Abstract
A constitutively active epidermal growth factor receptor (EGFR) mutant, EGFR variant III (EGFRvIII), has been detected at high frequencies in certain human cancers. This study evaluated transactivation and trafficking of erbB family members as a result of constitutive EGFR activity in a cancer cell line. Expression of EGFRvIII modulated erbB family members through different mechanisms; the erbB3 mRNA level was reduced, whereas wild-type EGFR (wtEGFR) and erbB2 protein levels were diminished, with no change in their mRNA levels, and there was no change in the erbB4 expression level. Both EGFR and erbB2 were internalized as a result of EGFRvIII's activity and redistributed to the cell surface upon addition of AG1478, an inhibitor of wtEGFR/EGFRvIII catalytic activity. Acute activation of EGFRvIII by removing AG1478 from cells increased phosphorylation of both wtEGFR and erbB2 and caused differential trafficking of EGFRvIII's activation partners; wtEGFR was directed primarily to lysosomal compartments and partially to recycling compartments, whereas erbB2 was directed primarily to recycling compartments and partially to lysosomal compartments. Our data demonstrate that the constitutive activity of EGFRvIII is sufficient to trigger endocytosis and trafficking of wtEGFR and erbB2, which may play a role in activating signaling pathways that are triggered during receptor endocytosis. (J Histochem Cytochem 58:529–541, 2010)
Keywords: EGFR, EGFRvIII, cancer, heterodimerization, endocytosis, trafficking, recycling, lysosome
The naturally occurring EGFR mutant, with a deletion of aa 6–273 in the extracellular domain, designated EGFR variant III (EGFRvIII, delta EGFR, or de2-7 EGFR) is expressed in a number of cancers, most notably glioblastoma (Lorimer 2002; Huang et al. 2007a; Sonabend et al. 2007). This mutant does not bind EGF (Moscatello et al. 1996); nevertheless, it is constitutively autophosphorylated (Nishikawa et al. 1994; Fernandes et al. 2001) and is present at the cell membrane (Wikstrand et al. 1997) and suffers from impaired downregulation, which is a result of decreased association with Cbl proteins and subsequently decreased polyubiquitination and degradation (Huang et al. 1997; Schmidt et al. 2003; Han et al. 2006; Grandal et al. 2007). The defective internalization and the constitutive phosphorylation of EGFRvIII seem to be contributors to persistence of signaling.
Ligand activation of wild-type EGFR (wtEGFR) leads to homo- or heterodimerization and transphosphorylation of other erbB family members (Yarden 2001). Thus, surface localization of an internalization-impaired, constitutively active EGFR mutant could similarly interact with other erbB members such as EGFR and erbB2. There is limited information in the literature about the interactions between EGFRvIII and erbB family members. There is evidence indicating that heterodimerization between EGFRvIII and either wtEGFR or erbB2 is possible. For example, a physical association between the EGFRvIII and an erbB2 ectodomain that is kinase deficient was previously reported, where it was demonstrated that ectodomain interactions were sufficient to form dimers (O'Rourke et al. 1998). Evidence for interaction of EGFRvIII with wtEGFR also exists; for example, activation of EGFRvIII by removal of its catalytic activity inhibitor, AG1478, restored tyrosine phosphorylation of both EGFRvIII and wtEGFR in the absence of an activating ligand (Zeineldin et al. 2006). Another study, which examined an EGFRvIII–heparin-binding EGF–wtEGFR autocrine loop generation in gliomas, raised the possibility that EGFRvIII may signal partly through wtEGFR (Ramnarain et al. 2006). It was found that expression of EGFRvIII upregulates specific gene products including the wtEGFR ligand heparin-binding EGF-like growth factor (HB-EGF), whereas use of a neutralizing antibody to HB-EGF blocked EGFRvIII-induced proliferation, thus raising the possibility that EGFRvIII signals partly through wtEGFR (Ramnarain et al. 2006). In addition, interaction of EGFRvIII with wtEGFR was demonstrated as EGFRvIII coimmunoprecipitated with wtEGFR, and wtEGFR in the complex was phosphorylated (Luwor et al. 2004). Another indication of heterodimerization was provided by examination of the efficacy of the therapeutic monoclonal antibody 528 (MAb 528) (Johns et al. 2007). The epitope for MAb 528 is within the extracellular domain of EGFR (Kawamoto et al. 1983); nevertheless, it binds both wtEGFR and EGFRvIII (Mishima et al. 2001). It was found that cells expressing only EGFRvIII and cells coexpressing either wtEGFR with EGFRvIII, or wtEGFR with the EGFRvIII mutant that is kinase active but autophosphorylation defective, all responded to MAb 528 therapy. On the other hand, cells coexpressing wtEGFR and a kinase-dead EGFRvIII were refractory to therapy. All this suggested that MAb 528 mediates its antitumor activity by disrupting interactions between EGFRvIII and wtEGFR when both forms of EGFR are expressed (Johns et al. 2007).
Although a limited number of studies report on heterodimerization and transphosphorylation of wtEGFR by EGFRvIII and possible association of erbB2 with EGFRvIII, there is a gap in knowledge regarding the fate of wtEGFR or erbB2. The impact of EGFRvIII on other erbB family members is not fully known, and for that reason, we evaluated the effect of the steady-state activity of EGFRvIII on erbB family member expression levels and endocytic trafficking.
Our hypothesis is that the constitutive activity of EGFRvIII leads to activation and internalization of both wtEGFR and erbB2. In this study, we chose to use OVCA 433 ovarian cancer cells as our model cell system because this cell line expresses low to moderate levels of the erbB family members, and it exhibits mitogenic responses to EGF. Although there are conflicting reports about the expression of EGFRvIII in ovarian cancer (Moscatello et al. 1995; Lassus et al. 2006; de Graeff et al. 2008; Steffensen et al. 2008), there are numerous EGF receptor mutations that have been identified in human tumors that alter receptor activity (Riese et al. 2007; Zandi et al. 2007; Ferguson 2008; Kumar et al. 2008) that have not been fully explored in ovarian tumors. Thus, EGFRvIII provides a model for the consequences of constitutive EGF receptor activation in ovarian cancer. We found that the steady-state activity of EGFRvIII caused downregulation and internalization of both wtEGFR and erbB2, while inhibition of EGFR/EGFRvIII's catalytic activity by AG1478 increased their protein levels and redistributed both receptors to the cell surface. Acute activation of EGFRvIII by removing cells from AG1478 resulted in activating wtEGFR and erbB2 and directing wtEGFR primarily to lysosomal degradation and partially to recycling compartments while directing erbB2 primarily to recycling compartments and partially to lysosomal degradation. Our findings show that downregulation of wtEGFR and erbB2 is a result of EGFRvIII expression and activity and that this is accompanied by differential trafficking.
Materials and Methods
Cell Culture and Treatments
Ovarian carcinoma cell line OVCA 433 was generously provided by Dr. Robert Bast, Jr., M.D. Anderson Cancer Center, Houston, TX. Cells were maintained at 37C under 5% carbon dioxide and grown in MEM supplemented with 10% (v/v) FBS, 1 mM sodium pyruvate, 2 mM l-glutamine, 0.5 units/ml penicillin, 0.5 μg/ml streptomycin. OVCA 433 cells stably expressing the constitutively active EGFRvIII (vIIIA1 cells) and the vector control cells (V) were described previously (Ning et al. 2005; Zeineldin et al. 2006) and were grown in the presence of 300 μg/ml G418. Unless otherwise indicated, cells were placed in MEM containing 0.1% (w/v) BSA for 24 hr, and then EGF was added at a final concentration of 20 nM. AG1478, which is a specific inhibitor of the catalytic activity of EGFR (IC50 = 3 nM in vitro) (Levitzki and Gazit 1995), was used at a final concentration of 5 μM. Stock solutions of inhibitors were prepared in DMSO and were diluted in growth medium, so DMSO did not exceed 0.02% of the final volume. Long-term inhibition involved adding the inhibitor to whole-growth medium.
Oligonucleotide Microarrays
RNA was isolated using an RNeasy isolation kit (Qiagen; Valencia, CA) according to the manufacturer's protocol. Sample preparation and microarray processing were carried out according to the Affymetrix expression analysis technical manual (Affymetrix; Santa Clara, CA). Briefly, 20 μg of RNA was reverse-transcribed by Superscript II reverse transcriptase using a T7-(dT)24 primer, which contains a T7 RNA polymerase promoter. The resulting double-stranded cDNA was then transcribed in vitro to generate biotinylated complementary RNA. Then, 10 μg of fragmented cRNA was hybridized to cancer chip microarray GeneChip human cancer G110 (HC-G110) array P/N 900257 (Affymetrix) for 16 hr at 45C with rotation at 60 rpm. The HC-G110 chip contains oligonucleotides for 1700 genes. Following hybridization, the microarray was washed and stained using an Affymetrix fluidics station and scanned using an argon ion confocal laser with a 488-nm emission and detection at 570 nm. Each cell line (vIIIA1 and V) was analyzed in five replicates. Data were normalized using a robust multiarray average to eliminate variations between chips arising from sample preparation and manufacturing and processing of arrays (Irizarry et al. 2003). Analysis of the data was performed using GeneSpring version 4.2.1 software (Silicon Genetics; San Carlos, CA), where an average was calculated for the five replicates of each cell line.
Immunoblot Analysis
Cells were washed with ice-cold PBS and harvested in lysis buffer [10 mM Tris-HCl (pH 7.4), 1% SDS, 5 mM EDTA, 0.1 mM dithiothreitol, 1 mM sodium orthovanadate, and 1 mM PMSF]. Fifteen micrograms of total cell lysates was resolved by electrophoresis using a 7.5% SDS-polyacrylamide gel, transferred to nitrocellulose membranes (Bio-Rad; Hercules, CA), and probed with the indicated antibodies. Primary antibodies were used at a 1:100 dilution and included anti-EGFR (Santa Cruz Biotechnology; Santa Cruz, CA), anti-erbB2 (Santa Cruz Biotechnology), anti-phospho-erbB2 (Upstate Biotechnology; Waltham, MA), anti-erbB3 (Ab-6) (Labvision; Fremont, CA), anti-erbB4 (Ab-2) (Labvision), anti-β− tubulin (Santa Cruz Biotechnology), and anti-GAPDH (Abcam; Cambridge, MA). Horseradish peroxidase-conjugated secondary antibodies (Promega; Madison, WI) were used at a 1:20,000 dilution. Bands were detected with a Super Signal chemiluminescent detection system (Pierce Corp.; Rockford, IL). Bands were quantitated using a Kodak Imaging Station 4000 (Carestream Health; New Haven, CT).
PCR Detection of mRNA
First-strand cDNA was generated using a High Capacity cDNA archive kit (Applied Biosystems; Foster City, CA) using 1 μg of total RNA in a 12.5-μl reaction mixture. PCR reaction mixtures contained 1 μl of cDNA, 10 pmol of each primer, 1× PCR buffer, 0.5 nmol of each deoxynucleotide triphosphate, 2.5 mM MgCl2, and 1 U of Taq polymerase in a total volume of 20 μl. The sets of primers used are listed in Table 1. EGFR 1037-bp products were attained after 40 cycles with an annealing temperature of 56C. erbB2 559-bp products were attained after 40 cycles with an annealing temperature of 60C. erbB3 365-bp products were attained after 25 cycles with an annealing temperature of 55C (Knowlden et al. 1998). erbB4 265-bp products were attained after 25 cycles with an annealing temperature of 55C (Knowlden et al. 1998). 18S 100-bp products were attained after 19 cycles with an annealing temperature of 55C. The thermal cycling started with an initial denaturation at 94C for 4 min and ended with a final extension at 72C for 7 min. Thermal cycling consisted of cycles of 94C for 30 sec, annealing temperature for 60 sec (30 sec for 18S), and 72C for 60 sec (30 sec for 18S). The PCR amplifications were performed with a PTC-200 Peltier thermal cycler (MJ Research, Inc.; Watertown, MA). Products were electrophoresed in 2% agarose gels and visualized by SYBR Green staining.
Table 1.
List of primers used in RT-PCR
| Gene product | Primer set (forward; reverse) |
|---|---|
| EGFR | 5′GGGGAATTCGCGATGCGACCCTCCGGG 3′; 5′GGGAAGCTTTCCGTTACACACTTTGCG 3′ |
| erbB2 | 5′CCCACGTCCGTAGAAAGGTA 3′; 5′TGAACAATACCACCCCTGTC 3′ |
| erbB3 | 5′GGTGCTGGGCTTGCTTTT 3′; 5′CGTGGCTGGAGTTGGTGTTA 3′ |
| erbB4 | 5′TGTGAGAAGATGGAAGATGGC 3′; 5′GTTGTGGTAAAGTGGAATGGC 3′ |
| 18S | 5′AAACGGCTACCACATCCAAG 3′; 5′CCTCCAATGGATCCTCGTTA 3′ |
Real-time PCR
The first-strand cDNA was generated as described above. Real-time PCR was performed using PreDeveloped TaqMan gene expression reagents for EGFR, erbB2, erbB3, erbB4, and GAPDH following the manufacturer's instructions (Applied Biosystems). The real-time PCR reactions were carried out using an Applied Biosystems 7900 HT Fast Real-Time PCR system. Evaluation of the amplification efficiency of EGFR, erbB2, erbB3, erbB4, and GAPDH was done according to Applied Biosystems' instructions and was found to be equivalent. The transcripts were quantitated using the ΔΔCT method according to Applied Biosystems, using GAPDH as the normalizer and OVCA 433 cells as the calibrator.
Immunofluorescence and Colocalization Analysis
Cells were seeded in a Lab Tek II chamber slide system (Nalge Nunc Int.; Naperville, IL). For staining, Lysotracker (Invitrogen; Carlsbad, CA) was added to cells during the last 30-min incubation at 37C. Cells were fixed for 7 min at room temperature in freshly prepared 3.7% (w/v) paraformaldehyde in PBS (1.37 M sodium chloride, 27 mM potassium chloride, 43 mM dibasic sodium phosphate, 15 mM monobasic potassium phosphate). Cells were permeabilized by incubation in 0.1% (w/v) Triton X-100 in PBS on ice for 4 min. For detection of EGFRvIII, blocking was carried out for 1 hr at 37C, using 10% (w/v) nonfat milk in PBS; otherwise, blocking was carried out for 1 hr at room temperature, using 3% (w/v) BSA in PBS for EGFR staining and 1% (w/v) BSA in PBS for erbB2 staining. Cells were then incubated overnight at 4C with the primary antibody, prepared in blocking solution at 1:100 dilution for all antibodies except for anti-EGFRvIII, which was prepared at 1:800 dilution. The primary antibodies used were chicken anti-EGFRvIII (Ab 1825; Aves Laboratory, Tigard, OR) (Zeineldin et al. 2006), mouse anti-EGFR (Ab-3, clone EGFR.1; Labvision), mouse anti-erbB2 (Santa Cruz Biotechnology), and rabbit anti-Rab11 (Abcam). This step was followed by incubation for 1 hr at room temperature in the dark with the secondary antibody diluted to 1:200 in the blocking buffer; except for detection of EGFRvIII, the incubation was carried out for 20 min at 37C with the secondary antibody diluted to 1:900. The secondary antibodies were anti-chicken-fluorescein (Millipore; Billerica, MA), anti-mouse-fluorescein (Millipore), anti-mouse-horseradish peroxidase (Tyramid kit; Invitrogen), and anti-rabbit-fluorescein (Millipore). The cells stained for erbB2 were stained according to the instructions for the Tyramid kit. Finally, cells were washed with PBS, and slides were mounted with Vectashield mounting medium (Vector Laboratories; Burlingame, CA). Confocal images were generated using a Zeiss LSM 510 confocal microscope (Carl Zeiss MicroImaging Inc.; Thornwood, NY), with excitation at 488 nm, using an argon laser, and at 543 nm, using helium neon 1, and LSM Image Browser software (Carl Zeiss MicroImaging Inc.). Images were compiled using Adobe Photoshop software (Adobe; San Francisco, CA). Analysis of colocalization was examined by determining Pearson's correlation coefficient using LSM image software (Carl Zeiss MicroImaging Inc.). Pearson's correlation coefficient values range from −1 to +1, where +1 indicates perfect colocalization, while −1 indicates total lack of colocalization, and a value of 0 indicates no correlation.
Statistical Analysis
For all analyses, including analysis of the microarrays data, differences between each two cell lines compared were evaluated using a nonparametric Welch's two-sample t test. Any p value below 0.05 was considered statistically significant.
Results
EGFRvIII Expression Selectively Modulated the mRNA or Protein Levels of erbB Receptors
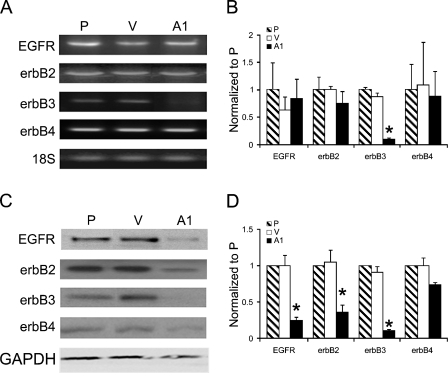
The effect of EGFRvIII steady-state activity on mRNA levels of several genes was examined by limited microarray analysis (using cancer chip HG-110; Affymetrix). Examination of the transcripts of the erbB family members, summarized in Table 2, revealed that erbB1, erbB2, and erbB4 were detected in parental (P), control (V), and EGFRvIII-expressing (vIIIA1) cells with no significant differences in their levels; whereas erbB3 (HER3) transcript was detected at a higher level in control cells than in vIIIA1 cells. Confirmation of these findings was carried out by RT-PCR (Figure 1A), and mRNA levels were quantified using real-time PCR (Figure 1B), which confirmed the microarray findings that the expression of erbB3 was highly downregulated in vIIIA1 cells but expression levels of EGFR, erbB2, and erbB4 showed no statistically significant changes.
Table 2.
Fold change number of erbB family members detected by the HC-G110 chip
| Gene product | Accession no.a | Affymetrix reference sequence | Fold change | p value |
|---|---|---|---|---|
| EGFR precursor mRNA | X00588 | 1537_at | ↓ 1.2 | NS |
| C-erb-B2 mRNA | X03363 | 1802_s_at | ↓ 1.8 | NS |
| HER3 mRNA complete cds | M34309 | 1585_at | ↓ 8.8 | 0.007 |
| erbB4 complete cds | L07868 | 1727_at | ↓ 1.5 | NS |
GenBank database.
Table data summarize the fold change number of erbB family members detected by the human cancer chip HC-G110 as a result of expressing EGFRvIII. NS, not statistically significant.
Figure 1.
Examination of erbB family member levels in EGFRvIII-expressing cells. (A) mRNA levels for erbB family members were evaluated by RT-PCR. 18S was amplified as an internal control. (B) mRNA levels were quantitated by TaqMan real-time PCR performed in triplicate. Bars = mean ± standard deviation (sd). Asterisk indicates statistically significant difference. (C) Total cell lysates (15 μg of protein) were fractionated by SDS-PAGE, and erbB family members were visualized by immunoblot analysis. GAPDH was detected as an internal loading control. P, parental OVCA 433 cells; V, vector control; A1, vIIIA1 cells. (D) Quantitation of band intensities obtained in immunoblot analysis using Kodak Imaging Station 4000. Band intensities were normalized to the internal controls. Bars = mean ± sd; n=4. Asterisk indicates statistically significant differences in a comparison of A1 cells to P and V cells.
Immunoblot analysis for the erbB family members (Figures 1C and 1D) correlated with the findings of the oligonucleotide microarrays for erbB3 and erbB4 but not for wtEGFR and erbB2. As for erbB4, although it was slightly reduced, there were no statistically significant differences in its protein levels in the EGFRvIII-expressing cells vs that of the parental or control cells. Nevertheless, the protein levels of wtEGFR, erbB2, and erbB3 detected by Western blot analysis were lower in vIIIA1 cells than in the parental and control cells (Figures 1C and 1D). The lower protein level of erbB3, but not wtEGFR or erbB2, in EGFRvIII-expressing cells correlated with the finding that erbB3 transcript was decreased. These findings suggested that the steady-state activity of EGFRvIII caused a reduction only of erbB3 at the mRNA level while reducing the protein levels of EGFR and erbB2. A possible explanation for the detection of lower EGFR and erbB2 protein levels without changes in their mRNA levels is that wtEGFR and erbB2 are downregulated from the cell surface because of constitutive activation by EGFRvIII.
EGFRvIII Steady-state Catalytic Activity Caused Downregulation of wtEGFR and erbB2
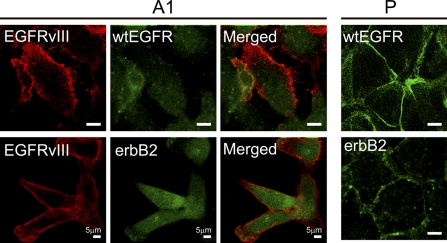
To examine the possibility that wtEGFR and erbB2 are downregulated as a result of EGFRvIII activity, we used confocal fluorescence microscopy to evaluate localization of EGFRvIII, wtEGFR, and erbB2 (Figure 2, left panels), which revealed that both wtEGFR and erbB2 were detected intracellularly in vIIIA1 cells, whereas both receptors were detected on the cell surface of the parental cell line (Figure 2, right panels). Conversely, EGFRvIII was detected in vIIIA1 cells at the cell surface with minor intracellular staining. This is consistent with previous reports demonstrating that EGFRvIII suffers from impaired internalization (Huang et al. 1997; Schmidt et al. 2003; Han et al. 2006; Grandal et al. 2007) and undergoes recycling to the cell surface (Grandal et al. 2007). We did not detect any intracellular colocalization of EGFRvIII with wtEGFR but did detect minimal intracellular colocalization of EGFRvIII with erbB2 that had an insignificant Pearson's correlation coefficient of 0.07. Our results indicated that the steady-state activity of EGFRvIII led to intracellular localization of wtEGFR and erbB2.
Figure 2.
Examination of cellular localization of EGFRvIII, wtEGFR, and erbB2 in EGFRvIII-expressing cells by confocal microscopy (left panels). Right panels show localization of wtEGFR and erbB2 in the parental cell line. P, OVCA 433 cells; A1, vIIIA1 cells. Bar = 5 μm.
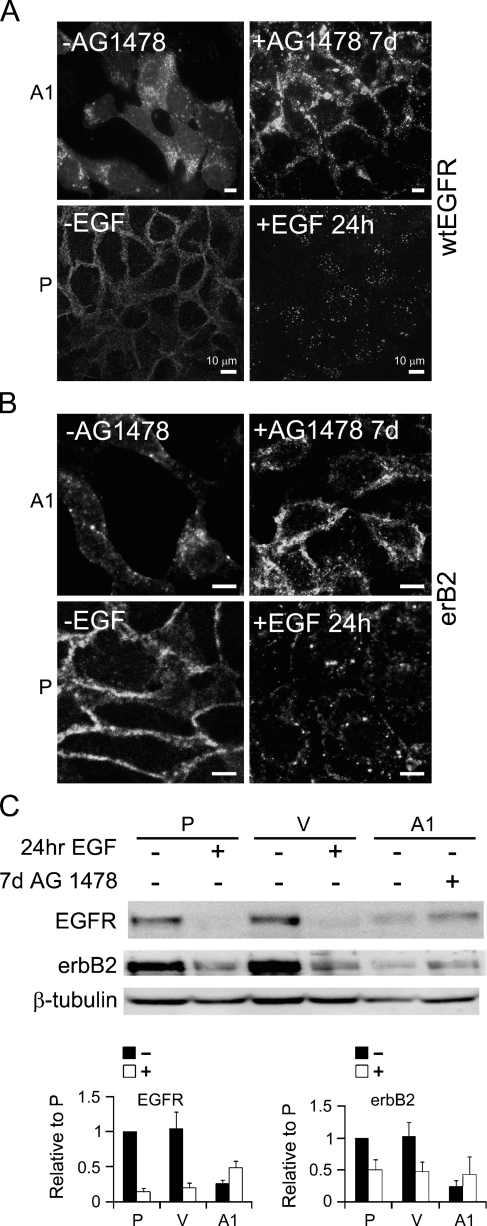
We further examined the localization of wtEGFR and erbB2 after inhibiting the catalytic activity of EGFRvIII, using AG1478. We found that inhibiting EGFRvIII's catalytic activity caused redistribution of wtEGFR and erbB2 mainly to the cell surface, with modest intracellular dot-like staining (Figures 3A and 3B). Furthermore, intracellular localization of wtEGFR and erbB2 in vIIIA1 cells was similar to that observed as a result of ligand activation of wtEGFR in the parental cell line (Figures 3A and 3B); and upon specific inhibition of EGFRvIII activity, both wtEGFR and erbB2 redistributed to the cell surface in a manner resembling that of inactive wtEGFR in parental cells (Figures 3A and 3B). Treatment of the parental cell line with AG1478 for 7 days did not change the localization of wtEGFR or erbB2, as they remained detected on the cell surface (data not shown). This demonstrated that the intracellular localization of wtEGFR and erbB2 was a result of EGFRvIII activity.
Figure 3.
Examination of cellular localization and protein levels of wtEGFR and erbB2 before (−AG1478) and after (+AG1478) treatment for 7 days and in parental (P) cell lines with (+EFG) and without (−EGF) stimulation with 20 nM EGF for 24 hr. Confocal images for detecting localization of (A) wtEGFR and (B) erbB2 in EGFRvIII-expressing cells. (C) Immunoblot analysis of wtEGFR and erbB2 in cells. β-Tubulin was detected as an internal loading control. Quantitation of band intensities was obtained by immunoblot analysis using Kodak Imaging Station 4000. Band intensities were normalized to the internal control and to the parental cell line. Bars = mean ± standard deviation, n=4. Control cells (V) are shown with (+EGF) and without (−EGF) stimulation with 20 nM EGF for 24 hr. vIIIA1 (A1) cells are shown with and without treatment with AG1478 for 7 days. P, OVCA 433 cells; V, vector controls; A1, vIIIA1 cells. Bar = 10 μm.
To examine the possible effect of the constitutive activity of EGFRvIII on the protein levels of wtEGFR and erbB2, we evaluated the effect of AG1478 on the protein levels of wtEGFR and erbB2. We demonstrated that adding this inhibitor increased wtEGFR and erbB2 protein levels in vIIIA1 cells, although not to the same levels as parental and control, untreated cells (Figure 3C), while removing it reduced their levels. Adding AG1478 for 7 days to the OVCA 433 parental cells had no effect on the levels of wtEGFR and erbB2 (data not shown). Thus, modulating the activity of EGFRvIII affected wtEGFR and erbB2 protein levels and redistributed them to the cell surface, which confirmed that this regulation is due to the catalytic activity of EGFRvIII.
Acute Activation of EGFRvIII Led to Activation and Internalization of wtEGFR and erbB2
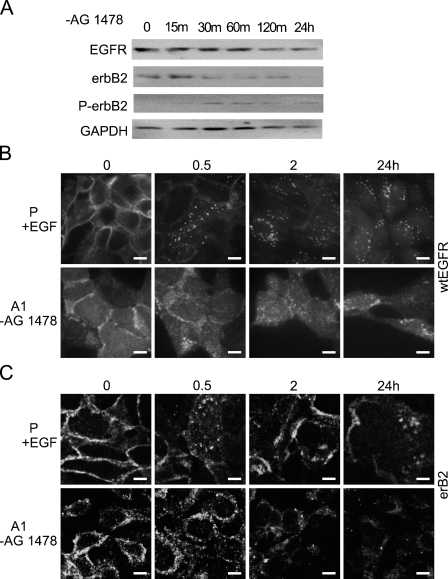
Examination of cellular localization of wtEGFR and erbB2 revealed that while EGFRvIII was detected on the cell surface of vIIIA1 cells, both wtEGFR and erbB2 were localized mainly intracellularly. Since we found that the steady-state catalytic activity of EGFRvIII caused downregulation of wtEGFR and erbB2, we wanted to determine whether restoring the activity of EGFRvIII after inhibiting its catalytic activity with AG1478 had a similar effect. For that reason, we incubated cells with the EGFR/EGFRvIII inhibitor AG1478 for 7 days and then removed the cells from the inhibitor and examined the protein levels and the cellular localization of wtEGFR and erbB2 at specific time points (Figure 4). We found that acute activation of EGFRvIII by withdrawal of its inhibitor caused downregulation of wtEGFR, as evident by immunoblot analysis (Figure 4A), and redistribution to the cytoplasm in a time-dependent manner, as detected by confocal microscopy, as early as 30 min after EGFRvIII activation (Figure 4B). This redistribution resembled activation of wtEGFR by its ligand EGF in control cells and reached its peak in vIIIA1 cells within 2 hr (Figure 4B). We previously reported the phosphorylation status of wtEGFR (Zeineldin et al. 2006), where we found that upon treating vIIIA1 cells with AG1478, tyrosine phosphorylation of both EGFRvIII and wild-type EGFR was inhibited and that ERK phosphorylation was reduced, while withdrawal of the inhibitor from the growth medium restored phosphorylation of EGFR, EGFRvIII, and ERK.
Figure 4.
Examination of protein levels and cellular localization of wtEGFR and erbB2 as a result of acute activation of EGFRvIII. vIIIA1 (A1) cells were treated with (+)AG1478 for 7 days, and then EGFRvIII was acutely activated by removing (−)AG1478 for the indicated times. (A) Immunoblot analysis of wtEGFR, erbB2, and phospho-erbB2 (P-erbB2) in cells. GAPDH was detected as an internal loading control. Confocal images show localization of (B) wtEGFR and (C) erbB2 in EGFRvIII-expressing cells after acute activation of EGFRvIII for the indicated times, compared with activation of wtEGFR in the parental (P) cell line for the indicated times. P, OVCA 433 parental cells; A1, vIIIA1 cells; m, min; h, hr. Bar = 10 μm.
Examination of erbB2 also showed similar results (Figures 4A and 4C), where activation of EGFRvIII led to downregulation of erbB2, with increased phosphorylation (Figure 4A) and accompanying intracellular redistribution, in a manner resembling our findings for wtEGFR (Figure 4C). Phosphorylated erbB2 was detected in cell lysates of EGFRvIII-expressing cells, though at a lower level than that of stimulated parental and control cells (data not shown), which may reflect downregulation of erbB2. Removal of vIIIA1 cells from AG1478 restored the phosphorylation of erbB2 (Figure 4A). In addition, the phospho-erbB2-to-total erbB2 ratio, as detected by Western blot analysis, was higher in untreated vIIIA1 cells than in AG1478-treated cells [ratio of 1.42 vs. 0.10, respectively (data not shown)]. On the other hand, treatment of the OVCA 433 parental cell line with AG1478 for 7 days followed by removal from the inhibitor had no effect on the levels of phospho-EGFR or phospho-erbB2 (data not shown). Our findings demonstrated that acute activation of EGFRVIII (by removal of its inhibitor) promoted the phosphorylation of wtEGFR and erbB2 and caused their downregulation.
Acute EGFRvIII Activation Caused Differential Trafficking of wtEGFR and erbB2
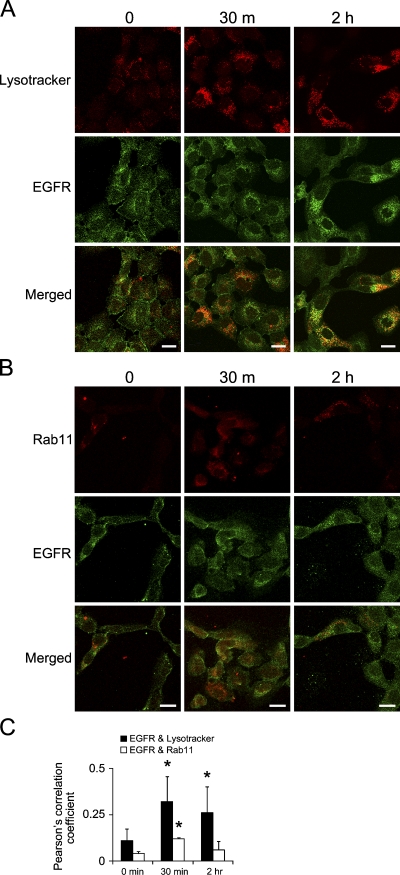
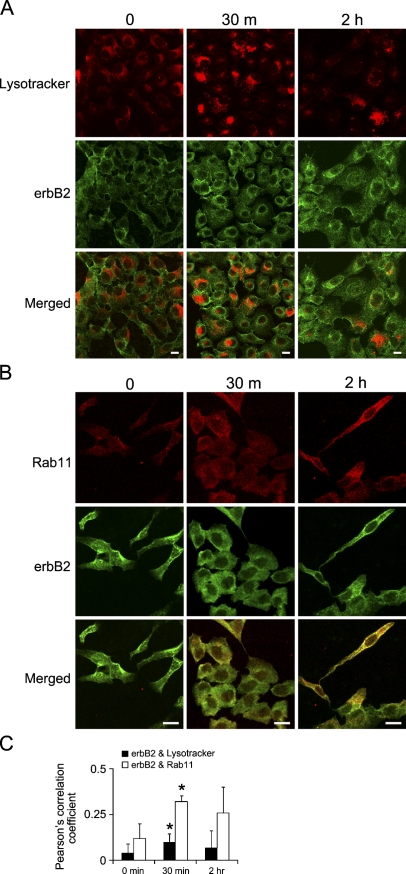
Since acute activation of EGFRvIII caused internalization of wtEGFR and erbB2 receptors, we investigated which compartments these receptors were directed to, a lysosomal or a recycling compartment. For this experiment, we used fluorescence confocal microscopy to evaluate colocalization of wtEGFR or erbB2, with lysosome markers (Lysotracker) or recycling endosomes (Rab11) after acutely activating EGFRvIII by removing the cells from AG1478. Examination of wtEGFR colocalization with lysosomes using fluorescein-labeled Lysotracker revealed that acute activation of EGFRvIII caused degradation of wtEGFR as we detected colocalized staining of lysosomes and wtEGFR as early as 30 min after EGFRvIII activation (Figure 5A). In addition, staining of the recycling endosome marker Rab11 demonstrated colocalization of wtEGFR and Rab11 (Figure 5B). Quantification of wtEGFR colocalization with Lysotracker and Rab11 was evaluated by calculating Pearson's correlation coefficient, as described in Materials and Methods and shown in Figure 5C, indicating significantly increased colocalization of wtEGFR with Lysotracker at 30 min after EGFRvIII activation (Pearson's correlation = 0.32) and also a slight but statistically significant increase of colocalization with Rab11 at 30 min (Pearson's correlation = 0.12). This showed that acute EGFRvIII activation caused internalization of wtEGFR, which was destined mainly for degradation and partially for recycling. In contrast, examination of the fate of erbB2 receptor by immunofluorescence colocalization revealed significantly increased colocalization with the recycling endosome marker Rab11 as early as 30 min after EGFRvIII activation (Pearson's correlation coefficient = 0.32) and a slight but statistically significant increase in colocalization with the degradation marker Lysotracker at 30 min (Pearson's correlation = 0.10), as shown in Figures 6A–6C. This indicated that acute EGFRvIII activation caused internalization of erbB2, which was destined mainly for recycling to the cell surface, while a smaller fraction was degraded.
Figure 5.
Trafficking of wtEGFR upon acute activation of EGFRvIII. vIIIA1 cells were treated with AG1478 for 7 days, then EGFRvIII was acutely activated by removing AG1478 for the indicated times. Confocal microcopy images show colocalization of wtEGFR with (A) Lysotracker, a lysosomal marker, and (B) Rab11, a recycling endosome marker. Bar = 20 μm. (C) Quantification of colocalization by Pearson's correlation coefficients. Bars = mean ± SD, n=3 to 8 (n refers to number of images, each with 6–26 cells). Asterisk indicates statistically significant difference comparing the Pearson's correlation coefficient at different time points to time 0 min.
Figure 6.
Trafficking of erbB2 upon acute activation of EGFRvIII. vIIIA1 cells were treated with AG1478 for 7 days, then EGFRvIII was acutely activated by removing AG1478 for the indicated times. Confocal microcopy imaging for detecting colocalization of erbB2 with (A) Lysotracker, a lysosomal marker, and (B) Rab11, a recycling endosome marker. Scale bar = 20 μm. (C) Quantification of colocalization by Pearson's correlation coefficients. Bars represent mean ± SD, n=4 to 8 (n refers to number of images, each with cells ranging from 6 to 26). Asterisk indicates statistically significant differences between the Pearson's correlation coefficient at different time points to time 0 min.
Discussion
EGFRvIII is a constitutively active receptor even in the absence of an activating ligand and is expressed in several tumors. Previous studies demonstrated interactions between the constitutively active EGFRvIII and wtEGFR (Luwor et al. 2004; Ramnarain et al. 2006; Zeineldin et al. 2006; Johns et al. 2007) and between EGFRvIII and erbB2 (O'Rourke et al. 1998). In this study, we demonstrated that expression of EGFRvIII in cancer cells results in activation and differential trafficking of wtEGFR and erbB2. The steady-state activity of EGFRvIII modulated three of four erbB family members via different mechanisms; erbB3 was decreased at the message level, while EGFR and erbB2 were reduced at the protein level (Figure 1). We focused on studying wtEGFR and erbB2, as our aim was to evaluate the effect of the constitutive activity of EGFRvIII on the activation and endocytosis of other erbB family members. The fact that erbB3 was suppressed as a result of EGFRvIII expression may affect dimerization of other erbB receptors, as there are reports demonstrating that inhibition or suppression of one member of the erbB family determines the erbB heterodimerization partners. For example, inhibition of EGFR promoted heterodimerization between erbB2 and erbB3 (Maegawa et al. 2007). Moreover, knockdown of erbB2 enhanced heterodimerization between EGFR and erbB3 (Hu et al. 2005). Based on this finding, in our study, the suppression of erbB3 as a result of EGFRvIII expression may contribute to enhancing interactions of EGFRvIII with wtEGFR and erbB2.
The steady-state activity of EGFRvIII caused reduction of protein levels and increased phosphorylation of wtEGFR (Zeineldin et al. 2006) and erbB2 (data not shown). In addition, acute activation of EGFRvIII by removal of a catalytic inhibitor led to decreased protein levels (Figure 4A) and increased phosphorylation of wtEGFR (Zeineldin et al. 2006) and erbB2 (Figure 4A). This reaction suggested the existence of a functional interaction between EGFRvIII and wtEGFR or erbB2. Although EGFRvIII lacks extracellular domains I and II and thus cannot assume an inactive tethered conformation (Hynes and Lane 2005), extracellular domains III and IV were reported to be sufficient to form dimers with erbB2 ectodomain (O'Rourke et al. 1998); thus, the same may apply to interactions between EGFRvIII and wtEGFR. In addition to extracellular domain interactions, the kinase domains or even transmembrane domains of dimers of EGFRvIII with wtEGFR or erbB2 may associate with each other. The kinase domain association was anticipated based on an analysis of potential interactions between wtEGFR and erbB2, using models of both molecules by molecular simulations and energy minimization (Murali et al. 1996). A recent report that found that the transmembrane sequences are important contributors to heterodimer formation such as wtEGFR–erbB2 heterodimers indicates that a similar contribution may exist for EGFRvIII–erbB2 and EGFRvIII–wtEGFR heterodimers (Samna Soumana et al. 2008). Recently, a report of enhanced heterodimerization of erbB2 with another EGFR mutant was implicated in defective downregulation of the EGFR mutant upon EGF stimulation (Shtiegman et al. 2007). Based on this finding, it is possible that heterodimerization of erbB2 with EGFRvIII contributes to defective downregulation of EGFRvIII. Further studies are needed to evaluate dimerization domains in the case of EGFRvIII and the kinetics of dimerization and internalization, in addition to evaluating the consequences of heterodimerization.
Evidence of EGFRvIII's ability to cross-activate other receptors is the fact that expression of EGFRvIII causes phosphorylation of c-Met receptor, while inhibition of EGFRvIII's catalytic activity by AG1478 reduces c-Met phosphorylation, indicating that this cross-activation is EGFRvIII dependent (Huang et al. 2007b). In our study, we found that EGFRvIII activated both wtEGFR and erbB2. Examining cellular localization demonstrated that while EGFRvIII was detected mainly at the cell surface, the wtEGFR and erbB2 proteins were detected intracellularly (Figure 2). The extracellular localization of EGFRvIII is expected, as this receptor suffers from impaired ubiquitination and internalization (Huang et al. 1997; Schmidt et al. 2003; Han et al. 2006; Grandal et al. 2007). We found no intracellular colocalization of wtEGFR with EGFRvIII and only minor colocalization of erbB2 with EGFRvIII, which may be the result of our detection of erbB2 in recycling endosomes, in which the small fraction of EGFRvIII has been reported to localize intracellularly (Grandal et al. 2007). Although EGFRvIII expression causes downregulation and internalization of both wtEGFR and erbB2, inhibition of EGFRvIII catalytic activity increased the levels of wtEGFR and erbB2 protein and increased their cell surface localization (Figure 3). Acute activation of EGFRvIII by removing the cells from AG1478 resulted in phosphorylation and downregulation of wtEGFR and erbB2 and their internalization (Figure 4). Examination of the fate of wtEGFR and erbB2 upon acute activation of EGFRvIII revealed their differential trafficking. wtEGFR was targeted mainly for degradation in lysosomes and partially to recycling to the cell surface (Figure 5), whereas erbB2 was targeted mainly for recycling to the cell surface and only partially for recycling endosomes (Figure 6). Our results show that the constitutive activity of EGFRvIII led to changes in steady-state levels and cellular localization of wtEGFR and erbB2. EGFRvIII itself is hypophosphorylated at the Y1045 residue, causing reduction of interaction with c-Cbl, impairing the following ubiquitination and degradation (Han et al. 2006). On the other hand, another tyrosine residue, Y1173, on EGFRvIII becomes phosphorylated to the same level as ligand-stimulated wtEGFR (Han et al. 2006). We report here that EGFRvIII activation was sufficient to trigger internalization and trafficking of both wtEGFR and erbB2, indicating that it is capable of phosphorylating residues on other erbB members that are involved in their internalization. The fact that EGFRvIII remains detected at the cell surface while both wtEGFR and erbB2 are phosphorylated and internalized into different endosomes indicates that the interactions between EGFRvIII and wtEGFR or erbB2 do not continue intracellularly.
Studies of the fate of ligand-activated wtEGFR indicate that it is downregulated and is predominantly destined for lysosomal degradation and partially for recycling to the cell surface (Sorkin and Goh 2008). In contrast, erbB2 is predominantly recycled to the cell surface, and it does not become downregulated through endocytosis as a result of its internalization-impaired carboxyl terminus (Sorkin et al. 1993; Baulida et al. 1996; Wang et al. 1999; Hendriks et al. 2003; Sorkin and Goh 2008). Our findings for wtEGFR activated by EGFRvIII show that wtEGFR was also directed to lysosomes and partly to recycling endosomes, whereas erbB2 was mainly recycled and partially degraded. Our finding indicate that erbB2 is degraded to some extent, as confirmed by colocalization with lysosomes by using confocal microscopy and by lower protein levels as detected by immunoblot analysis. Thus, EGFRvIII's constitutive activity may be distinct from EGF stimulation of wtEGFR, as it caused downregulation not only of wtEGFR but also of erbB2.
In conclusion, this study demonstrated that the steady-state activity of EGFRvIII affects erbB family members through different mechanisms. Although interactions between EGFRvIII and wtEGFR or erbB2 have been previously reported through studies of their phosphorylation or coimmunoprecipitation, this is the first report that demonstrates that low-level phosphorylation of EGFRvIII was sufficient to trigger internalization and trafficking of its dimerization partners.
Acknowledgments
This study was supported by National Institutes of Health Grants R01 CA-90492 and R01 CA-109545. Support was also provided by University of New Mexico Cancer Research and Treatment Center grant P30 CA118100.
Images in this article were generated by the University of New Mexico Cancer Center Fluorescence Microscopy Facility (supported as detailed on the webpage http://hsc.unm.edu/crtc/microscopy/Facility.html).
This article is distributed under the terms of a License to Publish Agreement (http://www.jhc.org/misc/ltopub.shtml). JHC deposits all of its published articles into the U.S. National Institutes of Health (http://www.nih.gov/) and PubMed Central (http://www.pubmedcentral.nih.gov/) repositories for public release twelve months after publication.
References
- Baulida J, Kraus MH, Alimandi M, Di Fiore PP, Carpenter G (1996) All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem 271:5251–5257 [DOI] [PubMed] [Google Scholar]
- de Graeff P, Crijns AP, Ten Hoor KA, Klip HG, Hollema H, Oien K, Bartlett JM, et al. (2008) The ErbB signalling pathway: protein expression and prognostic value in epithelial ovarian cancer. Br J Cancer 99:341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM (2008) Structure-based view of epidermal growth factor receptor regulation. Annu Rev Biophys 37:353–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes H, Cohen S, Bishayee S (2001) Glycosylation-induced conformational modification positively regulates receptor-receptor association: a study with an aberrant epidermal growth factor receptor (EGFRvIII/DeltaEGFR) expressed in cancer cells. J Biol Chem 276:5375–5383 [DOI] [PubMed] [Google Scholar]
- Grandal MV, Zandi R, Pedersen MW, Willumsen BM, van Deurs B, Poulsen HS (2007) EGFRvIII escapes downregulation due to impaired internalization and sorting to lysosomes. Carcinogenesis 28:1408–1417 [DOI] [PubMed] [Google Scholar]
- Han W, Zhang T, Yu H, Foulke JG, Tang CK (2006) Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer Biol Ther 5:1361–1368 [DOI] [PubMed] [Google Scholar]
- Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D (2003) Coregulation of epidermal growth factor receptor/human epidermal growth factor receptor 2 (HER2) levels and locations: quantitative analysis of HER2 overexpression effects. Cancer Res 63:1130–1137 [PubMed] [Google Scholar]
- Hu YP, Venkateswarlu S, Sergina N, Howell G, St Clair P, Humphrey LE, Li W, et al. (2005) Reorganization of ErbB family and cell survival signaling after knock-down of ErbB2 in colon cancer cells. J Biol Chem 280:27383–27392 [DOI] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, et al. (1997) The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem 272:2927–2935 [DOI] [PubMed] [Google Scholar]
- Huang PH, Cavenee WK, Furnari FB, White FM (2007a) Uncovering therapeutic targets for glioblastoma: a systems biology approach. Cell Cycle 6:2750–2754 [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, et al. (2007b) Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA 104:12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Lane HA (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 5:341–354 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- Johns TG, Perera RM, Vernes SC, Vitali AA, Cao DX, Cavenee WK, Scott AM, et al. (2007) The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res 13:1911–1925 [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sato JD, Le A, Polikoff J, Sato GH, Mendelsohn J (1983) Growth stimulation of A431 cells by epidermal growth factor: identification of high-affinity receptors for epidermal growth factor by an anti-receptor monoclonal antibody. Proc Natl Acad Sci USA 80:1337–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlden JM, Gee JM, Seery LT, Farrow L, Gullick WJ, Ellis IO, Blamey RW, et al. (1998) c-erbB3 and c-erbB4 expression is a feature of the endocrine responsive phenotype in clinical breast cancer. Oncogene 17:1949–1957 [DOI] [PubMed] [Google Scholar]
- Kumar A, Petri ET, Halmos B, Boggon TJ (2008) Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J Clin Oncol 26:1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassus H, Sihto H, Leminen A, Joensuu H, Isola J, Nupponen NN, Butzow R (2006) Gene amplification, mutation, and protein expression of EGFR and mutations of ERBB2 in serous ovarian carcinoma. J Mol Med 84:671–681 [DOI] [PubMed] [Google Scholar]
- Levitzki A, Gazit A (1995) Tyrosine kinase inhibition: an approach to drug development. Science 267:1782–1788 [DOI] [PubMed] [Google Scholar]
- Lorimer IA (2002) Mutant epidermal growth factor receptors as targets for cancer therapy. Curr Cancer Drug Targets 2:91–102 [DOI] [PubMed] [Google Scholar]
- Luwor RB, Zhu HJ, Walker F, Vitali AA, Perera RM, Burgess AW, Scott AM, et al. (2004) The tumor-specific de2–7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene 23:6095–6104 [DOI] [PubMed] [Google Scholar]
- Maegawa M, Takeuchi K, Funakoshi E, Kawasaki K, Nishio K, Shimizu N, Ito F (2007) Growth stimulation of non-small cell lung cancer cell lines by antibody against epidermal growth factor receptor promoting formation of ErbB2/ErbB3 heterodimers. Mol Cancer Res 5:393–401 [DOI] [PubMed] [Google Scholar]
- Mishima K, Johns TG, Luwor RB, Scott AM, Stockert E, Jungbluth AA, Ji XD, et al. (2001) Growth suppression of intracranial xenografted glioblastomas overexpressing mutant epidermal growth factor receptors by systemic administration of monoclonal antibody (mAb) 806, a novel monoclonal antibody directed to the receptor. Cancer Res 61:5349–5354 [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, Biegel JA, et al. (1995) Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res 55:5536–5539 [PubMed] [Google Scholar]
- Moscatello DK, Montgomery RB, Sundareshan P, McDanel H, Wong MY, Wong AJ (1996) Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene 13:85–96 [PubMed] [Google Scholar]
- Murali R, Brennan PJ, Kieber-Emmons T, Greene MI (1996) Structural analysis of p185c-neu and epidermal growth factor receptor tyrosine kinases: oligomerization of kinase domains. Proc Natl Acad Sci USA 93:6252–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Zeineldin R, Liu Y, Rosenberg M, Stack MS, Hudson LG (2005) Downregulation of integrin alpha2 surface expression by mutant epidermal growth factor receptor (EGFRvIII) induces aberrant cell spreading and focal adhesion formation. Cancer Res 65:9280–9286 [DOI] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ (1994) A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA 91:7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke DM, Nute EJ, Davis JG, Wu C, Lee A, Murali R, Zhang HT, et al. (1998) Inhibition of a naturally occurring EGFR oncoprotein by the p185neu ectodomain: implications for subdomain contributions to receptor assembly. Oncogene 16:1197–1207 [DOI] [PubMed] [Google Scholar]
- Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, et al. (2006) Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res 66:867–874 [DOI] [PubMed] [Google Scholar]
- Riese DJ 2nd, Gallo RM, Settleman J (2007) Mutational activation of ErbB family receptor tyrosine kinases: insights into mechanisms of signal transduction and tumorigenesis. Bioessays 29:558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samna Soumana O, Garnier N, Genest M (2008) Insight into the recognition patterns of the ErbB receptor family transmembrane domains: heterodimerization models through molecular dynamics search. Eur Biophys J 37:851–864 [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Furnari FB, Cavenee WK, Bogler O (2003) Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc Natl Acad Sci USA 100:6505–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtiegman K, Kochupurakkal BS, Zwang Y, Pines G, Starr A, Vexler A, Citri A, et al. (2007) Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene 26:6968–6978 [DOI] [PubMed] [Google Scholar]
- Sonabend AM, Dana K, Lesniak MS (2007) Targeting epidermal growth factor receptor variant III: a novel strategy for the therapy of malignant glioma. Expert Rev Anticancer Ther 7:S45–50 [DOI] [PubMed] [Google Scholar]
- Sorkin A, Di Fiore PP, Carpenter G (1993) The carboxyl terminus of epidermal growth factor receptor/erbB-2 chimerae is internalization impaired. Oncogene 8:3021–3028 [PubMed] [Google Scholar]
- Sorkin A, Goh LK (2008) Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res 314:3093–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen KD, Waldstrom M, Olsen DA, Corydon T, Lorentzen KA, Knudsen HJ, Jeppesen U, et al. (2008) Mutant epidermal growth factor receptor in benign, borderline, and malignant ovarian tumors. Clin Cancer Res 14:3278–3282 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang L, Yeung TK, Chen X (1999) Endocytosis deficiency of epidermal growth factor (EGF) receptor-ErbB2 heterodimers in response to EGF stimulation. Mol Biol Cell 10:1621–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD (1997) Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res 57:4130–4140 [PubMed] [Google Scholar]
- Yarden Y (2001) The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer 37(suppl 4):S3–8 [DOI] [PubMed] [Google Scholar]
- Zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS (2007) Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal 19:2013–2023 [DOI] [PubMed] [Google Scholar]
- Zeineldin R, Rosenberg M, Ortega D, Buhr C, Chavez MG, Stack MS, Kusewitt DF, et al. (2006) Mesenchymal transformation in epithelial ovarian tumor cells expressing epidermal growth factor receptor variant III. Mol Carcinog 45:851–860 [DOI] [PubMed] [Google Scholar]