Abstract
The chemokine receptor CXCR4 and its cognate ligand CXCL12 are pivotal for establishing metastases from many tumor types. Thus, CXCR4 may offer a cell surface target for molecular imaging of metastases, assisting diagnosis, staging and therapeutic monitoring. Further, Noninvasive detection of CXCR4 status of a primary tumor may provide an index of the metastatic potential of the lesion. Here, we report the development and evaluation of a positron-emitting analog of the stem cell mobilizing agent plerixafor, [64Cu]AMD3100, to image this receptor in human tumor xenografts preselected for graded expression of CXCR4. This imaging method was also evaluated in a lung metastases derived from human MDA-MB-231 breast cancer cells. Ex vivo biodistribution studies, performed to validate the in vivo imaging data, confirmed the ability of [64Cu]AMD3100 to image CXCR4 expression. Our findings demonstrate the feasibility of noninvasively imaging CXCR4 by positron emission tomography (PET) using a clinically approved agent as a molecular scaffold.
Keywords: Tumor microenvironment, chemokine receptor, metastasis, molecular imaging, brain tumor, breast cancer
Introduction
The ability of cancer cells to metastasize to distant sites is one of the most lethal aspects of cancer. CXCR4, a member of the surface G-protein–coupled seven-span transmembrane receptor class, plays a critical role in the homing of cancer cells to distant sites (1, 2), by binding to its ligand CXCL12, which is highly expressed in several sites where metastastic lesions are commonly observed (1, 3).
Increased CXCR4 expression is associated with an aggressive phenotype. Metastases frequently exhibit increased CXCR4 receptor expression compared to the primary (4–6). Overexpression of CXCR4 in primary tumors is directly related to the degree of lymph node metastasis (7, 8). Similarly, elevated CXCR4 expression in estrogen and progestin receptor negative breast cancers and triple negative breast cancers is closely associated with lymph node metastasis (9, 10). Inhibition of CXCR4-CXCL12 signaling either by antibodies, peptide analogs, small molecules or by siRNA knockdown has been found to reduce metastatic burden in various orthotopic and metastatic models of breast cancer (2, 11–15).
The critical role of CXCR4 in metastasis makes it an important biomarker to identify primary tumors that more likely to metastasize (7, 16–18). Preclinical and clinical studies have detected high concentrations of CXCR4 receptors in the primary brain tumors compared to normal brain parenchyma (19, 20). The invading regions of glioblastomas and satellite tumors, which are the primary reason for recurrence, have been observed to express high levels of CXCR4 (21). Tissue microarray analyses of patient biopsies have shown that nuclear staining for CXCR4 increases with tumor grade (5) and that elevated CXCR4 expression levels are associated with poor survival in patients with breast cancer (8, 22, 23). CXCR4 expression has therefore been proposed as a prognostic factor in several cancers including brain, breast, colon, prostate, melanoma, and osteosarcoma and considered a therapeutic target because of its role in tumor development, growth and metastasis (24, 25).
CXCR4 is expressed on the surface of several cell types including those of the central nervous, gastrointestinal and immune systems (26). In the immune system it is expressed on peripheral blood lymphocytes, monocytes, neutrophils, pre–B cells, mast cells, and CD34+ hematopoietic progenitor cells (27), and mediates leukocyte homing (28) and bone marrow homeostasis (29). Because of these expression patterns, extended or chronic use of CXCR4 targeted therapeutics may result in unwanted toxicity. A CXCR4 based imaging agent can be used to identify patients likely to respond to CXCR4 based therapies, there by reducing unnecessary toxicity. Attempts have been made to image CXCR4 expression in cancer models using 111In-labeled peptides and 125I-labeled monoclonal antibodies with SPECT/CT (30, 31). Synthesis and evaluation of the PET imaging agent [64Cu]AMD3100 was recently described in normal mice (32) but studies in tumor models have not been reported to date.
To image CXCR4 expression in tumors we utilized the ability of the cyclam function of the prototype CXCR4 inhibitor AMD3100 to form strong complexes with Cu to develop [64Cu]AMD3100 as an imaging agent for positron emission tomography (PET). AMD3100, is a high-affinity, specific CXCR4 antagonist that inhibits binding of the natural chemokine ligand CXCL12 (33–35). Previous studies have demonstrated that Cu bound to the cyclam increases the affinity of AMD3100 to the CXCR4 receptor by 6-fold (36). Here we present a detailed in vivo evaluation of [64Cu]AMD3100 in tumor models. In proof of principle studies, we evaluated CXCR4 expression in subcutaneous brain tumor models stably expressing CXCR4. We demonstrated that CXCR4 expression imaging is feasible in orthotopic and metastatic models of breast cancer by PET. Ex vivo biodistribution analyses were performed to further validate the in vivo studies.
Materials and Methods
Cell lines
The human glioblastoma cell line U87, and breast carcinoma epithelial cell line MDA-MB-231 were gifts from Drs. John Laterra and Zaver Bhujwalla, respectively. DU4475, a breast carcinoma epithelial cell line was purchased from American Type Culture Collection (Rockville, MD) and has been cultured for less than 6 months in our laboratory. Both U87 and MDA-MB-231 cell lines were authenticated and purchased from ATCC but have not been authenticated in our laboratory. U87 and breast cancer cell lines were maintained in MEM and RPMI media, respectively supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 mg/mL of streptomycin. A U87 cell line stably transfected with human CD4 and CXCR4 (U87-stb-CXCR4) was obtained from the NIH AIDS Research and Reference Reagent Program (Dr. HongKui Deng and Dr. Dan R. Littman) (37) and maintained in DMEM supplemented with 15%FBS, 1μg/mL puromycin, 300 μg/mL G418, 100 U/mL of penicillin, and 100 mg/mL of streptomycin. All cell lines were maintained in a humidified incubator with 5% CO2. All cell culture reagents were purchased from Invitrogen.
Radiopharmaceutical preparation
AMD3100 was purchased from Sigma-Aldrich (St. Louis, MO) and [64Cu]CuCl2 was obtained from Nordion or the University of Wisconsin. [64Cu]AMD3100 was prepared according to literature methods for the corresponding unlabeled material (36). Briefly, to 370–740 MBq (10–20 mCi) of [64Cu]CuCl2 buffered with 100 mM sodium acetate (pH, 5.5), was added 200 μg of AMD3100 and heated at 60°C for 40 min. [64Cu]AMD3100 (Fig. 1A) was purified on a reverse phase high-performance liquid chromatography (HPLC) system (Varian) using a C-18 column (Luna; 5 μm, 250 × 10 mm). The mobile phase consisting 89% water with 0.1% trifluoroacetic acid and 11% methanol with 0.1% trifluoroacetic acid was used at a flow rate of 5 mL/min. Ultraviolet absorbance was monitored at 266 nm and the radioactivity was detected by a model 105S single-channel radiation detector (Bioscan). A radioactive peak from 20–22 min containing the major product was collected and concentrated. The final product was then formulated in phosphate-buffered saline, sterile filtered and used for in vitro and in vivo experiments
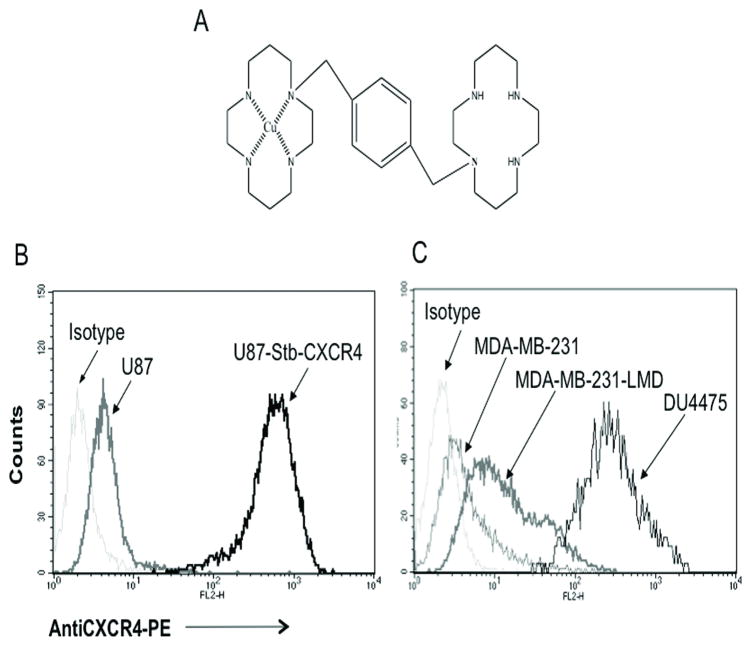
Figure 1. Characterization of CXCR4 expression.
A , structure of Cu-AMD3100; Flow cytometry evaluation of CXCR4 surface expression in brain (B) and breast cancer cell lines (C).
Receptor binding assays
U87, U87-stb-CXCR4, DU4475 and MDA-MB-231 cells seeded in 6 well plates at 60–80% confluence were used for receptor binding assays. Cells were incubated with 37 kBq (1μCi)/mL of [64Cu]AMD3100 at 4°C for 30 min in a binding buffer (PBS containing 5 mM MgCl2,1 mM CaCl2, 0.25% BSA, pH 7.4). After incubation, cells were washed quickly four times with 4°C binding buffer, detached using non-enzymatic buffer and cell associated radioactivity was determined in a gamma spectrometer (Pharmacia/LKB Nuclear, Inc.,). Radioactivity values were converted into percentage of incubated dose per million cells. Experiments were performed in triplicate and repeated three times.
Animal models
Procedures were conducted according to protocols approved by the Johns Hopkins Animal Care and Use Committee. Female NOD/SCID mice, six to eight weeks old, weighing between 20–25 g were purchased from the Johns Hopkins Immune Compromised Animal Core. Mice were implanted subcutaneously with U87 and U87-stb-CXCR4 brain tumor cells (3 × 106 cells/100 μL) in the left and right upper flanks, respectively. Brain tumor models were used when the tumor size was 400–500 mm3. For breast tumor imaging, CXCR4low MDA-MB-231 (3 × 106 cells/100 μL) and CXCR4high DU4475 (2 × 106 cells/100 μL) cells were inoculated in the left and right upper thoracic mammary fat pads, respectively. Tumors of 100–200 mm3 size were used for experiments to minimize necrosis. An experimental lung metastasis model was established by tail vein injection of 2×106 MDA-MB-231 cells in 200 μL of HBSS. NOD/SCID mice injected with HBSS alone were used as controls. Lung metastasis animals were used for experiments at 35 days after intravenous (IV) injection of tumor cells.
PET/CT imaging and analysis
An eXplore Vista small animal PET (GE, Piscataway, NJ) and X-SPECT small animal SPECT/CT system (Gamma Medica Ideas, Northridge, CA) were used for image acquisition. Prior to the injection of radiotracer mice were induced with 3% and maintained under 1.5% isoflurane. Dynamic imaging studies were performed in three mice harboring U87 and U87-stb-CXCR4 xenografts. After an intravenous infusion of [64Cu]AMD3100 (range: 8.2–13 MBq, mean: 10.5 MBq) a 60 min dynamic imaging sequence (14 frames: 2×30 s, 3×60 s, 3×120 s, 2×300s, 4×600 s) was acquired over the tumors followed by whole body imaging (2 bed positions, 15 minute emission/bed) starting at 90 min. To evaluate the specificity of the radiotracer, one mouse was subcutaneously injected with a blocking dose of 50 mg/kg of AMD3100 sixty minutes prior to [64Cu]AMD3100 injection and another mouse with [64Cu]CuCl2 alone. Whole body images on both mice were acquired at 90 min after injection. In case of breast orthotopic and lung metastasis models, two mice in each group underwent whole body imaging at 90 min after injection of [64Cu]AMD3100. Following a PET scan, CT imaging was acquired in 512 projections to allow anatomic co-registration. PET emission data were corrected for decay and dead time and reconstructed using the two dimensional ordered subsets-expectation maximization algorithm (2D OS-EM). Data evaluation was based on regions of interest (ROIs) drawn over transaxial slices of the PET images. ROIs were drawn over the whole tumor and the top 60% pixels were counted to minimize partial volume effects. The percentage of injected dose per gram (%ID/g) values were calculated based on a predetermined calibration factor using a known quantity of radioactivity. Time-activity values were averaged over three animals. Data was analyzed using AMIDE software (SourceForge) and volume rendered images were generated using Amira 5.2.0 software (Visage Imaging Inc., Carlsbad, CA).
Ex vivo biodistribution
NOD/SCID mice bearing either brain or breast tumors were injected IV with 740 kBq (20 μCi) of [64Cu]AMD3100 in 200 μL of saline. At 2, 10, 30, 60, 90, 120 and 240 min following injection, blood, lungs, liver, muscle, spleen, stomach, small intestine, kidney, and tumor samples were collected, weighed, and their radioactivity content was determined in an automated gamma spectrometer. For the breast tumor model bone and enlarged lymph nodes were also harvested. For the lung metastasis model only lungs, muscle and blood were collected. Aliquots of the injected radiotracer were counted along with the samples and served as standards for the calculation of the %ID/g.
Additionally, two separate groups of mice were used for binding specificity studies. One group received a blocking dose of 50 mg/kg AMD3100 subcutaneously 1 h prior to the injection of 740 kBq of [64Cu]AMD3100 and the second group received 740 kBq of [64Cu]CuCl2 alone. At 90 min after injection of the radiotracers ex vivo biodistribution was performed as described above. A minimum of four animals per time point were used for all biodistribution studies.
Cell extraction from lung metastases
Lungs from an additional group of mice were extracted and CXCR4 expression levels were analyzed by flow cytometry and immunoblot analysis. Briefly, lungs were cut into multiple small pieces and dissociated at 37°C for 30 min with 0.5 mg/mL of collagenase II in RPMI medium. The cell suspension was filtered through a cell strainer with 70 μm nylon mesh (Becton Dickinson Biosciences). The dissociation procedure was repeated three times to maximize cell yield. Cells collected by centrifugation were washed three times with RPMI fortified with 10% fetal bovine serum and cultured in RPMI medium as described above. The tumor extracted cells were seeded at 2 × 106 cells/mL in 100 mm plates and used for flow cytometry analysis after three or four passages.
Flow cytometry
Cells at 50–70% confluence were washed twice with PBS buffer (PBS, 2 mM EDTA, 0.5% FBS) and plates were placed on ice to prevent receptor internalization. Cells were detached using a nonenzymatic cocktail (Sigma). The expression levels of CXCR4 were determined by immunostaining with CXCR4 monoclonal antibody (Clone 12G5) conjugated to PE (R&D Systems) according to the manufacturer’s instructions. For receptor quantification, quantibrite beads (Becton Dickinson) were used. Cells were analyzed for CXCR4 expression on a FACSCan flow cytometer (Becton Dickinson). Ten thousand events were acquired in list mode and data were analyzed using Cellquest software (Becton Dickinson).
Immunohistochemistry
Whenever possible tumors from biodistribution experiments were used for histological examination. Sections of tumors were stained with hematoxylin and eosin, and immunohistochemistry was performed on tumor tissues using previously described rabbit polyclonal antibodies that recognize amino acids 328 to 338 of human CXCR4 (38) with minimum cross-reactivity to mouse CXCR4 (Imgenex) at a 1:100 dilution. Specific staining was visualized using biotin-SP-conjugated AffiniPure Goat Anti-Rabbit antibody (Jackson immunoresearch) followed by peroxidase-labeled streptavidin (LSAB+ System-HRP, DakoCytomation) and incubation with substrate-chromogen solution according to the manufacturer’s instructions.
Data analysis
Statistical analysis was performed using an unpaired two tailed t test. P-values < 0.05 for the comparison between CXCR4 high and CXCR4 low tumor uptake were considered to be statistically significant.
Results
Radiolabeling
The specific radioactivity of the [64Cu]AMD3100 after purification was typically 134±61 GBq/μmol (3.64±1.6 Ci/μmol), with radiochemical purity greater than 98% as determined by radio-HPLC (Suppl Fig. 1).
Receptor expression and in vitro radioligand binding
Flow cytometry analysis using 12G5 antibody revealed that U87-stb-CXCR4, U87, MDA-MB-231 and DU4475 cells to be > 95, 1–2, 10–18, and > 95 percent positive for CXCR4 expression, respectively (Fig. 1B and 1C). In accordance with the percentage of CXCR4 positive cells observed, [64Cu]AMD3100 showed specific binding in the order U87-stb-CXCR4>DU4475>MDA-MB-231>U87. Further receptor quantification analysis using quantibrite beads revealed that DU4475 and U87-stb-CXCR4 express 16,640 ± 5128 and 134,999 ± 20341 receptors/cell, respectively accounting for the differences in [64Cu]AMD3100 binding observed. A detailed analysis of receptor expression status and radioligand binding is presented in Table 1.
Table 1.
In vitro analysis of CXCR4 receptor expression and [64Cu]AMD3100 binding.
| Cell line | Receptors/Cell | % of incubated dose |
|---|---|---|
| U87-stb-CXCR4 | 134999±20341 | 13.1±1.6 |
| U87 | 3664±802 | 0.1±0.0 |
| DU4475 | 16640±5128 | 0.3±0.0 |
| MDA-MB-231 | 6833±1570 | 0.1±1.0 |
PET imaging
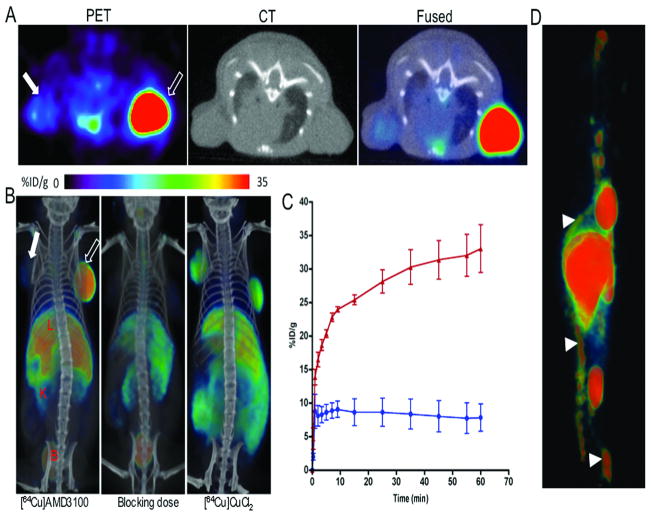
Delineation of CXCR4 specific tumor uptake and retention is evident in the subcutaneous, orthotopic and lung metastases models. The dynamic PET images acquired on the subcutaneous brain tumor models over 60 min showed a continuous accumulation of radioactivity in the U87-stb-CXCR4 tumor with %ID/g reaching 35 (Fig. 2A–C). Other than the U87-stb-CXCR4 tumor, whole body images at 90 min showed significant uptake in the liver, kidneys and bladder. Bone marrow uptake of [64Cu]AMD3100 was also evident (Fig. 2D). Blocking studies demonstrated greater than 80% reduction of radioactivity signal in both tumors. Relatively less radioactivity was also observed in liver and kidneys (Fig. 2B). To demonstrate the specificity of [64Cu]AMD3100 we also injected one mouse with [64Cu]CuCl2. The mouse receiving [64Cu]CuCl2 showed uniform distribution of radiotracer (Fig. 2B).
Figure 2. PET/CT imaging of CXCR4 expression in subcutaneous brain tumor xenografts with [64Cu]AMD3100.
NOD/SCID mice bearing U87 and U87-stb-CXCR4 glioblastoma xenografts on the left and right flanks, respectively, were given approximately 11.1 MBq (300 μCi) of 64Cu-labeled radiotracers via tail vein injection and PET/CT images were acquired. A, representative transaxial PET, CT and fused sections of both the tumors from a [64Cu]AMD3100 injected mouse at 90 min post-injection; B, volume rendered whole body images of [64Cu]AMD3100 (left panel), 50 mg/kg of AMD3100 blocking dose followed by [64Cu]AMD3100 (middle panel) and [64Cu]CuCl2 alone(right panel). All images were scaled to the same maximum threshold value. C, dynamic time-activity curves acquired over 60 min in mice injected with [64Cu]AMD3100. Data are means ± SEM of four animals. Specific accumulation of radioactivity in U87-stb-CXCR4 (red line) over U87 (blue line) is apparent. D, PET image of bone marrow uptake of [64Cu]AMD3100 at 90 min post-injection. Scale was adjusted for clear visualization of bone marrow. Solid arrow, U87 tumor; unfilled arrow, U87-stb-CXCR4 tumor; arrow head, bone marrow; L, liver; K, kidney; B, bladder.
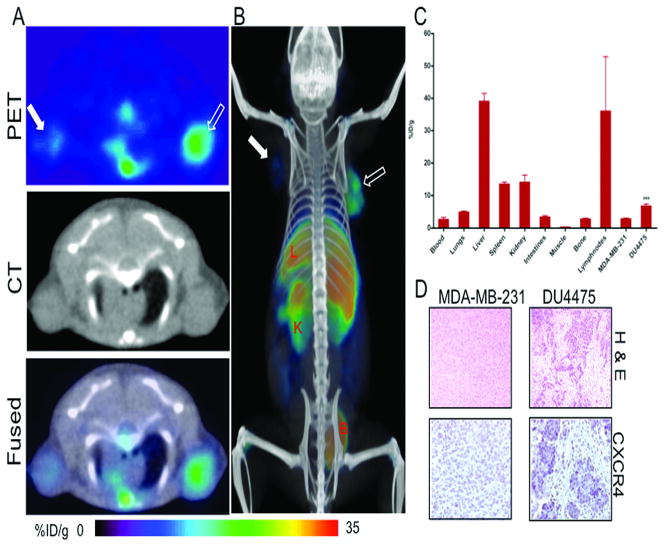
The 90 min whole body images in the breast tumor model demonstrated selective accumulation of activity in CXCR4high DU4475 tumors compared to CXCR4low MDA-MB-231 tumors (Fig. 3A-B). Accumulation of radioactivity was similarly observed in the lung metastasis model (Fig. 4A-B).
Figure 3. CXCR4 imaging in orthotopic breast tumor xenografts with [64Cu]AMD3100.
NOD/SCID mice harboring MDA-MB-231 and DU4475 orthotopic breast tumor xenografts in the upper thoracic mammary fat pad received ~11 MBq of [64Cu]AMD3100 via tail vein injection and whole body images were acquired at 90 min post-injection. A, transaxial PET, CT and fused sections of both the tumors; B, volume rendered whole body image showing clear accumulation of radioactivity in DU4475 tumor; C, biodistribution analysis of selected tissues from mice injected with 740 kBq of [64Cu]AMD3100 and sacrificed at 90 min post-injection. All radioactivity values were converted into percentage of injected dose per gram of tissue (%ID/g). Biodistribution data are means ± SEM of four to five animals; D, representative microscopy images of 10 μm-thick hematoxylin and eosin and CXCR4 stained sections obtained at ×10 magnification from both tumors. Significance is indicated by asterisks (*) and the comparative reference is MDA-MB-231 tumor uptake. ***P < 0.001. Solid arrow, MDA-MB-231 tumor; open arrow, DU4475 tumor; L, liver; K, kidney; B, bladder.
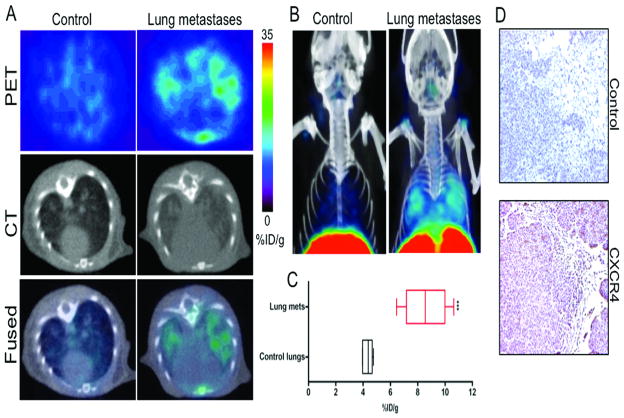
Figure 4. CXCR4 imaging of MDA-MB-231 derived lung metastases with [64Cu]AMD3100.
NOD/SCID mice that received either 2×106 MDA-MB-231 cells or HBSS were received ~11 MBq of [64Cu]AMD3100 at 35 days after inoculation. Whole body images were acquired at 90 min after injection. A, transaxial PET, CT and fused sections of lung metastasis and control mice; B, volume rendered whole body image showing clear accumulation of radioactivity in the lung metastases. Top slices of the volume rendered images were cut for clear visualization of lung uptake; C, box-and-whisker plot of the biodistribution analysis of lungs from mice injected with 740 kBq of [64Cu]AMD3100 at 90 min post-injection. All radioactivity values were converted into percentage of injected dose per gram of tissue (%ID/g) and are means ± SEM of four to five animals; D, representative microscopy images of 10 μm-thick CXCR4 and control antibody stained sections of the lung metastases obtained at ×10 magnification. Significance is indicated by asterisks (*) and the comparative reference is the lungs from mice injected with HBSS. ***P < 0.001.
Ex vivo biodistribution, Immunohistochemistry and Flow cytometry
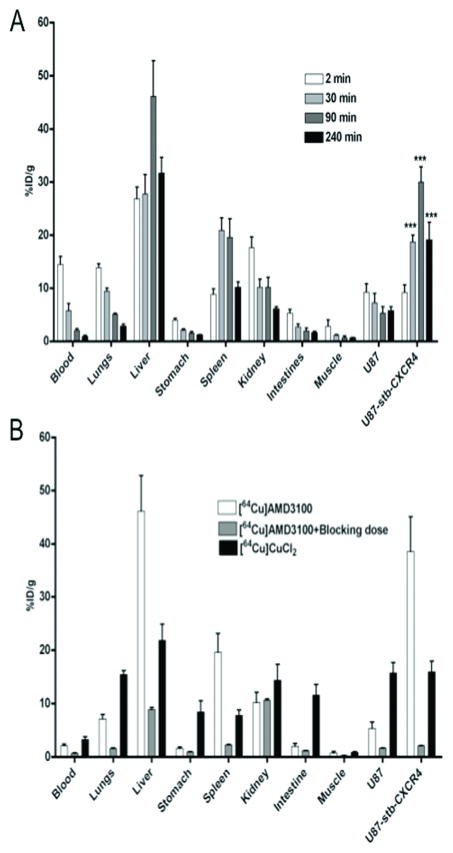
To quantify the degree of radiotracer uptake on a per organ basis, tissue and tumors were collected for up to 4 h after injection of [64Cu]AMD3100 from female NOD/SCID mice bearing subcutaneous brain tumors (Fig. 5A). The U87-stb-CXCR4 tumor showed consistently higher uptake relative to U87 tumors, except for the 2 min time point, in agreement with the imaging data (Fig. 5A). Both the imaging and biodistribution studies clearly show that 90 min after radiotracer injection is optimal for imaging these tumor models. The U87-stb-CXCR4 to U87 tumor ratios reached a maximum of 6.16 ± 1.37 at 90 min. Also, the tumor-to-muscle ratios reached a maximum of 47.36 ± 6.93 and 13.39 ± 4.20 for U87-stb-CXCR4 and U87, respectively. Similarly, the tumor-to-blood ratios were 16.93 ± 3.40 and 2.62 ± 0.52 for U87-stb-CXCR4 and U87, respectively. Liver was noted to have the highest accumulation of radioactivity at all times. Specificity was demonstrated in mice receiving either a 50 mg/kg blocking dose of AMD3100 followed by [64Cu]AMD3100 or [64Cu]CuCl2 alone. The administration of blocking dose resulted in > 90% reduction in radioactivity accumulation in U87-stb-CXCR4 tumors. There was also reduction of radioactivity accumulation in U87 tumors, blood and other tissues. In mice receiving blocking dose, the highest level of radioactivity was noted in kidneys, perhaps due to rapid clearance (Fig. 5B). Also, saturation of the plasma protein binding by unlabeled AMD3100 could have resulted in faster excretion. To investigate the role of Cu-transchelation, if any, we also injected another group of mice with [64Cu]CuCl2. While blocking dose studies clearly demonstrated the specificity of [64Cu]AMD3100, the radioactivity distribution in mice receiving [64Cu]CuCl2 was uniform, indicating that uptake observed could not be due to transchelated copper from the [64Cu]AMD3100.
Figure 5. Ex vivo biodistribution of [64Cu]AMD3100.
NOD/SCID mice bearing U87 and U87-stb-CXCR4 glioblastoma xenografts on the left and right flanks, respectively, were given approximately 750 kBq (20 μCi) of 64Cu-labeled radiotracers via tail vein and selected tissues were harvested at various time points. A. Biodistribution of [64Cu]AMD3100 at various time points; B, Comparison of various tissue uptake in [64Cu]AMD3100 injected mice with those of blocking dose or [64Cu]CuCl2 injected mice at 90 min post-injection. Significance is indicated by asterisks (*) and the comparative reference is U87 tumor uptake. ***P < 0.001.
Based on the above observations, biodistribution studies were carried out at 90 min post-injection in NOD/SCID mice harboring orthotopic MDA-MB-231 and DU4475 tumors and in MDA-MB-231 derived lung metastasis models. The tumor-to-muscle ratios were 26.24 ± 3.50 and 14.41 ± 0.73 for DU4475 and MDA-MB-231 tumors respectively. Similarly the tumor-to-blood ratios were 3.00 ± 0.39 and 1.93 ± 0.37 for the DU4475 and MDA-MB-231 tumors, respectively. There was a significant difference in the %ID/g, tumor-to-muscle and tumor-to-blood ratios between the tumor types (Fig. 3C). No significant differences were observed in other tissues compared to the brain tumor models. Representative H&E and CXCR4 stained sections showing the difference in CXCR4 expression is shown in Fig. 3D.
To evaluate if CXCR4 expression in metastases could be used for diagnostic purposes we tested the feasibility in an MDA-MB-231 derived experimental lung metastasis model. The %ID/g values clearly demonstrate a significant difference in radioactivity uptake between the lung metastasis and saline injected controls (Fig. 4C). The lung-to-muscle ratios at 90 min post-injection were 22.99 ± 2.50 and 14.32 ± 2.67 for lung metastasis and control animals, respectively. Representative CXCR4 stained section is shown in Fig. 4C. To validate the imaging and biodistribution results, we extracted lungs from a different group of mice and evaluated the CXCR4 expression levels in the lung metastases. Flow cytometry analysis demonstrated a 15–30% increase in the CXCR4 expression in the lung metastases derived cell lines (Fig. 1B and Suppl Fig. 2).
Discussion
Preclinical evaluation of [64Cu]AMD3100, a CXCR4 inhibitor known in unchelated form as Plerixafor, demonstrated the feasibility of PET imaging of CXCR4 expression in a glioblastoma cell line stably expressing CXCR4, and in breast orthotopic and lung metastasis models. PET imaging of CXCR4 expression using [64Cu]AMD3100 may provide a novel strategy for detecting tumors and metastases.
We initially characterized the kinetics and biodistribution of [64Cu]AMD3100 in subcutaneous brain tumor models stably expressing CXCR4. In a preclinical study, AMD3100 used at pharmacological doses caused growth inhibition of intracranial primary brain tumors, and resulted in a synergistic effect when combined with cytotoxic chemotherapy, in a CXCR4 expression dependent manner (39). Here, by comparing a glioblastoma cell line with high or low CXCR4 expression, we demonstrated that [64Cu]AMD3100 bound specifically to U87-stb-CXCR4 glioblastoma cells but not to the parental U87 cell line. The specificity in cells was validated by the in vivo uptake and retention in subcutaneous tumors derived from these cell lines. Dynamic PET imaging and biodistribution studies with [64Cu]AMD3100 exhibited a six-fold increase in tumor uptake in U87-stb-CXCR4 tumors compared to U87 tumors. This uptake was blocked by unlabeled AMD3100, demonstrating that the uptake was specific to the presence of CXCR4. Measurements of LogP for [64Cu]AMD3100 were found to be 0.52 ± 0.02 (Supp results) suggesting that [64Cu]AMD3100 may not be suitable for imaging CXCR4 beyond the blood-brain barrier.
We further validated our findings using established human breast cancer models and a model of experimental mestastasis. Most breast tumor tissue has some level of CXCR4 expression and more than 40% of breast tumors demonstrate elevated expression (5, 40). We imaged CXCR4 expression in two human breast cancer cell lines preselected for high (DU4475) and relatively low (MDA-MB-231) expression. Significantly higher [64Cu]AMD3100 binding was observed in DU4475 breast cancer cells compared to MDA-MB-231 cells. Both the imaging and ex vivo biodistribution data demonstrated CXCR4-specific uptake in the tumors. However, the uptake values in the DU4475 tumors were not as high as anticipated based on the receptor expression levels observed by flow cytometry. Histological analysis revealed considerable necrosis in those tumors even when small, accounting for the low uptake and retention observed. Bone marrow uptake was evident in the images, possibly due to CXCR4 expression on hematopoietic cells, although the collection of bone but not bone marrow alone may have reduced the total %ID/g values. Also, the enlarged lymph nodes exhibited increased accumulation of radioactivity indicating a potential role for [64Cu]AMD3100 PET imaging to identify lymph node metastasis. CXCR4 expression on immune cells and under inflammatory conditions may result in non-cancer specific uptake in lymph nodes, spleen and thymus. Similar non-cancer specific uptake can also be observed in tissues that have basal level of CXCR4 expression such as brain and pancreas (26), although elevated CXCR4 expression in tumors would result in increased uptake.
CXCR4 has been shown to play an important role in several steps of the metastatic cascade such as cancer cell migration, invasion and adhesion. Using gene expression profiling, Minn et al., demonstrated that in a subset of MDA-MB-231 cells with a propensity to metastasize to the lung, CXCR4 was one of the genes responsible for the metastatic phenotype (41). These data suggest that high levels of CXCR4 expression in cancer cells result in these cells homing to sites that are known to express high levels of the CXCR4 ligand, CXCL12.
One goal of our studies was to evaluate the use of [64Cu]AMD3100 to detect metastases. Both in vivo and ex vivo data clearly demonstrated detection of metastatic nodules in the lung, which were further confirmed by flow cytometry of single cell extracts from these metastases as well as immunohistochemical analysis. The increase in CXCR4 expression in metastases due to clonal selection of cancer cells that metastasize to the lungs has been previously reported (42) and was confirmed by our findings.
Although we demonstrated the ability to image CXCR4 expression in tumors and metastases using [64Cu]AMD3100, the liver and background tissue uptake observed should be discussed. Three major factors contribute to the accumulation of CXCR4 in normal tissue. One is the basal level of CXCR4 expression that produces a small background signal. The second is the moderate plasma protein binding observed with AMD3100. Nearly 58% of AMD3100 is plasma protein bound. Although that increases the background signal, the advantage is that it also enhances the probability of delivering the radiotracer to the tumor. Additionally, as observed in our results, the background slowly deceased with time while the tumor uptake increased until 90 min post-injection. This suggests that despite binding to plasma protein, [64Cu]AMD3100 was CXCR4 specific. The third is the instability of copper bound to the cyclam moiety and metabolism of [64Cu]AMD3100. Even though 64Cu-cyclam complexes are thermodynamically stable in vitro, several studies suggest that 64Cu may dissociate, and subsequently transchelate to superoxide dismutase, resulting in increased accumulation in the blood and liver (43–45). At 90 min, radioactivity in the blood of [64Cu]AMD3100 injected mice was 1.5-fold less compared to mice receiving [64Cu]CuCl2. In the case of transchelation, we would have detected similar blood radioactivity to that of [64Cu]CuCl2 injected mice, which was not observed. In addition, our metabolite studies using size exclusion columns demonstrated that 30–40% of radioactivity was protein bound at 90 min after injection (Suppl Fig. 3). Because of the plasma protein binding associated with AMD3100, it was difficult to further delineate the percentage of radioactivity bound to the cyclam moiety. However, the structurally similar bifunctional chelate CPTA (4-[(1,4,8,11-tetraazacyclotetradec-1-yl)methyl] benzoic acid) is routinely used for 64/67Cu chelation and blood analysis from radiolabled antibodies suggest that greater than 74% of activity is still associated with the antibody at 24 h post injection (46, 47). The increased liver uptake observed could also be attributed to the positive charge associated with the cyclam moiety. The positive charge on the cyclams has been identified as a cause of liver accumulation (48). In humans, AMD3100 elimination was primarily through kidneys, and nearly 70% of the drug was eliminated within 24 h as parent, indicating metabolic stability (49). Possible cyclam metabolites have been demonstrated to have low affinity to CXCR4 (34, 36) suggesting that uptake observed in our studies is due to intact [64Cu]AMD3100. Insights obtained from our studies and recent developments in bridged cyclam-based inhibitors may lead to the development of stable Cu-based CXCR4 imaging agents with optimized pharmacokinetics (50).
In summary, we have synthesized and evaluated a positron-emitting version of the prototype CXCR4 inhibitor AMD3100 in brain tumors stably expressing CXCR4. Our data with orthotopic breast xenografts and experimental metastases demonstrate that [64Cu]AMD3100 can be used to image graded levels of CXCR4 expression. The work presented here is a first step towards imaging a receptor directly involved in the metastatic process and provides a valuable strategy for noninvasive molecular characterization of tumors and metastases.
Supplementary Material
Acknowledgments
The authors would like to thank the University of Wisconsin team for Cu-64 production, Jianhua Yu, James Fox, Stephanie Davis, and Chris Endres, Ph.D. for assistance with imaging and image analysis, Ron Mease Ph.D., Catherine Foss Ph.D., and David L. Huso Ph.D. for helpful discussions and Lee Blosser and Ada Tam for assistance with flow cytometry. This work was partially supported by grants from the National Cancer Institute U24 CA92871 to M.G.P, NIH P50CA103175 (Z.M.B.) pilot grant (S.N.) and Michael Sholek translational research grant (S.N.) from American Brain tumor association.
References
- 1.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–7. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 2.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 3.Luker KE, Luker GD. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238:30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Oda Y, Yamamoto H, Tamiya S, et al. CXCR4 and VEGF expression in the primary site and the metastatic site of human osteosarcoma: analysis within a group of patients, all of whom developed lung metastasis. Mod Pathol. 2006;19:738–45. doi: 10.1038/modpathol.3800587. [DOI] [PubMed] [Google Scholar]
- 5.Salvucci O, Bouchard A, Baccarelli A, et al. The role of CXCR4 receptor expression in breast cancer: a large tissue microarray study. Breast Cancer Res Treat. 2006;97:275–83. doi: 10.1007/s10549-005-9121-8. [DOI] [PubMed] [Google Scholar]
- 6.Sun YX, Wang J, Shelburne CE, et al. Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem. 2003;89:462–73. doi: 10.1002/jcb.10522. [DOI] [PubMed] [Google Scholar]
- 7.Kato M, Kitayama J, Kazama S, Nagawa H. Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res. 2003;5:R144–50. doi: 10.1186/bcr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H, Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. The elevated level of CXCR4 is correlated with nodal metastasis of human breast cancer. Breast. 2005;14:360–7. doi: 10.1016/j.breast.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Woo S, Bae J, Kim C, Lee J, Koo B. A Significant Correlation between Nuclear CXCR4 Expression and Axillary Lymph Node Metastasis in Hormonal Receptor Negative Breast Cancer. Annals of Surgical Oncology. 2008;15:281–5. doi: 10.1245/s10434-007-9595-1. [DOI] [PubMed] [Google Scholar]
- 10.Chu QD, Panu L, Holm NT, Li BD, Johnson LW, Zhang S. High Chemokine Receptor CXCR4 Level in Triple Negative Breast Cancer Specimens Predicts Poor Clinical Outcome. J Surg Res. 2008 Oct 16; doi: 10.1016/j.jss.2008.09.020. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Liang Z, Wu T, Lou H, et al. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004;64:4302–8. doi: 10.1158/0008-5472.CAN-03-3958. [DOI] [PubMed] [Google Scholar]
- 12.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–71. [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MC, Luker KE, Garbow JR, et al. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–12. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 14.Tamamura H, Hori A, Kanzaki N, et al. T140 analogs as CXCR4 antagonists identified as anti-metastatic agents in the treatment of breast cancer. FEBS Lett. 2003;550:79–83. doi: 10.1016/s0014-5793(03)00824-x. [DOI] [PubMed] [Google Scholar]
- 15.Huang EH, Singh B, Cristofanilli M, et al. A CXCR4 antagonist CTCE-9908 inhibits primary tumor growth and metastasis of breast cancer. J Surg Res. 2009;155:231–6. doi: 10.1016/j.jss.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Cabioglu N, Sahin A, Doucet M, et al. Chemokine receptor CXCR4 expression in breast cancer as a potential predictive marker of isolated tumor cells in bone marrow. Clin Exp Metastasis. 2005;22:39–46. doi: 10.1007/s10585-005-3222-y. [DOI] [PubMed] [Google Scholar]
- 17.Cabioglu N, Yazici MS, Arun B, et al. CCR7 and CXCR4 as novel biomarkers predicting axillary lymph node metastasis in T1 breast cancer. Clin Cancer Res. 2005;11:5686–93. doi: 10.1158/1078-0432.CCR-05-0014. [DOI] [PubMed] [Google Scholar]
- 18.Kodama J, Hasengaowa, Kusumoto T, et al. Association of CXCR4 and CCR7 chemokine receptor expression and lymph node metastasis in human cervical cancer. Ann Oncol. 2007;18:70–6. doi: 10.1093/annonc/mdl342. [DOI] [PubMed] [Google Scholar]
- 19.Rempel SA, Dudas S, Ge S, Gutierrez JA. Identification and localization of the cytokine SDF1 and its receptor, CXC chemokine receptor 4, to regions of necrosis and angiogenesis in human glioblastoma. Clin Cancer Res. 2000;6:102–11. [PubMed] [Google Scholar]
- 20.Sehgal A, Keener C, Boynton AL, Warrick J, Murphy GP. CXCR-4, a chemokine receptor, is overexpressed in and required for proliferation of glioblastoma tumor cells. J Surg Oncol. 1998;69:99–104. doi: 10.1002/(sici)1096-9098(199810)69:2<99::aid-jso10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 21.Zagzag D, Esencay M, Mendez O, et al. Hypoxia- and vascular endothelial growth factor-induced stromal cell-derived factor-1alpha/CXCR4 expression in glioblastomas: one plausible explanation of Scherer’s structures. Am J Pathol. 2008;173:545–60. doi: 10.2353/ajpath.2008.071197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm NT, Byrnes K, Li BD, et al. Elevated Levels of Chemokine Receptor CXCR4 in HER-2 Negative Breast Cancer Specimens Predict Recurrence. J Surg Res. 2007;141:53–9. doi: 10.1016/j.jss.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Holm NT, Abreo F, Johnson LW, Li BD, Chu QD. Elevated chemokine receptor CXCR4 expression in primary tumors following neoadjuvant chemotherapy predicts poor outcomes for patients with locally advanced breast cancer (LABC) Breast Cancer Res Treat. 2009;113:293–9. doi: 10.1007/s10549-008-9921-8. [DOI] [PubMed] [Google Scholar]
- 24.Wong D, Korz W. Translating an Antagonist of Chemokine Receptor CXCR4: from bench to bedside. Clin Cancer Res. 2008;14:7975–80. doi: 10.1158/1078-0432.CCR-07-4846. [DOI] [PubMed] [Google Scholar]
- 25.Darash-Yahana M, Pikarsky E, Abramovitch R, et al. Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. Faseb J. 2004;18:1240–2. doi: 10.1096/fj.03-0935fje. [DOI] [PubMed] [Google Scholar]
- 26.Gupta SK, Pillarisetti K. Cutting edge: CXCR4-Lo: molecular cloning and functional expression of a novel human CXCR4 splice variant. J Immunol. 1999;163:2368–72. [PubMed] [Google Scholar]
- 27.Khan A, Greenman J, Archibald SJ. Small molecule CXCR4 chemokine receptor antagonists: developing drug candidates. Curr Med Chem. 2007;14:2257–77. doi: 10.2174/092986707781696618. [DOI] [PubMed] [Google Scholar]
- 28.Buckley CD, Amft N, Bradfield PF, et al. Persistent induction of the chemokine receptor CXCR4 by TGF-beta 1 on synovial T cells contributes to their accumulation within the rheumatoid synovium. J Immunol. 2000;165:3423–9. doi: 10.4049/jimmunol.165.6.3423. [DOI] [PubMed] [Google Scholar]
- 29.Ma Q, Jones D, Springer TA. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity. 1999;10:463–71. doi: 10.1016/s1074-7613(00)80046-1. [DOI] [PubMed] [Google Scholar]
- 30.Hanaoka H, Mukai T, Tamamura H, et al. Development of a 111In-labeled peptide derivative targeting a chemokine receptor, CXCR4, for imaging tumors. Nucl Med Biol. 2006;33:489–94. doi: 10.1016/j.nucmedbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Nimmagadda S, Pullambhatla M, Pomper MG. Immunoimaging of CXCR4 expression in brain tumor xenografts using SPECT/CT. J Nucl Med. 2009;50:1124–30. doi: 10.2967/jnumed.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson O, Weiss ID, Szajek L, Farber JM, Kiesewetter DO. 64Cu-AMD3100--a novel imaging agent for targeting chemokine receptor CXCR4. Bioorg Med Chem. 2009;17:1486–93. doi: 10.1016/j.bmc.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donzella GA, Schols D, Lin SW, et al. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–7. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 34.Gerlach LO, Skerlj RT, Bridger GJ, Schwartz TW. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J Biol Chem. 2001;276:14153–60. doi: 10.1074/jbc.M010429200. [DOI] [PubMed] [Google Scholar]
- 35.De Clercq E. The bicyclam AMD3100 story. Nat Rev Drug Discov. 2003;2:581–7. doi: 10.1038/nrd1134. [DOI] [PubMed] [Google Scholar]
- 36.Gerlach LO, Jakobsen JS, Jensen KP, et al. Metal ion enhanced binding of AMD3100 to Asp262 in the CXCR4 receptor. Biochemistry. 2003;42:710–7. doi: 10.1021/bi0264770. [DOI] [PubMed] [Google Scholar]
- 37.Bjorndal A, Deng H, Jansson M, et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–87. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zagzag D, Krishnamachary B, Yee H, et al. Stromal cell-derived factor-1alpha and CXCR4 expression in hemangioblastoma and clear cell-renal cell carcinoma: von Hippel-Lindau loss-of-function induces expression of a ligand and its receptor. Cancer Res. 2005;65:6178–88. doi: 10.1158/0008-5472.CAN-04-4406. [DOI] [PubMed] [Google Scholar]
- 39.Redjal N, Chan JA, Segal RA, Kung AL. CXCR4 inhibition synergizes with cytotoxic chemotherapy in gliomas. Clin Cancer Res. 2006;12:6765–71. doi: 10.1158/1078-0432.CCR-06-1372. [DOI] [PubMed] [Google Scholar]
- 40.Cabioglu N, Sahin AA, Morandi P, et al. Chemokine receptors in advanced breast cancer: differential expression in metastatic disease sites with diagnostic and therapeutic implications. Ann Oncol. 2009;20:1013–9. doi: 10.1093/annonc/mdn740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helbig G, Christopherson KW, 2nd, Bhat-Nakshatri P, et al. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278:21631–8. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 43.Smith-Jones PM, Fridrich R, Kaden TA, et al. Antibody labeling with copper-67 using the bifunctional macrocycle 4-[(1,4,8,11-tetraazacyclotetradec-1-yl)methyl]benzoic acid. Bioconjug Chem. 1991;2:415–21. doi: 10.1021/bc00012a006. [DOI] [PubMed] [Google Scholar]
- 44.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Copper chelation chemistry and its role in copper radiopharmaceuticals. Curr Pharm Des. 2007;13:3–16. doi: 10.2174/138161207779313768. [DOI] [PubMed] [Google Scholar]
- 45.Boswell CA, Sun X, Niu W, et al. Comparative in vivo stability of copper-64-labeled cross-bridged and conventional tetraazamacrocyclic complexes. J Med Chem. 2004;47:1465–74. doi: 10.1021/jm030383m. [DOI] [PubMed] [Google Scholar]
- 46.Novak-Hofer I, Zimmermann K, Maecke HR, Amstutz HP, Carrel F, Schubiger PA. Tumor uptake and metabolism of copper-67-labeled monoclonal antibody chCE7 in nude mice bearing neuroblastoma xenografts. J Nucl Med. 1997;38:536–44. [PubMed] [Google Scholar]
- 47.Rogers BE, Anderson CJ, Connett JM, et al. Comparison of four bifunctional chelates for radiolabeling monoclonal antibodies with copper radioisotopes: biodistribution and metabolism. Bioconjug Chem. 1996;7:511–22. doi: 10.1021/bc9600372. [DOI] [PubMed] [Google Scholar]
- 48.Jones-Wilson TM, Deal KA, Anderson CJ, et al. The in vivo behavior of copper-64-labeled azamacrocyclic complexes. Nucl Med Biol. 1998;25:523–30. doi: 10.1016/s0969-8051(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 49.Hendrix CW, Flexner C, MacFarland RT, et al. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob Agents Chemother. 2000;44:1667–73. doi: 10.1128/aac.44.6.1667-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan A, Nicholson G, Greenman J, et al. Binding optimization through coordination chemistry: CXCR4 chemokine receptor antagonists from ultrarigid metal complexes. J Am Chem Soc. 2009;131:3416–7. doi: 10.1021/ja807921k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.