Abstract
Surveys of transcription in many organisms have observed widespread expression of RNAs with no known function, encoded within and between canonical coding genes. The search to distinguish functional RNAs from transcriptional noise represents one of the great challenges in genomic biology. Here we report a next-generation sequencing technique designed to facilitate the inference of function of uncharacterized transcript forms by improving their coverage in sequencing libraries, in parallel with the detection of canonical mRNAs. We piloted this protocol, which is based on the capture of 3′ ends of polyadenylated RNAs, in budding yeast. Analysis of transcript ends in coding regions uncovered hundreds of alternative-length coding forms, which harbored a unique sequence motif and showed signatures of regulatory function in particular gene categories; independent single-gene measurements confirmed the differential regulation of short coding forms during heat shock. In addition, our 3′-end RNA-seq method applied to wild-type strains detected putative noncoding transcripts previously reported only in RNA surveillance mutants, and many such transcripts showed differential expression in yeast cultures grown under chemical stress. Our results underscore the power of the 3′-end protocol to improve detection of noncanonical transcript forms in a sequencing experiment of standard depth, and our findings strongly suggest that many unannotated, polyadenylated RNAs may have as yet uncharacterized regulatory functions.
Keywords: noncoding RNA, alternative polyadenylation, cryptic transcripts, stress response, next-generation sequencing, heat shock
INTRODUCTION
Recent advances in tiling microarray technologies (Stolc et al. 2004; David et al. 2006; Kapranov et al. 2007; Xu et al. 2009) and next-generation sequencing (Gowda et al. 2006; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Pan et al. 2008; Neil et al. 2009; Wang et al. 2009; Wilhelm and Landry 2009) in many species have shown that a large percentage of the eukaryotic genome is transcribed. Beside well-characterized mRNAs and structural and regulatory noncoding RNAs, the complex transcriptome of a given organism can include many molecular species of no known function, both putative noncoding RNAs and novel mRNA forms. A key challenge in genome biology is to determine which such transcripts are functionally relevant, motivated in part by the potential of RNAs as targets for treatment of human disease and as therapeutic agents themselves.
The extensive literature on regulatory noncoding RNAs has largely focused on short transcripts that mediate transcriptional and translational repression, most prominently microRNAs and Piwi-interacting RNAs (He and Hannon 2004; Kim 2006). In addition, detailed single-locus studies have revealed regulatory function by long and polyadenylated species (Brannan et al. 1990; Brown et al. 1991; Lee et al. 1999; Sotomaru et al. 2002; Martens et al. 2004; Willingham et al. 2005; Hongay et al. 2006; Camblong et al. 2007; Rinn et al. 2007; Thompson and Parker 2007; Uhler et al. 2007; Houseley et al. 2008). Genome-scale studies have identified characteristics of potential function at thousands of novel noncoding RNAs, including proximity to coding genes (Kapranov et al. 2007; Xu et al. 2009) and conservation across species (Guttman et al. 2009), suggesting that many additional biologically active RNAs remain to be uncovered (Mattick 2004).
Likewise, as classic studies of alternative splicing have given way to genome-scale investigation of coding transcripts, large data sets of novel splice forms have become available in higher organisms (Stolc et al. 2004; Wang et al. 2008). In addition, other mechanisms can give rise to noncanonical forms of mRNAs, including transcription initiation from internal promoters (Lochelt et al. 1994; Zhang et al. 1997; Yang et al. 1998; Scharf et al. 2007; Tsuchihara et al. 2009) and transcription termination at premature (Edwalds-Gilbert et al. 1997; Sparks et al. 1997; Sparks and Dieckmann 1998; Tian et al. 2005; Lutz 2008) or extended (True and Lindquist 2000; True et al. 2004; Shorter and Lindquist 2005) 3′ sites. Detailed single-gene studies have dissected the regulatory role of such noncanonical coding forms, but the generality of these mechanisms and their regulatory logic are not well understood.
We set out to develop a method that would facilitate the study of noncanonical transcript forms and mRNAs in parallel in a single experiment. A short-read sequencing approach for this purpose should minimize library complexity to detect and distinguish diverse transcript forms that are overlapping, while maximizing sensitivity for rare transcripts in a library of standard depth. We reasoned that sequencing only the end of each transcript would concentrate reads in a single peak rather than producing read peaks dispersed along the feature, reducing the tendency for internal fragments from long transcripts to populate sequencing libraries and freeing up depth in the library for rare or short species. As such, we developed a sequencing protocol that captures and reads only the 3′ ends of polyadenylated RNAs. We piloted this protocol in the model eukaryote Saccharomyces cerevisiae, and used it to discover noncanonical transcripts on a genome scale in yeast, under standard growth conditions and under treatment that destabilizes proteins to set off the unfolded protein response (Travers et al. 2000).
RESULTS
Strand-specific 3′-end RNA-seq
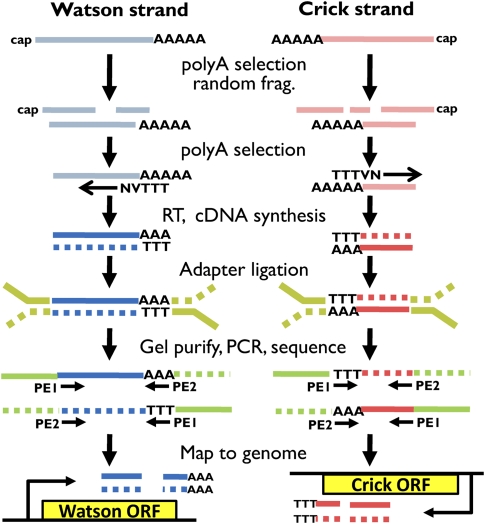
We developed a strategy for massively parallel, strand-specific sequencing of 3′ RNA ends (Fig. 1) by modifying the original mRNA-seq protocol (Nagalakshmi et al. 2008). Briefly, for a given sample of polyadenylated RNA, we fragmented the RNA and then reselected for polyadenylated fragments. We then used anchored oligo(dT) primers (Thomas et al. 1993) in a reverse transcription step, resulting in cDNA fragments of ∼100 base pairs (bp) with poly(A) tails of <20 bp. We constructed libraries with Illumina adapters, sequenced both ends of each fragment clone, and aligned the output sequences to the genome. Any mapped read with a stretch of As at the end was inferred to originate from the Watson strand of the annotated yeast genome from the Saccharomyces Genome Database (http://www.yeastgenome.org), while a stretch of Ts at the front of the read indicated a transcript originating from the Crick strand of the genome.
FIGURE 1.
Schematic of strand-specific 3′-end RNA-seq. Blue and red represent transcripts originating from the sense (Watson) and reverse complement (Crick) strands relative to the reference genome, respectively. RNA (light colors) was fragmented and filtered for polyadenylated species. Reverse transcription (RT) was primed with an anchored oligo(dT) primer (NVTTT: N = A,C,T,G and V = A,C,G) to yield double-stranded complementary DNA fragments (dark blue and dark red). Illumina paired-end adapters (green) were ligated to cDNA ends and amplified by PCR. Both ends of each strand of cDNA were sequenced to generate a paired-end read pair (PE1, PE2) and the reads were mapped to the reference genome. Read pairs mapping in an orientation such that the poly(A) stretch appears at the end were inferred to have originated from the sense strand relative to the reference, and pairs mapping such that poly(T) appears at the front were inferred to have originated from the reverse complement strand relative to the reference.
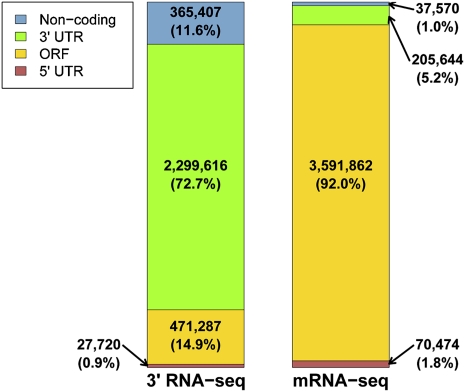
In piloting our 3′-end protocol, we sought to use it for the discovery of novel transcript forms and their regulation across changing conditions. As such, we grew yeast in rich media and under treatment with dithiothreitol (DTT), which disrupts protein structures by breaking disulfide bonds, initiating the environmental stress response (Gasch et al. 2000) and the unfolded protein response (Travers et al. 2000). We isolated RNA from yeast grown in each condition and sequenced libraries using both our new 3′-end protocol and standard mRNA-seq as in Nagalakshmi et al. (2008). In the last stage of 3′-end library construction, in which cDNA fragments ligated to adapters were amplified by the polymerase chain reaction (PCR), we tested two different PCR settings, P1 and P2 (see Materials and Methods). Analyzing the results of read mapping, we focused first on libraries from yeast grown in rich media. We compared expression levels of coding genes measured by mRNA-seq and by 3′-end RNA-seq and found good agreement (Supplemental Fig. S1), confirming the ability of the 3′-end protocol to quantitate gene expression. As expected, mRNA-seq libraries were populated by more reads from long genes than from short ones, while the 3′-end RNA-seq method eliminated this bias (Supplemental Fig. S2). We next categorized sequenced reads relative to open reading frame (ORF) definitions for both the 3′-end RNA-seq and mRNA-seq data (Fig. 2; Supplemental Table S1). As expected, most reads from the standard mRNA-seq method originated from inside ORFs (92% of mapped reads), whereas a majority of sequencing reads from our 3′-end RNA-seq procedure mapped within a few hundred base pairs downstream from ORF ends (Fig. 2; Supplemental Fig. S3; Supplemental Table S1). Noting that P1 3′-end RNA-seq libraries allowed the deepest sampling of intergenic and noncanonical transcripts (27% observed for P1; Supplemental Table S1), we used the P1 protocol in all further analyses. As a further check, we used the positions of reads from 3′ RNA-seq to infer 3′ UTR lengths for all expressed yeast genes and found good agreement with those previously reported from mRNA-seq (Nagalakshmi et al. 2008) and from computationally predicted poly(A) sites (Supplemental Fig. S4; Graber et al. 2002).
FIGURE 2.
The genomic distribution of mapped reads differs between 3′-end RNA-seq and standard mRNA-seq. Shown are the frequencies of uniquely mapped reads whose positions fell into the indicated genomic elements. Left, P1 3′-end RNA-seq library constructed as in Figure 1; right, mRNA-seq library constructed as in Nagalakshmi et al. (2008). For 3′-end RNA-seq, poly(A) (or poly(T)) positions were classified according to annotations on the same strand, while for mRNA-seq, start positions were classified without strand specificity. 5′ UTR definitions were taken from Nagalakshmi et al. (2008) and 3′ UTR lengths are defined in Materials and Methods. Noncoding, reads mapping to regions outside known ORFs, 5′ and 3′ UTRs, and known structural or regulatory RNAs.
Multiple length forms of coding transcripts
Apart from extensive coverage of 3′ UTRs, the data from our 3′-end sequencing protocol also revealed thousands of polyadenylated transcript ends mapping inside ORFs. Such sequencing reads, corresponding to 12%–15% of the total set of mapped reads from a given sample (Fig. 2; Supplemental Table S1), suggested the presence of alternative length forms of coding genes relative to canonical annotations in the yeast genome. To investigate the potential for biological function of these gene forms, we first defined transcript ends with respect to coherent groups of sequencing reads mapping close together, which we refer to as transcript units. We retained transcript units whose total read count in each library exceeded an expression-level dependent threshold for each gene (see Materials and Methods).
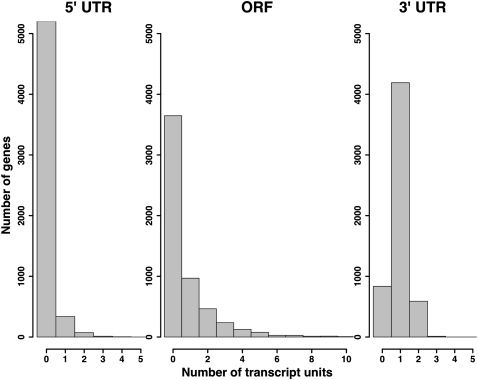
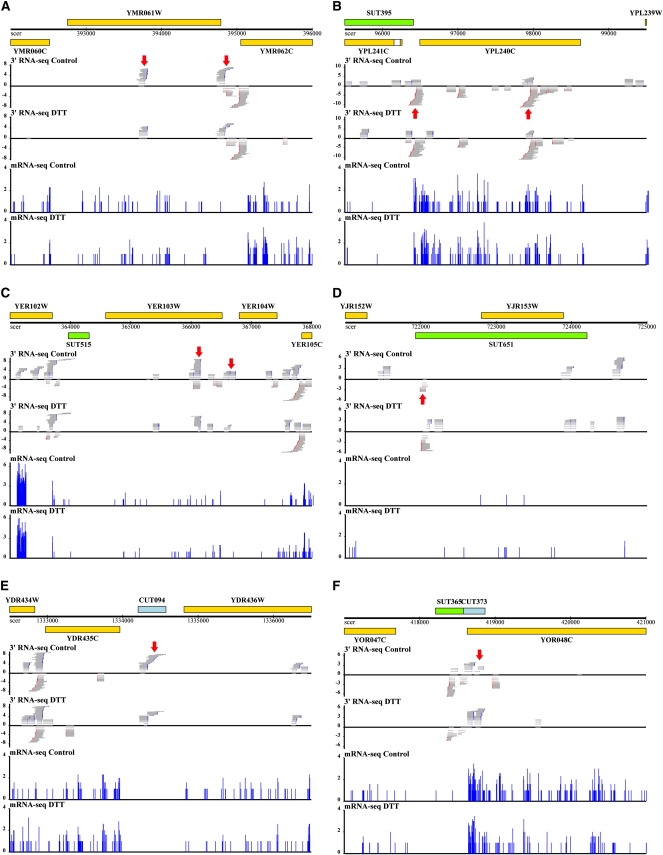
Tabulating the locations of all such transcript units with respect to gene annotations, we found that a majority of genes had no transcript units in the 5′ UTR or ORF and one transcript unit in the 3′ UTR (Fig. 3), conforming to canonical gene annotations. Strikingly, however, more than a third of all genes harbored >1 transcript unit inside the ORF, in RNA from cells grown either in rich media or under DTT treatment (Fig. 3). Figure 4 and Supplemental Figure S5 illustrate examples of such short coding forms, with the full list reported in Supplemental Table S2. As expected, we detected alternative-length coding transcripts from the yeast genes RNA14 and CBP1 (Fig. 4; Supplemental Fig. S5), whose condition-specific regulation by alternative termination has been previously reported (Sparks et al. 1997; Sparks and Dieckmann 1998). Our wide-ranging observation of alternative coding forms, beyond the latter well-studied examples, suggested their potential as a major regulatory strategy in yeast.
FIGURE 3.
Transcript ends cluster in gene elements. Each panel represents the frequency of genes containing dense clusters of transcript ends (transcript units, defined in Materials and Methods) from P1 3′-end RNA-seq, mapping to the indicated elements. (Left panel) 5′ UTRs; (middle panel) ORFs; (right panel) 3′ UTRs.
FIGURE 4.
Transcriptional profiles of example genes and intergenic RNAs. In each panel, genome annotations are shown at top: yellow, ORFs; green, SUTs (Xu et al. 2009); blue, CUTs (Xu et al. 2009). The bottom four plots in each panel report raw RNA-seq data. Gray horizontal bars represent reads from P1 3′ RNA-seq libraries constructed as in Figure 1, and blue vertical bars represent histograms of data from standard mRNA-seq as in Nagalakshmi et al. (2008). The x-axis reports the start position of a given read and the y-axis reports log2 of the number of reads mapping to the indicated position. For 3′-end RNA-seq libraries, reads mapping to the Crick strand are assigned negative counts; poly(A) tails are drawn in blue and reverse complement poly(T) tails in red. Control, RNA from cultures grown in rich media; DTT, RNA from cultures treated with dithiothreitol. Major transcript forms for genes and putative noncoding RNAs are indicated by red arrows. Panels represent genomic regions containing (A) YMR061W/RNA14, (B) YPL240C/HSP82, (C) YER103W/SSA4, (D) SUT651, (E) CUT094, and (F) CUT373.
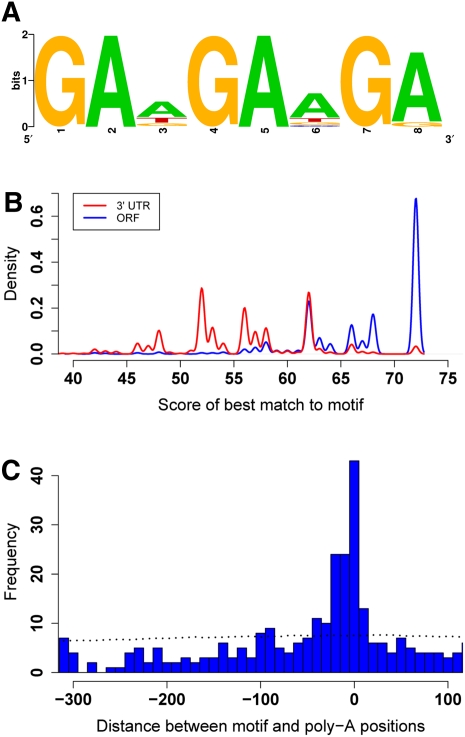
We reasoned that, if truncated mRNAs were the product of regulation by specific processing factors, the genes that harbor truncated forms would exhibit shared sequence elements. To test this, we identified 606 genes with highly expressed truncated mRNAs and used MEME (Bailey and Elkan 1994) to search for consensus sequence motifs within 50 bp upstream of truncated transcript ends. This analysis found a single, strongly enriched motif, GAAGAAGA (E-value < 3.1 × 10−103). Matches to this motif were more prevalent within ORFs than in full-length 3′ UTRs and recapitulated the end positions of truncated RNAs (Fig. 5), reflecting the specificity of this sequence element to the ends of truncated forms. While the GAAGAAGA motif does not match the target sequence of any yeast RNA-binding protein (Hogan et al. 2008), a putative binding function for this motif in yeast would echo the role of the GAAGAAGA sequence in mammals as an exonic splicing enhancer (Tacke et al. 1998; Sanford et al. 2009).
FIGURE 5.
A consensus sequence motif at the 3′ ends of alternative-length coding transcripts. Each panel represents results from the set of 606 genes with the most abundant truncated coding forms in cells grown in rich media. (A) The motif enriched at the 3′ ends of truncated coding forms. The sequence logo was visualized using the WebLogo program (Schneider and Stephens 1990; Crooks et al. 2004). (B) Frequency of match scores to the matrix in A across ORFs (blue line) or 3′ UTRs (red line). The x-axis reports the score of the sequence window in the indicated feature with the best match to the matrix in A, and the y-axis reports the frequency of such windows with a given score. (C) Frequency of positions of sequence matches inside ORFs to the matrix in A. The x-axis reports the distance between the position of the sequence window with the best match to the motif and the position of the end of the truncated transcript observed in P1 3′-end RNA-seq (blue bars) or a randomly selected position inside the ORF (black dotted line); the y-axis reports the frequency of best-match windows with a given distance.
Functional genomics of alternative-length coding forms
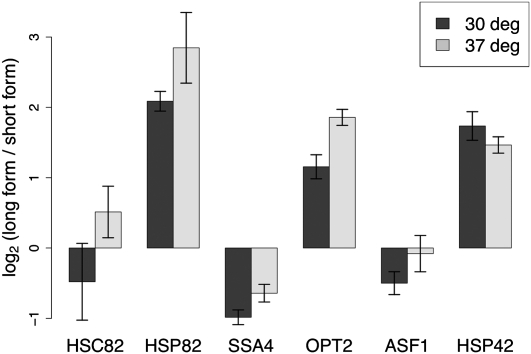
To further dissect the function of alternative-length coding transcripts, we took a genomic approach, testing Gene Ontology (GO) biological process terms for enrichment of genes harboring transcript ends inside ORFs. We set a significance cutoff for enrichment scores corresponding to a chance expectation of a single category in each analysis, and we examined all categories of functionally related genes whose enrichment for alternative length coding forms exceeded this criterion. The results, shown in Table 1, revealed a strong signal for a number of categories. In our 3′-end RNA sequencing data from yeast growing mitotically in rich media, we observed enrichment of alternative-length forms in gene categories involved in stress response and meiosis (Table 1). In contrast, in RNA from yeast treated with DTT, short coding forms were enriched among chromatin remodeling and protein catabolism genes; we also observed short coding forms in several broad categories annotated in positive regulation, which contain genes mediating transcription elongation, ribosomal RNA processing, and other housekeeping processes (Table 1). Thus, in each case, we detected enrichment of short coding forms among genes whose products were unlikely to be in high demand in the cell. Such a pattern is suggestive of a general regulatory function for truncated coding transcripts: under this model, short coding forms are produced and degraded when the gene product is not of immediate use, while a full-length transcript becomes the major form in conditions when the gene product is required.
TABLE 1.
Gene Ontology terms are enriched for genes with truncated coding transcripts
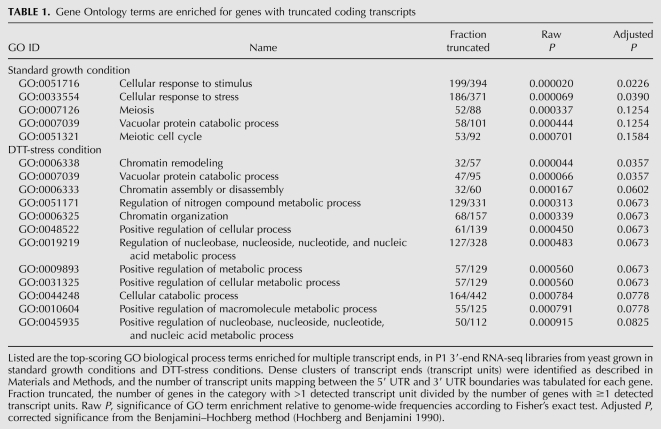
We focused on the results from cultures grown in rich media, and in particular on the hypothesis that truncated coding forms of stress response transcripts (Table 1) reflected a means to repress stress genes in nutrient-replete conditions. To test this hypothesis, we selected for further study six genes from the stress response GO category: four heat-shock genes (HSP82, HSC82, SSA4, and HSP42), the transporter OPT2, and the stress-sensitive chromatin regulator ASF1 (see Fig. 4; see Supplemental Fig. S5). We reasoned that, if truncated coding forms represented a mechanism for repression in standard culture conditions, this repression would be relieved during heat shock, shifting the balance from short to full-length mRNA forms. We grew yeast in standard culture conditions at 30°C and during heat shock at 37°C, and using RNA from each culture we measured the abundance of transcript forms with quantitative PCR. As shown in Figure 6, for five of the six genes, after heat shock the full-length transcript increased in abundance relative to the short coding form. For negative control genes unrelated to the heat shock pathway, the abundance of alternative length forms showed no change upon heat treatment (Supplemental Fig. S6), confirming the specificity of these regulatory changes. Results for one heat shock gene, HSP42, were not consistent with our hypothesis, showing a modest increase in the abundance of its short form in RNA from cells grown at high temperatures; this discrepancy may be related to the basal function of HSP42p as a chaperone even in standard conditions (Haslbeck et al. 2004). Overall, however, our results suggest a regulatory function for short coding transcript forms among heat shock genes, lending further support to a model in which alternative length forms serve as a general regulatory strategy in yeast.
FIGURE 6.
Transcript length forms change in heat shock. Shown are transcript abundances measured by quantitative PCR with two primer sets per gene, one at the annotated ORF end (interrogating the long form of the RNA) and the other at the position of an internal 3′ alternative transcript end (interrogating the short form) inferred from 3′-end RNA-seq data. The y-axis reports the log2 fold-change in abundance between the two amplicons in RNA from cultures grown at 30°C (dark gray) and at 37°C (light gray); negative values correspond to loci at which the short form of the RNA is more abundant than the long form. Cultures were grown to ∼3.5 × 107 cells/mL for RNA used to quantitate HSP82 and HSC82, and ∼1.0 × 107 cells/mL for all other genes.
Intergenic transcripts and induction by chemical stress
We next set out to assess the utility of our 3′-end sequencing protocol in the study of putative noncoding transcripts, which we took to encompass all RNAs expressed from intergenic regions lacking functional annotation. As expected, in our sequencing results, reads from coding transcripts dominated library composition for both the standard mRNA-seq protocol and our 3′-end procedure (Fig. 2). However, while only 1% of mapped reads were intergenic in the former, 12%–15% met this criterion in the P1 3′-end libraries (Fig. 2; Supplemental Table S1), reflecting a higher sensitivity for these transcripts and illustrating the promise of our protocol for the study of noncoding, polyadenylated RNAs.
We sought to compare the observed transcription at intergenic loci from our 3′-end protocol with intergenic RNAs annotated in a previous report (Xu et al. 2009). The latter used yeast strains mutant for RNA surveillance pathways to identify capped, polyadenylated, putatively noncoding RNAs, known as cryptic unstable transcripts (CUTs), in parallel with a study of wild-type strains that detected stable unannotated transcripts (SUTs). We defined transcript units, clusters of transcript ends mapping close together, within CUT and SUT boundaries and only analyzed those transcript units containing ≥5 read counts in each library. As expected, our 3′-end sequencing protocol detected a majority of previously defined SUTs (616 out of 847 of all SUTs) (Supplemental Fig. S7). Remarkably, though our analysis was confined to wild-type strains with no defect in RNA surveillance, our 3′-end RNA-seq data also detected 675 previously defined CUTs (73% of all CUTs) (Supplemental Fig. S7), reflecting the extensive coverage of such features in our data. We compared the positions of sequenced reads from our libraries to previously defined CUT and SUT annotations (Xu et al. 2009) and found good agreement (Supplemental Fig. S8), with most peaks of sequencing reads located within 100 bp upstream of CUT or SUT ends. Figure 4 and Supplemental Figure S9 show several examples of intergenic expression from our 3′-end RNA-seq data, including transcripts with a continuous pattern of 3′ ends throughout the length of the annotated feature (CUT094, CUT311, CUT577, and CUT695) and those with a more punctuated pattern of discrete transcript units (SUT168, SUT573, SUT651, and CUT578).
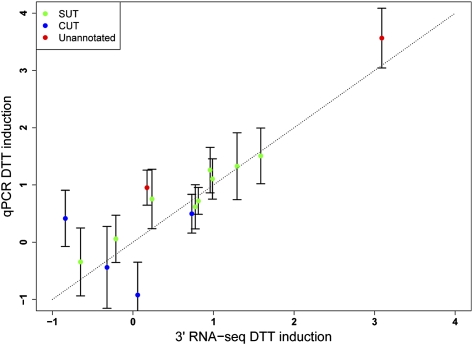
As a preliminary investigation into the regulation of intergenic, putative noncoding RNAs during the yeast unfolded protein response, we used the previous definitions of CUTs and SUTs (Xu et al. 2009) to examine changes in noncoding expression between cells grown in rich media and those treated with DTT. The full list of expression changes is given in Supplemental Table S3, and several dramatic examples are shown in Figure 4: For SUT651, encoded antisense to the coding gene YJR153W/PGU1, and for CUT373, antisense to the coding gene RAT1/YOR048C, expression of the noncoding RNA was anti-correlated with the level of mRNA from the opposite strand. In addition, manual inspection revealed a handful of previously unreported intergenic RNAs induced in DTT (Supplemental Fig. S9). As an independent means to confirm the expression-level changes of noncoding transcripts, we designed quantitative PCR assays for a subset of CUTs, SUTs, and unannotated putative noncoding RNAs with expression changes in DTT treatment predicted from our 3′ RNA-seq data. As shown in Figure 7, we observed good agreement between quantitative PCR and RNA-seq measures of differential expression in DTT, underscoring the power of our 3′-end protocol to detect reproducible and robust condition-specific changes in expression among noncoding transcripts. Taken together, our observations of noncoding expression reinforce the notion that the massive genome-wide production of low-abundance noncoding species is a characteristic of wild-type organisms, and that the potential regulatory function of these transcripts can be studied without perturbing RNA degradation pathways.
FIGURE 7.
Comparison of measures of expression changes of intergenic RNAs under dithiothreitol treatment. Each point represents an intergenic RNA with expression changes during DTT treatment. For each RNA, the y-axis reports the log2 fold-change of abundance measured by quantitative PCR using primers that interrogated the 3′ boundary of the feature. The x-axis reports the induction fold-change in DTT measured by P1 3′-end RNA-seq, defined as the log2 of the normalized sum of read counts in transcript units at the 3′ boundary of the feature in libraries from control samples, subtracted from the analogous quantity from DTT-treated cultures. Error bars are calculated as one standard deviation from four biological replicates.
DISCUSSION
In the study of unannotated “dark matter” transcription, distinguishing biologically relevant molecular species from transcriptional noise represents one of the great challenges in genomic biology (Struhl 2007). In many cases, inferring function for uncharacterized RNAs will require techniques that allow observation of low-abundance transcript forms and canonical mRNAs in the same experiment. Sequencing-based analysis of transcript ends can serve as a powerful method for the study of transcriptomes (Eveland et al. 2008; Nagalakshmi et al. 2008; Neil et al. 2009; Ozsolak et al. 2009) but can be hampered by relatively low throughput and/or the necessity for genetic perturbations. In this work, we report a 3′-end RNA-seq protocol that provides the sequences and counts of diverse transcript forms with high sensitivity, enabling coverage of intergenic transcripts and alternative and canonical forms of mRNAs in libraries sequenced at standard depth.
We have harnessed the sensitivity of 3′-end RNA-seq to survey intergenic, putative noncoding RNAs in yeast. By virtue of its focus on polyadenylated transcripts, our procedure is well-suited for the study of noncoding RNAs transcribed by RNA polymerase II. Polyadenylated molecular species comprise the vast majority of intergenic RNAs in yeast (David et al. 2006) and are also abundant in mammals (Kapranov et al. 2007; Guttman et al. 2009). Tour-de-force molecular dissection experiments have studied the regulatory function of a number of long, polyadenylated noncoding RNAs (Brannan et al. 1990; Brown et al. 1991; Lee et al. 1999; Sotomaru et al. 2002; Martens et al. 2004; Willingham et al. 2005; Hongay et al. 2006; Camblong et al. 2007; Rinn et al. 2007; Thompson and Parker 2007; Uhler et al. 2007; Houseley et al. 2008). Such regulators exert their effects by a range of mechanisms, from cis-acting repression via transcription interference, as with the antisense regulator of the yeast meiosis factor IME4 (Hongay et al. 2006), to trans-acting interactions with protein factors, as observed for the human noncoding transcript NRON, a nuclear trafficking regulator (Willingham et al. 2005). The pace of continued discovery of these regulatory RNAs suggests that many more remain as yet unidentified, underscoring the importance of genomic methods designed to quantitate noncoding RNA expression.
We have also used our 3′-end sequencing protocol in a genome-wide survey of alternative-length coding transcripts, which exhibit a shared sequence motif and are expressed preferentially in genes of particular functional categories. On a single-gene scale, the molecular biology literature has described several main mechanisms that generate truncated mRNA forms. Short coding transcript forms can result from transcription initiation from promoters internal to ORFs (Lochelt et al. 1994; Zhang et al. 1997; Yang et al. 1998; Scharf et al. 2007; Tsuchihara et al. 2009), although in yeast, most such cryptic initiation sites appear to be suppressed (Cheung et al. 2008). Likewise, many studies have observed premature termination and polyadenylation at individual genes (Edwalds-Gilbert et al. 1997; Sparks et al. 1997; Sparks and Dieckmann 1998; Tian et al. 2005; Lutz 2008). In a number of cases, condition-specific use of alternative 3′ ends has been identified as a means of gene regulation (Peterson and Perry 1989; Berger and Meselson 1994; Edwalds-Gilbert et al. 1997; Sparks et al. 1997; Santos et al. 1998; Sparks and Dieckmann 1998; Hoopes et al. 2000; Quesada et al. 2003; Simpson et al. 2003; Xing et al. 2004; Arigo et al. 2006; Lutz 2008). These include the flowering time regulator FCA in Arabidopsis, at which alternative polyadenylation within an intron governs the balance of functional and nonfunctional forms (Quesada et al. 2003; Simpson et al. 2003), and the yeast genes CBP1 and RNA14, whose full-length forms, generated during fermentation, are translated to functional proteins while short forms transcribed during respiration are not (Sparks et al. 1997; Sparks and Dieckmann 1998). Interestingly, at the Drosophila heat shock factor HSP70, truncated forms are produced by paused RNA polymerase II in the uninduced state (Rougvie and Lis 1988) while long forms appear upon heat shock (Berger and Meselson 1994), consistent with the length regulation we observe among yeast heat shock mRNAs. Given the known control of heat shock gene expression at the level of transcriptional initiation (Pirkkala et al. 2001; Hahn et al. 2004), condition-specific mRNA length forms are expected to function as an additional point of input for regulation in this complex pathway. The picture emerging from our work and that of others suggests that truncation and extension of coding transcripts may be a major regulatory strategy, in heat shock and in other pathways, that dovetails with regulation of transcriptional initiation and splicing.
On a genomic scale, what predominant mechanisms are likely to govern the abundance of truncated mRNA forms? One model would predict a major role for polymerase pausing, which is widespread in yeast and other eukaryotes (Brodsky et al. 2005; Kim et al. 2005; Muse et al. 2007; Pelechano et al. 2009) and is known to generate truncated mRNAs at a range of genes beside those involved in heat shock (Fort et al. 1987; Reines et al. 1987; Haley and Waterfield 1991; London et al. 1991; Kash et al. 1993). Under an alternative model, termination and polyadenylation sites may be regulated specifically in response to environmental change. Consistent with the latter idea, heat shock of HeLa cells induces an interaction between the heat shock transcription factor HSF1 and symplekin, a scaffolding protein necessary for polyadenylation (Xing et al. 2004). Likewise, environmental stress in yeast regulates the association of several RNA-binding proteins with pre-mRNAs (Krebber et al. 1999; Henry et al. 2003), and these factors in some cases appear to dictate the balance between long and short mRNA forms (Minvielle-Sebastia et al. 1998; Bucheli et al. 2007). The full complement of length form regulators could also involve splicing factors, which are known to influence polyadenylation (Lutz et al. 1996; Licatalosi et al. 2008; Wang et al. 2008); our identification, in truncated yeast mRNAs, of a motif nearly identical to a mammalian splicing enhancer leaves open the possibility that splicing-related proteins may regulate coding length forms on a genomic scale. Future work will be necessary to distinguish between the models for the regulation of mRNA forms. In parallel, genomic approaches will continue the search for regulatory function among unannotated transcripts, both genic and intergenic.
MATERIALS AND METHODS
Yeast growth and treatment
All experiments used yeast strain BY4716 (isogenic to s288c, MATα lys2Δ0). For RNA-seq, we grew two cultures, biological replicates 1 and 2, to early log-phase (OD = 0.6–0.9) at 30°C in YPD medium, and treated each with 4 mM DTT (or an equal volume of distilled water as control) for 1 h. Standard mRNA-seq library construction was applied to biological replicate 2. For the heat shock experiment, BY4716 was grown in YPD medium at 30°C to OD of 0.3 or 0.6, split into two flasks, and then grown at either 30°C or 37°C for 3 h. In all cases, RNA was harvested using the hot phenol method (Ausubel et al. 1995) and purified of genomic DNA with TurboDNase (Ambion).
3′-end RNA-seq
Polyadenylated RNA was isolated from total RNA samples using Dyna oligo(dT) magnetic beads (Invitrogen) and subjected to zinc-catalyzed fragmentation (Ambion) for 2–3 min. Polyadenylated RNA fragments were reisolated using Dyna oligo(dT) magnetic beads. First-strand cDNA was synthesized using anchored oligo(dT) primers (NVT20, N = A,C,G,T, V = A,C,G; Invitrogen) and Superscript II reverse transcriptase (Invitrogen). The complementary second strand was synthesized using RNase H (Invitrogen) and Escherichia coli DNA polymerase I (Invitrogen) to generate a double-stranded cDNA library. Paired-end DNA adapters (Illumina) were ligated onto both ends of each fragment using Quick DNA Ligase (New England Biolabs). cDNA fragments of ∼250 bp were isolated by 2% agarose gel electrophoresis to remove remaining adapters, and the resulting library was enriched by PCR using Phusion polymerase (New England Biolabs). Libraries made from biological replicates 1 and 2 used PCR conditions P1 and P2, where the number of PCR cycles was 23 and 17, respectively. Samples were sequenced using 36 bp paired-end modules on an Illumina 2G Genome Analyzer. In each sequencing run, an mRNA-seq library on the same flow cell was used as the control lane. A more detailed method for 3′-end RNA-seq library preparation is provided as supplemental material.
Data analysis
Output sequence reads (4–7 million reads per library; Supplemental Table S1) were aligned to the S. cerevisiae reference genome from the Saccharomyces Genome Database (http://www.yeastgenome.org) using Bowtie (Langmead et al. 2009) as follows. The reverse complements of each set of paired-end reads were first generated, and this set was used as the input for Bowtie with the options “–phred64-quals -l 20 -n 3 -e 300 –rf.” We retained each pair of uniquely mapped reads mapping within 250 bp of one another and filtered these for consecutive series of Ts at the front or As at the end (>1 bp). To ensure that the reads were from polyadenyl tails and not from stretches of As and Ts in the genome, we identified reads whose stretch of As or Ts contained <50% mismatches to the reference genome and removed these reads from further analysis. We also removed all reads that had a stretch of poly(T) or poly(A) at both the front and end. For each read, the poly(A) site was tabulated as the genomic position of the mapped nucleotide immediately adjacent to the poly(A) tail.
Filtered reads from each library were sorted according to the genome coordinate of the inferred poly(A) sites and grouped into transcript units as follows. For each chromosome, we set the 5′ boundary of the first transcript unit to be the position of the first read, and sequentially added reads to this unit based on their mapped position, setting the 3′ boundary when the next read mapped >d bp from the last read assigned to the unit; subsequent transcript units were assembled similarly. We found a regime in which the number of called transcript units was relatively insensitive to the d parameter (Supplemental Fig. S10), and within this regime, we chose d = 30 bp. Filtering such groups to retain all with ≥5 sequence reads in libraries from control and DTT-treated cultures, we identified 29,725 and 21,642 units in the former and the latter, respectively. The median across the poly(A) positions of all reads in a transcript unit was taken as the position of the transcript unit.
For analysis of coding transcription, we estimated 3′ UTR lengths for Figure 2 and Supplemental Figures S3 and S11 as follows. For each gene we examined the region from the ORF end to the start of the 5′ UTR of an adjacent gene or noncoding feature annotation, using for this purpose 5′ UTR lengths from Nagalakshmi et al. (2008). Within this region we identified the transcript unit with the maximum number of counts. We took the genome coordinate of this transcript unit as the end of the 3′ UTR. For all subsequent analyses, we used these estimated 3′ UTR definitions and 5′ UTR definitions from Nagalakshmi et al. (2008).
To normalize for the tendency for improved detection of noncanonical forms among highly expressed genes, we applied a further filtering step in the analysis of transcript units within genes. For each gene, we tabulated the number of reads n3UTR mapping to the 3′ UTR as defined above. For each transcript unit i in the gene comprised of ni reads in each library, we calculated the ratio ni/nUTR and eliminated the transcript unit from further consideration if this quantity was <0.25 in any library. The number of transcript units in 5′ UTRs, ORFs, and 3′ UTRs emerging from our data with this filtering scheme is shown in Figure 3 for the control sample and listed in Supplemental Table S2 for both samples. For all analyses of coding genes, we only used transcript units that passed this filtering scheme. A comparison of results from this scheme with those from an alternative strategy, in which we filtered transcript units in genes according to the absolute number of reads in each unit and did not correct for the respective gene's expression level, is shown in Supplemental Figure S11.
Expression fold-change of putative noncoding RNAs in DTT was calculated as follows. In each library, we identified all transcript units with ≥5 reads that fell within the boundaries of CUTs and SUTs taken from Xu et al. (2009). For each such feature in each library, we summed the read counts; for DTT-treated samples, this sum was normalized by the ratio of the total number of uniquely mapped reads in control and DTT-treated libraries. The difference between this normalized sum from the DTT sample and the read count sum from the control sample yielded the induction measures listed in Supplemental Table S3. In Figure 7, data were treated analogously except that, for each putative noncoding RNA, the measured expression level was taken as the sum of read counts within the boundaries defined by feature ends and the positions of primers used for quantitative PCR.
Consensus sequence motif search
To search for sequence motifs in truncated gene forms, we used 3′-end RNA-seq data from cultures grown in rich media to identify the transcript unit of highest expression mapping inside each ORF, and we compiled the 606 genes in which this major internal transcript unit comprised >50 sequence reads. For each of these 606 transcript units, we retrieved the strand-specific DNA sequence 50 bp upstream of the poly(A) position of the transcript unit, and we used this set as the input to MEME (Bailey and Elkan 1994), searching for a motif with a length from 4 to 8 bp.
To score matches to the GAAGAAGA motif in Figure 5B, we used the 8-position position-weight matrix output by MEME. We scanned gene elements with a sliding window of 8-mer and a step size of 1 bp; given the nucleotide at each site in a window, we defined the score as the probability of observing that nucleotide according to the MEME motif, and we defined the score for a given window as the sum of such probabilities across all sites in the window. For each gene element, we identified the window of highest score, reporting its score, multiplied by 10, in Figure 5B and its position in Figure 5C. To calculate the null distribution of expected motif positions in Figure 5C, for each gene we calculated the distance between the location of the best-scoring window and a random location inside the ORF, and we took the average of 1000 such simulations.
For comparison to the results from MEME, we also implemented an independent means to identify sequence motifs in truncated mRNA forms as follows. Using the set of 606 genes with highly abundant truncated forms as above, we extracted the 50 bp upstream of the poly(A) position of the major transcript unit inside the ORF for each gene and tabulated the frequency of each possible hexamer (4096 total hexamers) and 8-mer (65,536 total 8-mers) across this sequence set. We then repeated this frequency analysis on a null set in which we randomly selected a 50-mer from inside the ORF of each of the 606 genes. We observed strong, significant enrichment of GAAGAAGA and slight variants in truncated transcript ends relative to null sequences (data not shown), confirming the signal for this motif as reported in the main text. This analysis also detected enrichment of the TATATA element, which is a yeast polyadenylation signal (Graber et al. 2002), suggesting a mechanism of specific sequence regulation of polyadenyl tails for truncated forms (data not shown).
Gene Ontology term analysis
We used the Biological Process categories from the yeast gene and GO term associations (Ashburner et al. 2000) maintained by the Saccharomyces Genome Database (http://downloads.yeastgenome.org/literature_curation/gene_association.sgd.gz). For a given category, we only considered genes with >50 read counts mapping anywhere in their respective loci, and categories with fewer than six genes meeting this criterion were not considered further. Category pairs with identical sets of genes were combined. For each GO category, we identified the number of genes j harboring >1 transcript unit from the start of the 5′ UTR to the end of the 3′ UTR, as defined above, and likewise the number of genes k harboring exactly 1 transcript unit. Finally, we formulated a 2 × 2 table to compare j and k for each category to the sums of these quantities across all categories, and we assessed the difference using Fisher's exact test. The computed P-values were corrected for multiple testing using the Benjamini–Hochberg method (Hochberg and Benjamini 1990). A total of 1130 GO terms were analyzed for the library from cultures grown in rich media and 1082 from cultures treated with DTT, such that the expected number of false positive categories in Table 1 was 0.000701 × 1130 = 0.79 and 0.000915 × 1082 = 0.99, respectively.
Quantitative PCR
For each gene studied by quantitative PCR we designed two primer sets, where each set consists of forward and reverse primers designed to amplify around transcript ends detected by 3′-end RNA-seq. Each primer set was placed ∼100 bp upstream of one detected polyadenylation position. Primers were designed using Primer3 Plus (Untergasser et al. 2007), checked for hairpins and dimers using BeaconDesigner Web Edition (PREMIER Biosoft International), and synthesized by Elim Biopharmaceuticals. Primers used are listed in Supplemental Table S4. Single-stranded cDNA was synthesized from total RNA using oligo(dT) primer (Invitrogen) and Superscript III reverse transcriptase (Invitrogen). The cDNA was amplified using the DyNamo qPCR HS (Finnzymes) for 40 cycles on a Stratagene MX3000P qPCR machine.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Ellen Kittler and Leath Tonkin for expertise in Solexa sequencing; Hana Lee for RNA for Solexa libraries; and Don Rio for helpful discussions and comments on the manuscript. This work was supported by a Burroughs-Wellcome Career Award at the Scientific Interface, an Ellison Medical Foundation New Scholar Award in Aging, and NIH R01 GM087432-01 to R.B.B., and a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research to O.K.Y.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2038810.
REFERENCES
- Arigo JT, Carroll KL, Ames JM, Corden JL 2006. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent B, Kingston RE, Moore DD 1995. Current protocols in molecular biology Wiley, New York [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Berger SL, Meselson M 1994. Production and cleavage of Drosophila hsp70 transcripts extending beyond the polyadenylation site. Nucleic Acids Res 22: 3218–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM 1990. The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA 2005. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol 6: R64 doi: 10.1186/gb-2005-6-8-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44 [DOI] [PubMed] [Google Scholar]
- Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S 2007. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA 13: 1756–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277 doi: 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM 2006. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C 1997. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res 25: 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, McCarty DR, Koch KE 2008. Transcript profiling by 3′-untranslated region sequencing resolves expression of gene families. Plant Physiol 146: 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P, Rech J, Vie A, Piechaczyk M, Bonnieu A, Jeanteur P, Blanchard JM 1987. Regulation of c-fos gene expression in hamster fibroblasts: Initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res 15: 5657–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M, Li H, Alessi J, Chen F, Pratt R, Wang GL 2006. Robust analysis of 5′-transcript ends (5′-RATE): A novel technique for transcriptome analysis and genome annotation. Nucleic Acids Res 34: e126 doi: 10.1093/nar/gkl522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, McAllister GD, Smith TF 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res 30: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24: 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley JD, Waterfield MD 1991. Contributory effects of de novo transcription and premature transcript termination in the regulation of human epidermal growth factor receptor proto-oncogene RNA synthesis. J Biol Chem 266: 1746–1753 [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J 2004. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Henry MF, Mandel D, Routson V, Henry PA 2003. The yeast hnRNP-like protein Hrp1/Nab4 accumulates in the cytoplasm after hyperosmotic stress: A novel Fps1-dependent response. Mol Biol Cell 14: 3929–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y 1990. More powerful procedures for multiple significance testing. Stat Med 9: 811–818 [DOI] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6: e255 doi: 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745 [DOI] [PubMed] [Google Scholar]
- Hoopes BC, Bowers GD, DiVisconte MJ 2000. The two Saccharomyces cerevisiae SUA7 (TFIIB) transcripts differ at the 3′-end and respond differently to stress. Nucleic Acids Res 28: 4435–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kash SF, Innis JW, Jackson AU, Kellems RE 1993. Functional analysis of a stable transcription arrest site in the first intron of the murine adenosine deaminase gene. Mol Cell Biol 13: 2718–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN 2006. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev 20: 1993–1997 [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber H, Taura T, Lee MS, Silver PA 1999. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev 13: 1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochelt M, Flugel RM, Aboud M 1994. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J Virol 68: 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London L, Keene RG, Landick R 1991. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol Cell Biol 11: 4599–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS 2008. Alternative polyadenylation: A twist on mRNA 3′ end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10: 325–337 [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Mattick JS 2004. RNA regulation: A new genetics? Nat Rev Genet 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Beyer K, Krecic AM, Hector RE, Swanson MS, Keller W 1998. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J 17: 7454–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM 2009. Direct RNA sequencing. Nature 461: 814–818 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Jimeno-Gonzalez S, Rodriguez-Gil A, Garcia-Martinez J, Perez-Ortin JE, Chavez S 2009. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet 5: e1000614 doi: 10.1371/journal.pgen.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ML, Perry RP 1989. The regulated production of μs and μm mRNA is dependent on the relative efficiencies of μs poly(A) site usage and the cμ4-to-M1 splice. Mol Cell Biol 9: 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson GG 2003. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J 22: 3142–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Wells D, Chamberlin MJ, Kane CM 1987. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol 196: 299–312 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos BC, Chevaile A, Kojima R, Gullans SR 1998. Characterization of the Hsp110/SSE gene family response to hyperosmolality and other stresses. Am J Physiol 274: F1054–F1061 [DOI] [PubMed] [Google Scholar]
- Scharf S, Zech J, Bursen A, Schraets D, Oliver PL, Kliem S, Pfitzner E, Gillert E, Dingermann T, Marschalek R 2007. Transcription linked to recombination: A gene-internal promoter coincides with the recombination hot spot II of the human MLL gene. Oncogene 26: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S 2005. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 6: 435–450 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C 2003. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Sotomaru Y, Katsuzawa Y, Hatada I, Obata Y, Sasaki H, Kono T 2002. Unregulated expression of the imprinted genes H19 and Igf2r in mouse uniparental fetuses. J Biol Chem 277: 12474–12478 [DOI] [PubMed] [Google Scholar]
- Sparks KA, Dieckmann CL 1998. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res 26: 4676–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks KA, Mayer SA, Dieckmann CL 1997. Premature 3′-end formation of CBP1 mRNA results in the downregulation of cytochrome b mRNA during the induction of respiration in Saccharomyces cerevisiae. Mol Cell Biol 17: 4199–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306: 655–660 [DOI] [PubMed] [Google Scholar]
- Struhl K 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Tacke R, Tohyama M, Ogawa S, Manley JL 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93: 139–148 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Hesse SA, McKie AT, Farzaneh F 1993. Sequencing of cDNA using anchored oligo dT primers. Nucleic Acids Res 21: 3915–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483 [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431: 184–187 [DOI] [PubMed] [Google Scholar]
- Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hashimoto S, Matsushima K, Mizushima-Sugano J, Yamashita R, Nakai K, et al. 2009. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res 37: 2249–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ 2007. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci 104: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35: W71–W74 doi: 10.1093/nar/gkm306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Landry JR 2009. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48: 249–257 [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD 2004. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem 279: 10551–10555 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Niu Z, Cohen AJ, Nah HD, Adams SL 1997. The chick type III collagen gene contains two promoters that are preferentially expressed in different cell types and are separated by over 20 kb of DNA containing 23 exons. Nucleic Acids Res 25: 2470–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]