommodation, this work has revealed the presence of reversible structural fluctuations associated with the incoming aa-tRNA, a specific ordering of conformational events and entropic contributions of the 3′-CCA end of aa-tRNA.
The present study also predicts a multistate mechanism of accommodation. The predicted mechanism is robust across simulation methods and protocols employed, suggesting that the structure of the accommodation corridor imposes stringent limitations on the accessible pathways. Specifically, elbow accommodation always precedes 3′-CCA accommodation, though the exact timing of arm accommodation can vary slightly. The feasibility of independent movements of different parts of the tRNA molecule provides a structural framework for interpreting experimental measurements.
Two examples of experimental results that may be re-interpreted in terms of a multistep accommodation mechanism can be found in studies of aa-tRNA dissociation (Dale et al. 2009) and antibiotic-dependent ribosome stalling (Vazquez-Laslop et al. 2008). It was recently shown for Escherichia coli ribosomes that the dissociation rate from the A site is comparable for deacylated-tRNA and acetylated-tRNAs that carry a Phe amino acid, when the tRNA has a tight-binding anticodon (GAC) (Vazquez-Laslop et al. 2008). In contrast, when the anticodon is weak-binding (GAA), acetylating the tRNA with Phe reduces the rate of dissociation by about an order of magnitude. Further, for weak-binding anticodons, deacyl-tRNA dissociation is tRNA body-dependent though aa-tRNA dissociation rates are body-independent. Since our simulations demonstrate the possibility of a multistep accommodation (association) process, it is likely that dissociation also involves multiple steps, each of which has the potential to limit the overall rate. Thus, for weak-binding anticodons, tRNA dissociation may be rate limited by 3′-CCA end release (which may also be coupled to a conformational rearrangement in the PTC, as the authors suggest).
A second ribosomal function for which a multistep, multiple-pathway mechanism may be relevant is drug-dependent ribosome stalling. While it has previously been shown that the PTC is the focal point of erythromycin-dependent stalling of ribosomes from E. coli (Vazquez-Laslop et al. 2008), the atomic mechanism of stalling is still an active area of investigation. Stalled complexes were shown to be insensitive to puromycin, suggesting the aa-tRNA occludes the A site of the PTC in stalled complexes. Our simulations suggest that stalled complexes may be composed of a fully accommodated elbow and arm, and that the 3′-CCA end need not be fully accommodated, but only be close enough to the PTC to prevent puromycin binding. More recently, experimental measurements have revealed that the composition of the A-site codon also contributes to stalling (A Mankin, pers. comm.). One interpretation of this finding is that in stalled complexes, the PTC is structurally perturbed, such that some amino acids are allowed to enter the PTC (and form peptide bonds), while others are not. This is consistent with there being multiple PTC entry pathways; each amino acid has a favorable path and rearrangements in the PTC affect each path differently.
In contrast to macroscopic machines, where tightly coupled rigid parts operate deterministically, experiments can now directly quantify the stochastic dynamics of molecular machines through a variety of single-molecule methods. These rapid advances are necessitating a new set of theoretical and computational methods to explore such functions. Here, in the context of tRNA accommodation, we show how an integrated approach involving multiple simulation techniques and smFRET may begin to unravel the atomic details of functional motions in the ribosome.
MATERIALS AND METHODS
Details of the structure-based Hamiltonian
To simulate accommodation in the T. Thermophilus 70S ribosome (Supplemental Fig. S1), we use an all-atom structure-based model. All heavy (nonhydrogen) atoms are included as spherical beads (140,743 atoms). A potential energy function based on the A/A conformation was used (i.e., the A/A conformation is the global energetic minimum). Each simulation was started in the A/T conformation and was terminated when the A/A conformation was reached (defined as R3′ < 8 Å). When in the A/T state, many A/A intermolecular contacts are not formed and many intramolecular contacts and dihedrals angles are perturbed from their lowest energy configurations. As the tRNA accommodates, these strained interactions relax and provide stabilizing energy to the system. The Hamiltonian employed in this work has been well-characterized elsewhere (Whitford et al. 2009a,b) and is fully described in the Supplemental material.
Simulation details
Structure-based simulations
All simulations were performed using the Gromacs (Berendsen et al. 1995; Lindahl et al. 2001) software package, with force-field files that were generated in-house.7 Reduced units were used (see Supplemental material; Frenkel and Smit 1996) (Gromacs v4 Manual). A time step of 0.0005 was employed. Stochastic dynamics was used with a coupling time of 1.0. Simulations were performed in 5,000,000 time-step segments, and were continued until the A/A structure was reached (5–120 M total time steps). The dynamic load balancing utility of Gromacs was employed (Hess et al. 2008), which allowed each simulation to be performed using 64 and 128 cores with minimal reduction in performance due to intercore communication.
Explicit-solvent simulations
Simulations were performed using Gromacs (Berendsen et al. 1995) v4.0 with the Amber99 force field (Wang et al. 2000).8 All systems were fully solvated with [MgCl2] = 7 mM and [KCl] = 100 mM. A/T simulations included 977,217 water molecules for a total of 3,178,833 atoms (ribosome + ions + water) in each simulation. A/A simulations included 976,610 water molecules for a total of 3,177,012 atoms. The simulation parameters and equilibration protocol are described in the Supplemental material.
Targeted molecular dynamics (TMD)
TMD was implemented as described elsewhere (Schlitter et al. 1994). Each TMD simulation was performed in combination with a structure-based force field and was allowed up to 12 μs to fully accommodate, though simulations typically accommodated in ∼8 μs.
Experimental details
Single-molecule experiments
Sample preparation is described in the Supplemental material. Single-molecule FRET measurements were performed using a prism-based total internal reflection (TIR) fluorescence microscope, where fluorescently labeled molecules were spatially immobilized near a quartz surface following previously published procedures (Munro et al. 2007). Ribosome complexes bearing (Cy3-s4U8) tRNAfMet were excited with monochromatic light at 532 nm (Ventus750mW Nd:YAG, Laser Quantum) by the evanescent wave generated by TIR. Stop-flow delivery of the ternary complex (Cy5-acp3U47) Phe-tRNAPhe, EF-Tu, and GTP to surface-immobilized ribosomes results in FRET due to the binding of ternary complex and spatial localization of Cy3 and Cy5 fluorophores within the Forster radius (20–80 Å). Fluorescence emission was collected using a 1.2 NA water-immersion objective (CFI Plan Apochromat, Nikon) and optically split by a dichroic mirror and focused (Dual View, Photometrics) onto a CCD-chip (Cascade 512B, Photometrics) where images were acquired at a frames rate of 25/sec using commercially available software (MetaMorph, Universal Imaging Corp). All experiments were performed in Tris-polymix buffer (pH 7.5) (Munro et al. 2007) containing oxygen-scavenging and triplet state quenching compounds to extend fluorophore lifetime and to minimize blinking (Dave et al. 2009) (1 unit/μL glucose oxidase, 10 unit/μL catalase, 0.1% v/v glucose, 1 mM Trolox, 1 mM Cyclooctatetraene, and 1 mM nitrobenzyl alcohol). Data analysis is described in the Supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
P.C.W. thanks Dr. Changbong Hyeon and Jeffrey Noel for discussions regarding RNA simulations and all-atom modeling, as well as Haripriya Ramu and Dr. Alexander Mankin for discussion regarding ribosome stalling. We thank Yanan Yu for porting TMD into Gromacs. We acknowledge all members of the Blanchard laboratory and the Onuchic group for feedback throughout the course of this study and during manuscript preparation. K.Y.S. is grateful to M. Sheats, C. Ahrens, M. Vernon, B. Bergen, M. Perks, and S. Swaminarayan for cell processor code development. This work was supported by the Center for Theoretical Biological Physics, sponsored by the NSF (Grant PHY-0822283), with additional support from NSF-MCB-0543906, the LANL LDRD program, NIH Grant R01-GM072686, National Institute of General Medical Sciences Grant 5R01GM079238-03, and NSF Career Award 0644129. We also thank the New Mexico Computing Applications Center for computing time on the Encanto Supercomputer and Los Alamos National Laboratory for computing time on the Roadrunner Supercomputer.
Footnotes
Crystallographic B-factors were converted to spatial root-mean-squared displacement (RMSD) values via the relation Bi = [8π2/3(RMSDi)2] (Garcia et al. 1997), where Bi and RMSDi are the B-factor and RMSD associated with atom i.
In a two-state system, k ∝ exp(−βΔG). Given two measured rates, ki and kj, the corresponding difference in ΔG is given by ΔΔG = ΔGj − ΔGi = kBT ln(ki/kj). Thus, a factor of 100 difference in rates corresponds to a difference in ΔΔG of ∼4.6 kBT. Even if one estimates the uncertainty in simulated time to be an order of magnitude, then ΔΔG would be between 2.3 and 6.9 kBT.
Since these simulations are “kinetic” (i.e., each simulation is terminated once the A/A conformation is reached), the sampling does not represent a complete canonical ensemble. Accordingly, entropic signatures may be present in the probability distributions, but precise quantification of the thermodynamics requires more exhaustive sampling techniques.
These models are freely available at http://smog.ucsd.edu.
Based on the observation of irreversible changes in RNA backbone conformation, questions regarding the backbone dynamics in the Amber99 force field have been raised. Here, consistent with a previous study (Garcia and Paschek 2008), we see the backbone α and γ dihedral angles make reversible rearrangements.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2035410.
REFERENCES
- Adcock SA, McCammon JA 2006. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem Rev 106: 1589–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J 2008. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon I, Bashan A, Zarivach R, Yonath A 2005. Symmetry at the active site of the ribosome: Structural and functional implications. Biol Chem 386: 833–844 [DOI] [PubMed] [Google Scholar]
- Agmon I, Bashan A, Yonath A 2006. On ribosome conservation and evolution. Isr J Ecol Evol 52: 359–374 [Google Scholar]
- Aitken CE, Petrov A, Puglisi JD 2010. Single ribosome dynamics and the mechanism of translation. Annu Rev Biophys 39: 491–513 [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, van der Spoel DJ, van Drunen R 1995. GROMACS—a message-passing parallel molecular-dynamics implementation. Comput Phys Commun 91: 43–56 [Google Scholar]
- Blaha G, Stanley RE, Steitz TA 2009. Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science 325: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC 2009. Single-molecule observations of ribosome function. Curr Opin Struct Biol 19: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD 2004. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Bryngelson JD, Wolynes PG 1989. Intermediates and barrier crossing in a random energy-model (with applications to protein folding). J Phys Chem 93: 6902–6915 [Google Scholar]
- Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG 1995. Funnels, pathways, and the energy landscape of protein-folding—a synthesis. Protein Struct Funct Genet 21: 167–195 [DOI] [PubMed] [Google Scholar]
- Clementi C, Nymeyer H, Onuchic J 2000. Topological and energetic factors: What determines the structural details of the transition state ensemble and ‘en-route’ intermediates for protein folding? An investigation for small globular proteins. J Mol Biol 298: 937–953 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R 2005. An active role for tRNA in decoding beyond codon: Anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale T, Fahlman RP, Olejniczak M, Uhlenbeck OC 2009. Specificity of the ribosomal A site for aminoacyl-tRNAs. Nucleic Acids Res 37: 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave R, Terry DS, Munro JB, Blanchard SC 2009. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J 96: 2371–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M 2005. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579: 959–962 [DOI] [PubMed] [Google Scholar]
- Freddolino PL, Liu F, Gruebele M, Schulten K 2008. Ten-microsecond molecular dynamics simulation of a fast-folding WW domain. Biophys J 94: L75–L77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel D, Smit B 1996. Understanding molecular simulation: From algorithms to applications, 2nd ed Elsevier, San Diego, CA [Google Scholar]
- Garcia AE, Paschek D 2008. Simulation of the pressure and temperature folding/unfolding equilibrium of a small RNA hairpin. J Am Chem Soc 130: 815–817 [DOI] [PubMed] [Google Scholar]
- Garcia AE, Krumhansl JA, Frauenfelder H 1997. Variations on a theme by Debye and Waller: From simple crystals to proteins. Protein Struct Funct Genet 29: 153–160 [PubMed] [Google Scholar]
- Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K 2009. Regulation of the protein-conducting channel by a bound ribosome. Structure 17: 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E 2008. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447 [DOI] [PubMed] [Google Scholar]
- Hills RD, Brooks CL III 2009. Insights from coarse-grained Go models for protein folding and dynamics. Int J Mol Sci 10: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE 2009. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol 19: 120–127 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel DJ 2001. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model 7: 306–317 [Google Scholar]
- Malhotra A, Harvey SC 1994. A quantitative model of the Escherichia coli 16S RNA in the 30S ribosomal subunit. J Mol Biol 240: 308–340 [DOI] [PubMed] [Google Scholar]
- Malhotra A, Tan RK-Z, Harvey SC 1990. Prediction of the 3-dimensional structure of Escherichia coli 30S ribosomal-subunit—a molecular mechanics approach. Proc Natl Acad Sci 87: 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L, Sorin EJ, Tieleman DP, Pande VS, Colombo G 2008. Molecular simulation of multistate peptide dynamics: A comparison between microsecond timescale sampling and multiple shorter trajectories. J Comput Chem 29: 1740–1752 [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Conner N, Blanchard SC 2007. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell 25: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone PM, Snow CD, Lucent D, Pande VS 2008. Side-chain recognition and gating in the ribosome exit tunnel. Proc Natl Acad Sci 105: 16549–16554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AN, Meskauskas A, Roshwalb SC, Dinman JD 2008. Yeast ribosomal protein L10 helps coordinate tRNA movement through the large subunit. Nucleic Acids Res 36: 6187–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus DL, Cho SS, Hyeon C, Thirumalai D 2009. Minimal models for proteins and RNA: From folding to function. In Molecular biology of protein folding (ed. Conn PM), Vol. 84, pp. 203–250 Elsevier Academic, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- Roberts E, Sethi A, Montoya J, Woese RC, Luthey-Schulten Z 2008. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci 105: 13953–13958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MA, Wintermeyer W 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung C-S 2005. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci 102: 15854–15859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitter J, Engels M, Kruger P 1994. Targeted molecular-dynamics—a new approach for searching pathways of conformational transitions. J Mol Graph 12: 84–89 [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV 4th, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette J-C, Murphy FV 4th, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. 2009. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moore PB 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited. RNA 6: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F, Valle M, Frank J, Brooks CL 2003. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci 100: 9319–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trylska J, Tozzini V, McCammon J 2005. Exploring global motions and correlations in the ribosome. Biophys J 89: 1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CS, Sanbonmatsu KY 2004. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophys J 87: 2714–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30: 190–202 [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. 2009. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci 106: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cieplak P, Kollman PA 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem 21: 1049–1074 [Google Scholar]
- Wang Y, Rader AJ, Bahar I, Jernigan RL 2004. Global ribosome motions revealed with elastic network model. J Struct Biol 147: 302–314 [DOI] [PubMed] [Google Scholar]
- Whitford PC, Noel JK, Gosavi S, Schug A, Sanbonmatsu KY, Onuchic JN 2009a. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical force fields. Prot Struct Func Bioinfo 75: 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY 2009b. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys J 96: L7–L9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]