Abstract
Small nuclear and small nucleolar RNAs (snRNAs and snoRNAs) are critical components of snRNPs and snoRNPs and play an essential role in the maturation of, respectively, mRNAs and rRNAs within the nucleus of eukaryotic cells. Complex and specific pathways exist for the assembly of snRNPs and snoRNPs, involving, for instance, nucleocytoplasmic transport of snRNAs and intranuclear transport between compartments of snoRNAs. The phosphorylated adaptor for nuclear export (PHAX) is required for nuclear export of snRNAs in metazoans and also involved in the intranuclear transport of snoRNAs to Cajal bodies. PHAX contains a conserved single-stranded nucleic acid binding domain (RNA_GG_bind domain) with no sequence homology with any other known RNA-binding module. Here, we report NMR and X-ray crystallography studies that elucidate the structural basis for RNA recognition by the PHAX RNA-binding domain (PHAX-RBD). The crystal structure of the RNA_GG_bind domain from the parasite Cryptosporidium parvum (Cp RBD) forms well-folded dimers in solution in the absence of any ligand. The human PHAX-RBD is monomeric and only adopts a tertiary fold upon RNA binding. The PHAX-RBD represents a novel helical fold and binds single-stranded RNA with micromolar affinity without sequence specificity. RNA recognition by human PHAX-RBD is consistent with mutational analysis that affects RNA binding and PHAX-mediated nuclear export. Our data suggest that the PHAX-RBD mediates auxiliary RNA contacts with the snRNA and snoRNA substrates that are required for transport and/or substrate release.
Keywords: nuclear export, PHAX, protein–RNA, snRNA, structural biology
INTRODUCTION
Distinct nucleocytoplasmic transport pathways are known for different RNAs such as tRNA, miRNA, snRNA, mRNA, and rRNA (Ohno et al. 2002; Kohler and Hurt 2007). The RNA substrates are directed to their respective transport pathways by direct binding to export receptors (e.g., tRNA) (Arts et al. 1998; Cook et al. 2009) or to adaptor proteins (e.g., snRNA) (Ohno et al. 2000), which recognize specific features of these RNAs, such as length, sequence motifs, or three-dimensional structure (Ohno et al. 2002; Masuyama et al. 2004). Here we focus on the phosphorylated adaptor for nuclear export, PHAX (Ohno et al. 2000), which is involved in the nuclear export of small nuclear RNAs (snRNAs). The snRNAs and their corresponding small ribonucleoprotein particles (snRNPs) are essential components of the spliceosome (Wahl et al. 2009). SnRNAs are transcribed by Pol II in the nucleus and acquire a 7-methyl guanosine (m7G) 5′ cap. In metazoa, they are then exported to the cytoplasm to be assembled into snRNPs (Kohler and Hurt 2007). The export of snRNAs is mediated by an assembly of five proteins: the heterodimeric nuclear cap binding complex (CBC) (Izaurralde et al. 1995), PHAX (Ohno et al. 2000), CRM1, the nuclear export receptor of the importin β family, and RanGTP (Fornerod et al. 1997; Izaurralde et al. 1997). CBC and PHAX are adaptor proteins linking the snRNA cargo to the export machinery. CBC recognizes and binds the m7G cap of the snRNAs, and PHAX interacts directly with the CBC and the snRNA (Segref et al. 2001). A stable pre-export complex is initially formed between CBC, PHAX, and the snRNA. Phosphorylation of PHAX by CK2 kinase is a prerequisite for binding of the nuclear export signal (NES) of PHAX to CRM1-RanGTP leading to nuclear export of the CBC-PHAX-snRNA-CRM1-RanGTP complex (Ohno et al. 2000; Kitao et al. 2008). In the cytoplasm, RanGTP hydrolysis and dephosphorylation of PHAX by phosphatase 2A (Ohno et al. 2000; Kitao et al. 2008) lead to the release of CRM1 and RanGDP. Disassembly of PHAX and CBC occurs simultaneously with binding of the survival of motor neuron (SMN) protein complex to snRNA (Massenet et al. 2002).
Somewhat less well understood is the intranuclear transport of proteins and RNAs. Some of the factors involved in nuclear export of snRNAs, such as CBC, PHAX, and CRM1, are also involved in intranuclear transport of a subset of small nucleolar RNAs (snoRNAs) (U3, U8, and U13) into and between nuclear compartments such as speckles, Cajal bodies, and nucleoli (Boulon et al. 2002; Lamond and Sleeman 2003). These transport processes are linked to the assembly of snoRNPs, which are essential for the maturation of ribosomal RNA (Verheggen et al. 2002; Boulon et al. 2004). PHAX accumulates in Cajal bodies (Frey and Matera 2001), and PHAX and CRM1 are involved in distinct steps in the transport of U3 snoRNA to the nucleoli (Boulon et al. 2004; Watkins et al. 2004). RNAi-mediated depletion of PHAX specifically blocks the nuclear export of newly synthesized U snRNA and leads to nuclear mislocalization of U snRNPs and snoRNPs (Lemm et al. 2006; Watkins et al. 2007).
Interestingly, PHAX is not involved in mRNA export and seems to be restricted to the intranuclear transport or nuclear export of small structured, noncoding Pol II (and hence capped) RNA transcripts. The features of these RNAs that differentiate them from mRNAs have been investigated (Ohno et al. 2002; Masuyama et al. 2004).
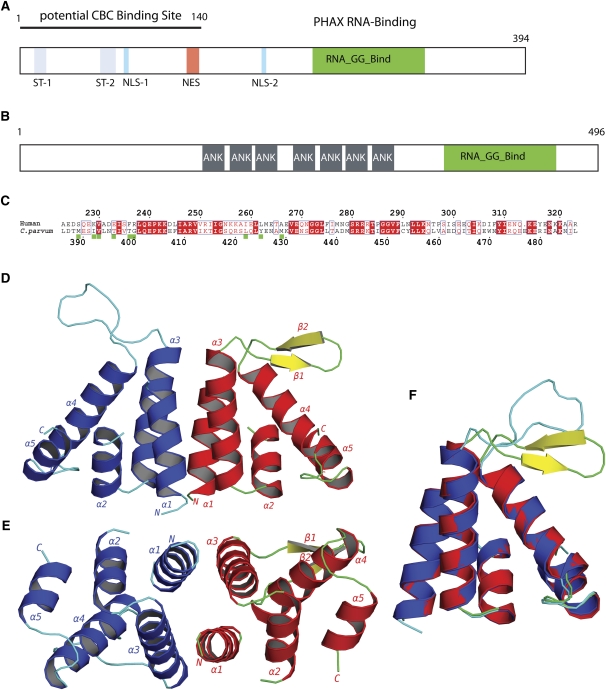
PHAX is a conserved, metazoan specific protein, which in human comprises 394 residues (Fig. 1A). Although biochemical studies have mapped functional regions (Ohno et al. 2000; Segref et al. 2001), little is known about the three-dimensional structure of the protein. The most evolutionary conserved region of PHAX is a stretch of about 90 residues in the second half of the protein, which has been shown to have nonspecific ssRNA-binding activity (Segref et al. 2001). Mutations of conserved residues in this PHAX RNA-binding domain (RBD) (PHAX-RBD) abolish U snRNA export to the cytoplasm, showing that RNA binding by PHAX is essential for transport (Segref et al. 2001). The PHAX-RBD is a member of the RNA_GG_bind domain family (PFAM10258), which is characterized by two absolutely conserved double glycines GGX12GG. The domain has been identified in three distinct architectural frameworks: in metazoan PHAX homologs, in seryl-tRNA synthetase from Oryza sativa (rice), and in a family of ankyrin-repeat-containing proteins of unknown function from single-celled parasites such as Plasmodium, Toxoplasma, and Cryptosporidium (Fig. 1B,C; Supplemental Fig. 1).
FIGURE 1.
The PHAX RNA-binding domain. (A) Schematic functional organization of human PHAX based on biochemical data showing phosphorylation sites (ST-1, ST-2), bipartite nuclear import sequence (NLS-1, NLS-2), nuclear export sequence (NES), and location of the RNA-binding domain (green) and the CBC binding site. (B) Schematic domain organization of C. parvum protein Q7YZ62, showing the location of ankyrin repeats (gray), and the RNA-GG-bind domain (green). (C) Sequence alignment of human PHAX-RBD and CpGGBD. Green boxes: Residues that are involved in the dimer interface of CpGGBD. (D) Cartoon representation of the AB dimer of the RNA_GG_bind domain from C. parvum. (E) Dimer rotated by 90°. (F) Overlay of monomers A (blue and cyan) and molecule B (red, green, and yellow).
Here we report crystal and NMR studies that elucidate the structural basis for the recognition of RNA by the conserved PHAX-RBD. We also determined the crystal structure of the RNA_GG_bind domain from Cryptosporidium parvum. This protein forms well-folded dimers in solution in the absence of any ligand. In contrast, the isolated human PHAX RNA-binding domain is monomeric but unstructured in the absence of RNA. It adopts a tertiary structure upon binding to single-stranded RNA. RNA recognition involves mainly contacts with the sugar-phosphate backbone of the RNA consistent with the nonspecific binding. The structure explains the effect of mutations that impair RNA binding and PHAX-mediated snRNA export.
RESULTS
The PHAX RNA-binding domain
For structural studies, we cloned and expressed various constructs encompassing the human PHAX-RBD as well as the RNA_GG_bind domain of the ankyrin-repeat-containing protein (Uniprot entry Q7YZ62) from the parasite C. parvum (Fig. 1A,B; Bankier et al. 2003). These two domains are 39% identical over 98 residues. Gel filtration and dynamic light scattering (DLS) experiments revealed that the C. parvum RNA_GG_bind domain (CpGGBD) is dimeric, while the human PHAX-RBD is monomeric in solution (data not shown). Unexpectedly, 1H,15N correlation NMR spectra of human PHAX-RBD indicated that the protein alone is poorly folded, while the CpGGBD adopts a stable tertiary fold in solution (Supplemental Fig. 2A).
Structure of the RNA_GG_bind domain from C. parvum
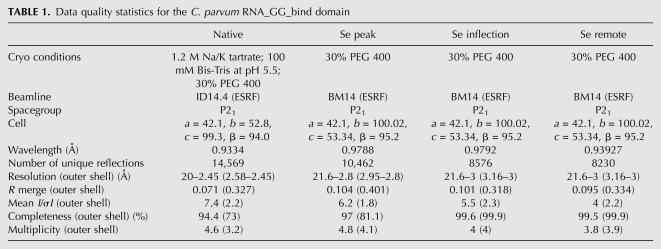
We solved the crystal structure of the CpGGBD using selenomethionyl substituted protein by the MAD method at 2.8 Å in one P21 crystal form. The native structure was refined at 2.45 Å in a second P21 crystal form to an R-factor (R-free) of 0.194 (0.257) (Tables 1, 2). Both crystal forms contain four molecules in the asymmetric unit associated to form two dimers. The structures of both dimers are equivalent in both crystal forms, and only the AB dimer in the higher-resolution native structure will therefore be described (Fig. 1D,E). Residues 387–475 are clearly seen in the electron density, whereas the loop 403–405 is poorly ordered and the charged C-terminal tail (476–496) absent from the density.
TABLE 1.
Data quality statistics for the C. parvum RNA_GG_bind domain
TABLE 2.
Refinement and structure quality statistics for the C. parvum RNA_GG_bind domain
The CpGGBD structure is mainly helical as expected from secondary structure predictions and circular dichroism experiments (data not shown). The monomer comprises five α-helices linked by short loops (1–4 residues) apart from helices α3 and α4, which are linked by 13 residues (435–447) that take up different conformations in each subunit of the dimer (Fig. 1F). The fold is unique to the RNA_GG_bind domain; no significant structural homology was found with other RNA-binding domains or proteins using the programs DALI, Dejavu, or MSFOLD (Kleywegt and Jones 1997; Krissinel and Henrick 2004; Holm et al. 2008).
The molecules A and B are almost related by a twofold axis at the dimer interface with an angle between rotation axis and the centroid vector of 89.2° (Fig. 1D). The interface is formed through the interaction of helix α1 of one molecule with the helix α3 of the other, and the total buried surface area of the interface is 1567 Å2 (PISA server, http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (Krissinel and Henrick 2007). Hydrophobic residues such as Met389, Ile392, Val393, Thr396, Thr399 (helix α1) and Leu422, Tyr425, and Met429 (helix α3) participate in this interface. In contrast, the equivalent residues in the human PHAX-RBD are generally more hydrophilic (Met389–Asp226, Ile392–Lys230, Thr396–Glu234). Also, Gly400 in CpGGBD, which allows the large Met389 to be in the interface, is replaced by the charged Arg238 in human PHAX (Fig. 1C). Although the sequences of the RNA_GG_bind domains are overall rather similar (∼40% identical), the key diagnostic differences between metazoan and parasite PHAX-RBDs appear to be Thr (or valine or serine) at the equivalent to position 396 of the dimeric parasite proteins, compared to Glu, Asp, or rarely Gln at the equivalent of position 234 in monomeric metazoan PHAX proteins; Gly, Ala, or Ile at position 400, compared to Arg/Lys at position 238; and Ile, Leu, or Val at position 392 compared to Lys, Asp, or Glu at position 230. These differences are conserved in comparing the sequences of parasite proteins to the metazoan PHAX proteins (Supplemental Fig. 1), suggesting that the former are most likely all dimers, and the latter are most likely all monomers.
The region between helices α3 and α4, which is enclosed by the two strictly conserved pairs of glycines 435–436 and 449–450, is a random coil in the A and C molecules, while it forms a β-hairpin in the B and D molecules. The glycines clearly impart a high flexibility to this region, and changes in the backbone torsions of particularly glycines 435–436 lead to a rigid body–like movement of the entire loop (Fig. 1F). Both conformations are stabilized and blocked by the crystal contacts. Below it is shown the PHAX-RBD–RNA complex that this loop closes over the bound RNA.
Nucleic acid binding by the human PHAX-RBD
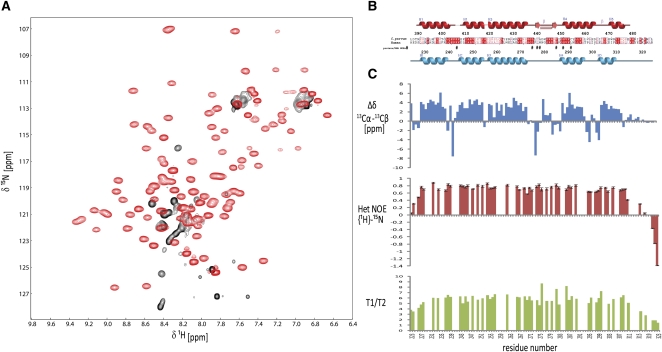
We next tested whether the fold of the human PHAX-RBD might be stabilized by RNA binding. Addition of a U9 RNA oligonucleotide to the human PHAX-RBD (residues 223–332) dramatically improved the NMR spectra. Homogeneous line widths and good spectral dispersion indicate that the PHAX-RBD adopts a tertiary structure in the presence of this RNA (Fig. 2A). Circular dichroism experiments show that the free domain has already some helical conformation, which increases upon RNA binding (Supplemental Fig. 2A). Thus, the free protein seems to adopt a molten globule-like conformation, where secondary structure elements exist but no defined tertiary structure is formed. The secondary 13C chemical shifts obtained after assigning the NMR frequencies and 15N relaxation data indicate that residues 230–311 adopt a globular structure comprising five α-helices (Fig. 2B,C). The 13-residue loop connecting helices α3 and α4 (residues 275–287) is not mobile on subnanosecond timescales as judged from the NMR relaxation data. The trimmed mean average 15N T1/T2 ratio corresponds to a tumbling correlation time of τc = 8.3 nsec, indicating that the PHAX-RBD/U9 RNA complex is monomeric in solution.
FIGURE 2.
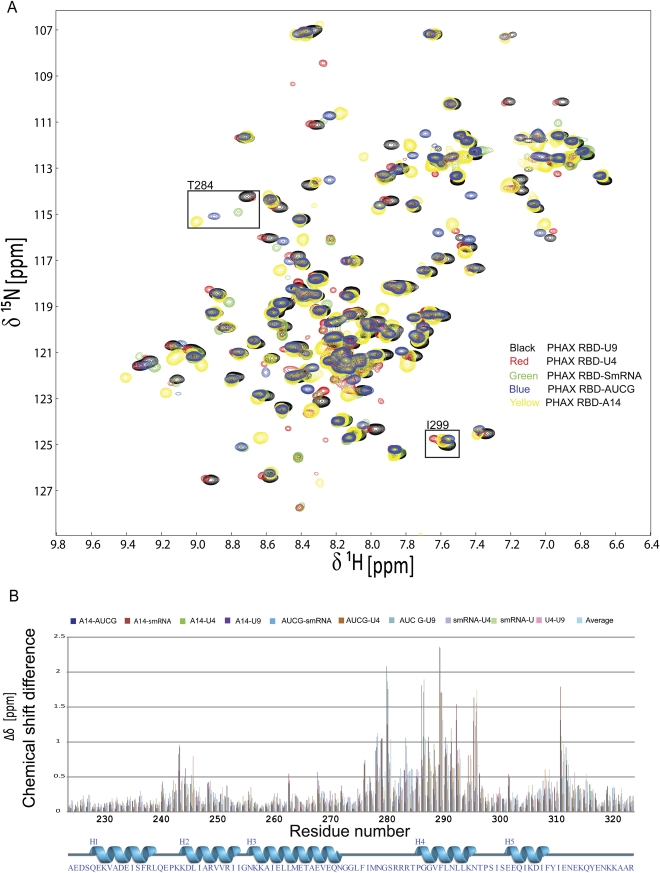
NMR analysis of the human PHAX-RBD domain. (A) 1H,15N NMR spectra of human PHAX-RBD in the absence (black) and presence (red) of a U9 RNA oligonucleotide. (B) Sequence alignment and secondary structure of the human PHAX and C. parvum RNA_GG_bind domains. (#) Residues in human PHAX-RBD that show intermolecular NOEs with the AUCG RNA ligand. (C) Secondary chemical shifts and 15N relaxation data of the human PHAX-RBD bound to U9 RNA. 1H,15N heteronuclear NOE and 15N T1, T2 relaxation data indicate that the protein–RNA complex tumbles as a monomer in solution.
To investigate the RNA-binding specificity, we monitored binding of PHAX-RBD to several different RNA oligonucleotides (U9, U4, AUCG, A14, and an oligo that resembles the Sm protein-binding site in snRNAs, SmRNA–AAUUUGUGG) by 1H,15N NMR correlation experiments. All RNA ligands induce folding of the PHAX-RBD domain (Fig. 3). In contrast, single-stranded DNA oligonucleotides (T9 and ATCGA) bind PHAX-RBD with strongly reduced affinity and induce folding of the domain only at much higher molar ratios compared to the RNA ligands (Fig. 3; Supplemental Fig. 2C,D). As the unbound protein is unstructured, residues important for RNA binding were determined by comparing pairwise amide (1H,15N) chemical shifts of the RBD bound to the different RNA ligands (Fig. 3). For the PHAX-RBD/U9 and PHAX-RBD/AUCG complexes, we performed complete backbone and side chain chemical shift assignments. These chemical shifts were then used to aid assignment of the NMR signals of the other complexes by comparing amide frequencies derived from 1H,15N correlations and 15N-edited NOESY spectra. The NMR signals of some residues have very similar chemical shifts, independent of the bound ligand, while others have significant chemical shift differences, suggesting that these residues are involved in direct contacts with the RNA. This analysis indicates that the RNA-binding site comprises residues in helix α4 and in the loop connecting helices α3 and α4.
FIGURE 3.
RNA binding by the human PHAX-RBD. (A) 15N-labeled human PHAX-RBD (0.2 mM) was titrated with different RNA ligands: U9 (black), U4 (red), AUCG (yellow), A14 (blue), SmRNA–AAUUUGUGG (green). Saturation is observed at a 1:1 molar ratio. (B) 1H,15N pairwise chemical shift differences Δδ = {[Δδ(1H)]2×5) + [Δδ(15N)]2}1/2 of the data shown in A versus primary sequence.
The nucleic acid binding affinities were determined by isothermal titration calorimetry with dissociation constants (Kd) of 6.5 μM and 4.2 μM for the AUCG and U4 RNA oligos, respectively. As expected from the NMR titrations, ssDNA has significant reduced binding affinity with Kd = 148 μM for T9 (Supplemental Fig. 3).
Structure of PHAX-RBD in complex with AUCG
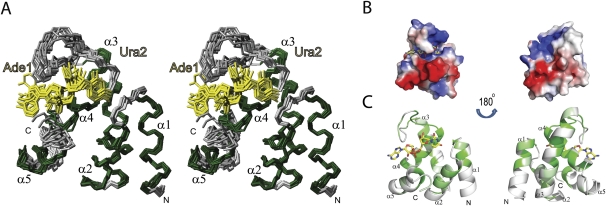
To understand molecular details of the RNA recognition, we determined the solution structure of PHAX-RBD bound to an AUCG tetranucleotide (Fig. 4; Table 3). The three-dimensional structure of the PHAX-RBD/AUCG complex was calculated based on 40 intra-RNA and 32 intermolecular NOE restraints, observed for the first two nucleotides. The overall fold of the human PHAX-RBD (when bound to AUCG) resembles the CpGGBD (Supplemental Fig. 4). Backbone atom coordinates superimpose with a root-mean-square deviation (RMSD) of 3.3/3.6 Å to CpGGBD-A/B, respectively. This mainly reflects the different conformations of the 13-residue linker connecting helices α3 and α4. In addition, helices α4 and α5 have a slightly different orientation, presumably linked to RNA binding, which brings the α3–α4 loop and helix α4 together. When the CpGGBD and human PHAX-RBD structures are superimposed omitting the α3–α4 loop and helix α5, the backbone RMSD drops to 1.3 Å.
FIGURE 4.
Solution structure of the human PHAX-RBD bound to AUCG RNA. (A) Stereo view of the 10 lowest energy structures of PHAX-RBD bound to AUCG. Residues 224–311of PHAX-RBD and the first two bases of the RNA, Ade1 and Ura2, are shown. Helical secondary structure elements are colored in green and loops in gray. RNA bases are colored in yellow. (B) Electrostatic surface representation of PHAX-RBD. (C) Sequence conservation of the PHAX-RBD in metazoan species plotted onto a surface representation of the structure. Colors from white to green correspond to increasing conservation.
TABLE 3.
Structural statistics for the PHAX-RBD/AUCG RNA complex
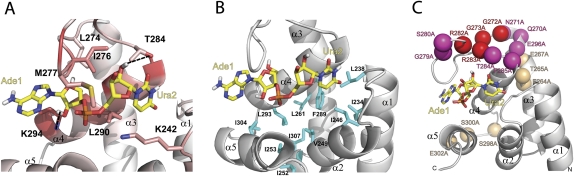
The structure of the human PHAX-RBD/RNA complex suggests how the tertiary fold of the RBD is stabilized by the RNA ligand. Residues in the α1–α2 and α3–α4 loops as well as residues in helix α4 form direct contacts with the RNA (Fig. 5A). Additional interactions that involve hydrophobic side chains are presumably indirectly stabilized by the presence of the RNA. For example, Phe289 in helix α4 is a central residue located in the hydrophobic core of the RBD fold. It contacts Ile234 and Leu238 in helix α1, Ile246 and Ile253 in helix α2, Leu261 in helix α3, and Leu293 in helix α4 (Fig. 5B). Helix α5 packs against helix α4 by contacts of Ile304–Ile293, respectively, and to helix α2 by contacts involving Ile307–Val249 and Ile252, respectively (Fig. 5B).
FIGURE 5.
RNA recognition by the human PHAX-RBD. (A) Zoomed view of the binding pocket highlighting side-chain residues that show NOE contacts to the RNA. The backbone ribbon is colored by chemical shift perturbation (Fig. 3) from white to red. Potential hydrogen bonds are indicated with black dotted lines. (B) Side chains of residues in the hydrophobic core of the human PHAX-RBD are highlighted and colored in blue. (C) Mutations in human PHAX that affect RNA binding (Segref et al. 2001) are mapped onto the three-dimensional structure. Colors from light orange to magenta to red scale with the reduction in RNA-binding affinity compared to the wild-type protein.
RNA recognition by the human PHAX-RBD
A close-up view of the RNA-binding pocket (Fig. 5A) shows details of the RNA recognition by PHAX-RBD. Most of the 32 intermolecular NOEs involve contacts of hydrophobic side chains to the sugar backbone of the first two nucleotides of the AUCG RNA oligonucleotide (Supplemental Fig. 5). The Ade1 ribose shows intermolecular NOEs with residues Leu290, Leu274, and Gly287. The ribose of Ura2 shows intermolecular NOEs with the side chains of Ile276, Met277, Thr284, and Leu290, while its base contacts Lys242 and Leu290. As a result the conformation of Ura2 is well-defined in the ensemble of structures. The discrimination against DNA likely depends on recognition of the 2′-OH in the ribose of the RNA ligands. There are two potential hydrogen bonds involving the side chains of conserved residues Lys294 and Thr284 and the 2′-OH of Ade1 and Ura2, respectively (Fig. 5A). This is supported by the large chemical shift perturbations observed for these amino acids.
Analysis of the electrostatic surface potential shows that the RNA-binding region is positively charged (Fig. 4B). This surface overlaps very well with the RNA-binding region mapped by the differential chemical shift perturbations (Fig. 5A). Residues that are important for RNA recognition are conserved in insects and vertebrates but not in other organisms, including C. parvum (Fig. 2B; Supplemental Fig. 1). For example, Leu290, which is located in the center of the RNA-binding site and shows NOEs to both RNA bases, is replaced by Cys453 in C. parvum. Further amino acid differences are Ile276 and Met277 in human PHAX, which are substituted by Thr439 and Ala440, respectively, in the parasite ortholog. In contrast, residues that stabilize the hydrophobic core are extremely well conserved across species, with the exception of residues that stabilize the packing of helix α5. This reflects the small structural difference between the parasite and the human PHAX-RBD in that region.
A previous study has identified residues in PHAX that are important for RNA binding (Segref et al. 2001). Many mutations that severely impair RNA binding and nuclear export involve residues that directly contribute to RNA recognition. Mutations of residues in the α3–α4 loop strongly reduce RNA binding. For example, the Arg282Ala–Arg283Ala double mutant (Fig. 5C, red) removes positively charged side chains in the proximity of the RNA ligand. In addition, Arg283 contributes to stabilization of the α3–α4 loop by a potential hydrogen bond to the Glu269 side chain and additional hydrophobic interactions with Leu274 (loop) and V288 (α4) (Supplemental Fig. 6). The Arg282 side chain shows NOE contacts to the aromatic side chain of Phe275, presumably stabilizing the loop conformation (Supplemental Fig. 6). Mutation of the first, absolutely conserved, double glycine motif (Gly272Ala–Gly273Ala) in the PHAX-RBD disrupts RNA binding. As these residues are not directly in contact with the RNA, the most likely explanation is that the mutation reduces the conformational flexibility of the α3–α4 loop and allows for a specific backbone conformation. Consistent with this, Gly272 adopts an unusual backbone (φ = +82°), which is not compatible with an alanine at this position. On the other hand, mutations of residues that are remote from the binding site, such as Glu264Ala–Thr265Ala–Glu267Ala (in the middle of helix α3) or Ser298Ala–Ser300Ala–Glu302Ala (helix α4), have no severe effect on RNA binding (Fig. 5C, light orange). Mutations that have a mild effect on RNA binding are located in the proximity of the RNA-binding site (Fig. 5C, shown in magenta). Overall, the mutational analysis is fully consistent with the mode of RNA recognition seen in the PHAX-RBD/RNA complex.
DISCUSSION
A novel RNA-binding fold
Structural similarity searches indicate that the arrangement of the five helices in the RNA_GG_bind domain represents a new fold. There are only few nucleic acid binding domains known that have an entirely α-helical fold; these include the Pumilio protein, SAM domains (Auweter et al. 2006), and dimeric viral proteins such as Rop, the influenza virus NS1 protein, or the viral B2 protein, an inhibitor of RNA interference (Chao et al. 2005; Lingel et al. 2005). However, the structure and RNA recognition employed are distinct. The PHAX-RBD thus represents a novel α-helical fold for single-stranded RNA binding.
Both of the two conserved GG motifs in the PHAX-RBD are important for RNA binding. Gly272 in the first GG motif (G272–G273) exhibits an unusual backbone torsion angle that allows the α3–α4 loop to close over the RNA (see above). The second GG motif (Gly286–Gly287) allows a close approach of the Ura2 base in the RNA ligand to the protein backbone. This is somewhat reminiscent of the role of the glycine residues in the conserved GxxG loop of KH domains for recognition of single-stranded RNA, which allow a close approach of the RNA ligand and support a unique backbone conformation of this loop (Supplemental Fig. 7; Liu et al. 2001; Auweter et al. 2006).
Consistent with non-sequence-specific RNA binding, the PHAX-RBD exhibits a positively charged surface comprised of arginine and lysine residues that mediate electrostatic interactions with the RNA ligand. Hydrophobic interactions are made with the RNA riboses. No base-specific contacts are observed, but potential hydrogen bonds by positively charged side chains with 2′-hydroxyl groups may explain the preferred binding to RNA over DNA. A surprising finding is that the human PHAX-RBD is misfolded in solution and that the tertiary fold is stabilized by RNA binding. Although we cannot rule out the possibility that the PHAX-RBD might be stabilized in the context of the full-length PHAX protein, this observation still indicates that RNA binding leads to a stabilization of the PHAX-RBD structure.
The most common RNA-binding domains such as RRM and KH domains are thought to originate from simple and ancient protein folds and are conserved structurally and in their nucleic acid binding properties from prokaryotes to eukaryotes. In contrast, RNA-binding properties of the PHAX-RBD are only conserved in metazoan orthologs. Consistently, some residues that are involved in RNA recognition by the human PHAX-RBD are not conserved in the CpGGBD. This suggests that RNA-binding capability has only been acquired during the more recent evolution. The biological function of the RNA_GG_bind domain in the ankyrin repeat protein of C. parvum is not known, although we note that the related Plasmodium flaciparum protein XP-001347497 has 2290 residues (with ankyrin repeats between 1411 and 1612 and the RNA_GG_bind from 1716 to 1789) and is annotated as the 10b antigen (Franzen et al. 1989).
A model of the PHAX export complex
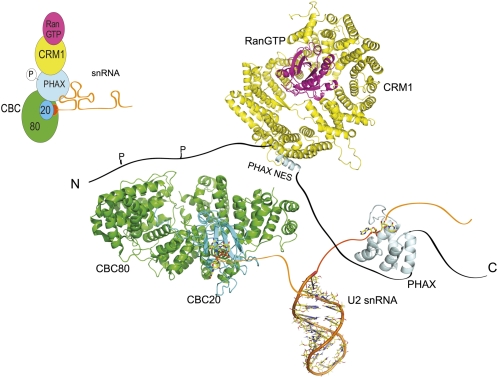
Currently available structural and biochemical data suggest a model for PHAX-mediated RNA transport. As PHAX binds RNA without sequence specificity and moderate affinity, targeting of specific RNAs may involve contacts with other proteins, which also may depend on phosphorylation of PHAX. For export of U snRNAs, the N terminus of PHAX needs to contact the CBC, when it is already bound to the RNA cap structure (Ohno et al. 2000). This leads to an increased affinity of PHAX to CBC resulting in the formation of the pre-complex. The formation of the export complex is completed only after PHAX phosphorylation and subsequent binding of CRM1 and RanGTP (Fig. 6; Ohno et al. 2002; Kitao et al. 2008). Apart from the RBD, PHAX comprises large stretches of unstructured regions, which are presumably recognized by its binding partners in the export complex. It will be important to determine the quaternary arrangement of these proteins together with RNA, starting from the available structural data (Mazza et al. 2002; this study) to further elucidate the molecular details of the nuclear export of snRNAs.
FIGURE 6.
Structural model of snRNA export. Schematic representation of the quaternary structure of the snRNA export with CRM1-RAN-GTP complex (PDB 3GJX), stem I of U2 snRNA (PDB 2O32), nuclear cap binding complex (CBC 80 + 20) in complex with a cap analog m7GpppG (PDB 1H2T) and the human PHAX-RBD (PDB 2KC7).
MATERIALS AND METHODS
Cloning, expression, and purification
Human PHAX-RBD
The region of human PHAX (SWISS-PROT Q9H814) encoding residues 223–323 was cloned into a modified pETM11 vector with an N-terminal six-histidine tag followed by a GB1 fusion protein and a TEV protease site. Expression was carried out on Escherichia coli BL21 cells for 16 h at 30°C after induction with 0.5 mM IPTG when cells were at O.D. = 0.7. For the preparation of isotope-labeled proteins, cells were grown in M9 minimal medium supplemented with [13C6] glucose and/or 15NH4Cl. Cell lysates were incubated with Ni-NTA Superflow beads (QIAGEN). After several wash steps with buffer containing 20 mM Tris (pH 8.5), 500 mM NaCl, 5% glycerol, and 10 mM Imidazol, elution was made with a similar buffer containing 350 mM Imidazol. Before overnight cleavage of the fusion protein with tobacco etch virus protease, the buffer was exchanged to 20 mM Tris (pH 8.5), 500 mM NaCl, and 1 mM β-mercaptoethanol. A second nickel affinity purification step was performed followed by concentration and purification over a gel filtration column 26/60 Sephadex II (GE Healthcare). A final buffer exchange was made to 20 mM NaPi (pH 6.5), 50 mM NaCl (NMR buffer), and the protein was concentrated to 0.2–0.8 mM for NMR experiments. For measurements in D2O, the protein was lyophilized and redissolved in D2O. Extensive crystallization trials of the human PHAX-RBD, with and without various RNAs, were never successful.
C. parvum RNA_GG_bind (CpGGBD)
The genomic DNA for C. parvum strain Iowa was bought from the ATCC (PRA-67D). The E. coli strains used were XL2blue for DNA isolation and Origami (DE3) (Novagen) for protein expression. All strains harboring plasmids were grown in Luria broth media supplemented with the appropriate antibiotic. The putative RNA-binding domain of the C. parvum protein Q7YZ62 (residues 386–496) was cloned in-frame with an N-terminal hexahistidine tag and the tobacco etch virus (TEV) protease cleavage site of the prokaryotic expression vector pProEXHTb (Invitrogen). Cells were grown in Luria broth media supplemented with 15 μg/mL kanamycin and tetracyclin and 100 μg/mL ampicillin at 37°C until OD600nm ∼0.8 before induction with 0.15 mM IPTG overnight at 18°C. The selenomethionyl-substituted domain was expressed using the Doublie protocol (Doublie 1997). Following expression, the domain was purified by standard chromatographic procedures using the sequential application of Ni2+ affinity chromatography (chelating Sepharose FF; Amersham), cation exchange chromatography (monoS 5/5; Amersham), and gel size exclusion chromatography (Tricorn S75 10/30; Amersham). At each step, the protein purity was assessed by SDS-PAGE using a 15% gel. The domain was concentrated on Microsep 5K (Vivascience) to a final concentration of 2.5–4 mg/mL in 20 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM TCEP.
Structure determination
Crystallization and data collection of CpGGBD
All crystals were obtained by the hanging-drop vapor diffusion method using 2-μL drops containing a 50:50 (v/v) mix of the protein and the reservoir solution at 20°C. The RNA-binding domain of C. parvum was crystallized using 1.3–1.5 M Na/K tartrate as precipitant and 100 mM Bis-Tris buffer (pH 5.5). Five micromolars of DTT and 2 mM of L-cysteine were added for the selenomethionyl-substituted domain. Clusters of thin plates appeared from precipitate after 3–4 d. Preceding data collection, the crystals were transferred in a cryoprotecting solution (30% Peg 400, 1.2 M Na/K tartrate, 100 mM Bis-Tris buffer at pH 5.5 for the native CpGGBD or 30% Peg 400 only for the selenomethionyl-substituted CpGGBD) prior to mounting in a rayon fiber loop and being placed in a boiling nitrogen stream at 100 K. All data were collected from a single crystal. Native data were collected at 2.45 Å in the space-group P21 with the following unit cell: a = 42.18, b = 52.86, c = 99.31, and β = 94.05° at ESRF ID14-4 using an ADSC detector. The MAD data set for the selenomethionylated domain was collected at 2.8 Å in the space-group P21 with the following unit cell: a = 42.1, b = 100.017, c = 53.338, and β = 95.18° at ESRF BM-14 using a MARCCD detector. The wavelength for the SeMet MAD experiment was chosen by scanning through the absorption edge of the crystal and optimizing the wavelength for the f″ signal from the selenium. Native data were processed and reduced with XDS and XSCALE (Kabsch 1993), while MOSFLM and SCALA were used for the MAD data (Leslie 1992). All further computing was performed using the CCP4 suite (Collaborative Computational Project Number 4 1994), unless otherwise stated.
Structure determination of CpGGBD
Initial phases were determined by the MAD method and subsequently improved through noncrystallographic symmetry (NCS) averaging. The P21 form could have possessed from 2 to 4 molecules in the asymmetric unit. Twelve selenium sites were found using the anomalous differences at 4.6 Å with the program SHELXD, indicating that there were actually four molecules in the asymmetric unit (Schneider and Sheldrick 2002). Only seven of the eight first sites with an occupancy of >0.5 were maintained. Those sites were introduced in autoSHARP, and another five selenium sites were found (De La Fortelle and Bricogne 1997; Bricogne et al. 2003). A total of 12 heavy-atom positions were refined to give an initial figure of merit of 0.32 without solvent flattening. Solvent flattening with DM and an estimate of 38.5% solvent content resulted in excellent and easily interpretable maps with an overall figure of merit of 0.797 (Cowtan and Zhang 1999). RESOLVE was used for building a first model and for phase improvement including twofold NCS averaging with a matrix-derived automatically from the Se positions, giving an overall figure of merit of 0.86 (0.7 at 2.8 Å) (Terwilliger and Berendzen 1999). The two other NCS operators were found after examination of the model. A partial model of the dimer was used to solve the native structure in the related P21 space group at 2.45 Å by molecular replacement using Molrep (Vagin and Teplyakov 1997). For the refinement of each structure, 5% of the observations were set aside for cross-validation analysis (Brünger 1992) and were used to monitor various refinement strategies during maximum likelihood refinement using REFMAC program (Murshudov et al. 1997). Manual corrections of the model using Coot (Emsley and Cowtan 2004) were interspersed with cycles of maximum-likelihood and TLS refinement (Winn et al. 2003). Water molecules were added manually. The stereochemical quality of the model was assessed with the program PROCHECK (Laskowski et al. 1993). Details of the data and model quality are given in Tables 1 and 2.
NMR spectroscopy
NMR data were acquired at 297 K on Bruker DRX500, DRX600, and AV900 spectrometers equipped with cryogenic triple-resonance probes. The buffer contained 20 mM NaPO4 (pH 6.5) and 50 mM NaCl at concentrations of 0.2 and 0.8 mM for the NMR titrations and structure calculation, respectively. Spectra were processed with NMRPipe (Delaglio et al. 1995) and processed with NMRView (Johnson and Blevins 1994). Chemical shift assignments were obtained from standard triple resonance NMR experiments (Sattler et al. 1999). Distance restraints were derived from 15N- and 13C-resolved three-dimensional, 1H homonuclear two-dimensional, and 14N,12C filtered NOESY experiments with NOE mixing times of 120 msec. Stereospecific assignments of leucine and valine methyl groups were obtained using a 10% 13C-enriched sample as described (Neri et al. 1989). Restraints for the backbone angles ψ and φ were derived from TALOS (Cornilescu et al. 1999). 15N relaxation experiments (T1, T2, and heteronuclear {1H}-15N NOE) were measured with a 0.3 mM 15N-labeled PHAX-RBD sample in complex with U9 (1:1) at 297 K on a 500-MHz spectrometer equipped with cryoprobe, as described (Farrow et al. 1994). Analysis of the data was performed using Python scripts written by Bernd Simon (EMBL Heidelberg).
Only manually assigned NOEs and dihedrals angle restraints were applied in a simulated annealing protocol using ARIA (Linge et al. 2001) and CNS (Brünger et al. 1998). Water refinement was performed on the final ensemble of NMR structures (Linge et al. 2003). Structural quality was evaluated using ProcheckNMR (Laskowski et al. 1996). The ensemble representation was prepared using MOLMOL (Koradi et al. 1996). Ribbon representations and the electrostatic surface potential were prepared with PYMOL (DeLano Scientific).
Isothermal titration calorimetry
ITC experiments were performed using a ITC200 instrument (MicroCal) at 24°C. RNA (1 mM; purchased from BioSpring) and DNA oligonucleotides (2 mM; purchased from Eurofins) were titrated to 0.1 mM PHAX-RBD. The lyophilized RNA was dissolved in the same buffer as the protein, 20 mM sodium phosphate, 50 mM NaCl (pH 6.5). The heat of dilution was measured by titrating AUCG RNA into buffer. This value was only 1% of the total observed in the experiment. The titration protocol consists of one initial injection of 0.4 μL followed by 38 injections of 1 μL of the ligand into the protein sample with intervals of 120 sec, allowing the titration peak to reach the baseline. The dissociation constant, binding entropy, enthalpy, and stoichiometry parameters were calculated using the program Origin v7.0 (MicroCal). Measurements were repeated twice.
Accession codes
The C. parvum RNA_GG_bind domain PDB entry is: 2W4S. The human PHAX-RBD/AUCG complex is: 2KC7.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Bernd Simon, Vladimir Rybin, Catherine Mazza, Delphine Guilligay, and Lars Ferbitz for help; Gunter Stier for expression vectors; the European Synchrotron Radiation Facility–European Molecular Biology Laboratory (ESRF-EMBL) Joint Structural Biology Group for access to ESRF beamlines; and the BM14 staff members for providing support during data collection. A.M. acknowledges support through a PhD fellowship (SFRH/BD/22323/2005) from the Portuguese Foundation for Science and Technology (FCT). This work was supported by the European Commission (3D Repertoire, LSHG-CT-2005-512028).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2009910.
REFERENCES
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17: 7430–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH 2006. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res 34: 4943–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankier AT, Spriggs HF, Fartmann B, Konfortov BA, Madera M, Vogel C, Teichmann SA, Ivens A, Dear PH 2003. Integrated mapping, chromosomal sequencing, and sequence analysis of Cryptosporidium parvum. Genome Res 13: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Basyuk E, Blanchard JM, Bertrand E, Verheggen C 2002. Intranuclear RNA trafficking: Insights from live cell imaging. Biochimie 84: 805–813 [DOI] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16: 777–787 [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W 2003. Generation, representation, and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Brünger AT 1992. Free R-Value—a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355: 472–475 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12: 952–957 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr D50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E 2009. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Cowtan KD, Zhang KY 1999. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol 72: 245–270 [DOI] [PubMed] [Google Scholar]
- De La Fortelle E, Bricogne G 1997. Maximum-likelihood heavy-atom refinement parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In Methods in enzymology (ed. Abelson JN et al. ), vol. 276, pp. 472–494 Academic, New York: [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A 1995. NMRPipe: A multidimensional spectral processing system based on UNIX Pipes. J Biol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Doublie S 1997. Preparation of selenomethionyl proteins for phase determination Academic, New York: [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Franzen L, Wahlin B, Wahlgren M, Aslund L, Perlmann P, Wigzell H, Pettersson U 1989. Enhancement or inhibition of Plasmodium falciparum erythrocyte reinvasion in vitro by antibodies to an asparagine-rich protein. Mol Biochem Parasitol 32: 201–211 [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG 2001. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 154: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A 2008. Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16: 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA 1994. NMRView: A computer program for the visualization and analysis of NMR data. 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Kabsch W 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kitao S, Segref A, Kast J, Wilm M, Mattaj IW, Ohno M 2008. A compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export from the nucleus. Mol Cell Biol 28: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA 1997. Detecting folding motifs and similarities in protein structures. Methods Enzymol 277: 525–545 [DOI] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K 1996. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Sleeman JE 2003. Nuclear substructure and dynamics. Curr Biol 13: R825–R828 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM 1993. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Lührmann R 2006. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 17: 3221–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4/ESF-EACMB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- Linge JP, O'Donoghue SI, Nilges M 2001. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol 339: 71–90 [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M 2003. Refinement of protein structures in explicit solvent. Proteins 50: 496–506 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M 2005. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep 6: 1149–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G 2002. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol 22: 6533–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K, Taniguchi I, Kataoka N, Ohno M 2004. RNA length defines RNA export pathway. Genes Dev 18: 2074–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S 2002. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J 21: 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neri D, Szyperski T, Otting G, Senn H, Wuthrich K 1989. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28: 7510–7516 [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198 [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Kuersten S, Mattaj IW 2002. Identity elements used in export of mRNAs. Mol Cell 9: 659–671 [DOI] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34: 93–158 [Google Scholar]
- Schneider TR, Sheldrick GM 2002. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Segref A, Mattaj IW, Ohno M 2001. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J 1999. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A 1997. MOLREP: An automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 21: 2736–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Lührmann R 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 16: 789–798 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Lührmann R 2007. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol Cell Biol 27: 7018–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 374: 300–321 [DOI] [PubMed] [Google Scholar]