SUMMARY
Dominant RUNX1 inhibition has been proposed as a common pathway for CBF-leukemia. CBFβ-SMMHC, a fusion protein in human acute myeloid leukemia (AML), dominantly inhibits RUNX1 largely through its RUNX1 high-affinity binding domain (HABD). However, the type I CBFβ-SMMHC fusion in AML patients lacks HABD. Here we report that the type I CBFβ-SMMHC protein binds RUNX1 inefficiently. Knock-in mice expressing CBFβ-SMMHC with a HABD deletion developed leukemia quickly, even though hematopoietic defects associated with Runx1-inhibition were partially rescued. A larger pool of leukemia initiating cells, increased MN1 expression, and retention of RUNX1 phosphorylation are potential mechanisms for accelerated leukemia development in these mice. Our data suggest that RUNX1 dominant inhibition may not be a critical step for leukemogenesis by CBFβ-SMMHC.
INTRODUCTION
A chromosome 16 inversion, inv(16)(p13q22), is associated with almost all cases of human acute myeloid leukemia (AML) subtype M4Eo (Liu et al., 1995). A chimeric gene CBFB–MYH11 is generated by this inversion, which encodes a fusion protein between CBFβ and smooth muscle myosin heavy chain (SMMHC) (Liu et al., 1993). In a knock-in mouse model CBFβ-SMMHC blocks embryonic definitive hematopoiesis as well as differentiation of adult hematopoietic stem cells (HSCs) to myeloid and lymphoid lineages (Castilla et al., 1996). This phenotype is similar to those in the Runx1−/− or Cbfb−/− mice (Okuda et al., 1996; Wang et al., 1996), suggesting that CBFβ-SMMHC is a dominant repressor of Runx1/CBFβ function. Furthermore, mutagenesis studies using Cbfb+/MYH11 knock-in mice indicated that CBFβ-SMMHC is necessary but not sufficient for leukemogenesis; additional genetic events are required (Castilla et al., 1999; Castilla et al., 2004).
Normally CBFβ binds to RUNX1 via its heterodimerization interface (Warren et al., 2000), which is retained in CBFβ-SMMHC. Moreover, the fusion between the coiled-coil rod region of SMMHC and CBFβ creates a novel binding site for RUNX1, called Runx1 high affinity binding domain (HABD) (Lukasik et al., 2002). As a result, CBFβ-SMMHC binds RUNX1 at two sites, which is associated with higher binding affinity. This high affinity RUNX1 binding has been proposed to explain the dominant nature of CBFβ-SMMHC function over wildtype CBFβ (Lukasik et al., 2002).
Clinically 10 different types of CBFβ-SMMHC fusion can be produced by the chromosome 16 inversion mainly due to the variation of inversion breakpoint locations in the MYH11 gene (van Dongen et al., 1999). Nine of the 10 fusion types contain the HABD. However, one fusion type, type I, which contains the shortest fragment of SMMHC among all 10 fusion types, does not contain the HABD. We decided to study this fusion type and a similar deletion construct to determine if HABD is required for leukemogenesis by CBFβ-SMMHC.
RESULTS
Type I CBFβ-SMMHC has highly reduced binding affinity for RUNX1
The type I CBFB-MYH11 fusion junction results in the deletion of amino acids 134 through 236 of the CBFβ-SMMHC protein (CBFβ-SMMHCd134-236) (Dissing et al., 1998; Van der Reijden et al., 2001). The deletion encompasses the entire HABD (aa 179 through 221) as well as the sequence encoded by Cbfb exon 5 (Figure 1A), which results in the deletion of a significant segment of the C-terminal helix in the CBFβ portion of the fusion protein (aa 134–138). A deletion in this helix will likely disrupt the fold of CBFβ, which is predicted to result in severe reduction in RUNX1 binding.
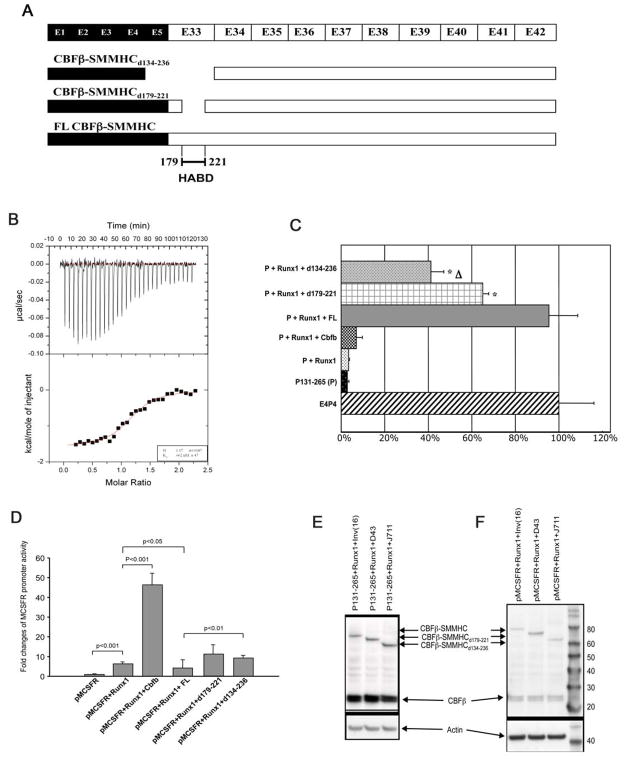
Figure 1. The type I CBFβ-SMMHC fusion variant is very inefficient in binding and repressing RUNX1.
(A) Diagrammatic representation of CBFβ-SMMHC fusion variants. CBFβ-SMMHCd134-236, generated by type I fusion, misses the HABD as well as CBFβ residues encoded by exon 5. CBFβ-SMMHCd179-221 misses only the HABD and has been described before (Lukasik et al., 2002).
(B) ITC measurement of interactions between 200 μM CBFβ-SMMHCd134-236 and 7.5 μl injections of 15 μM RUNX1 Runt domain. The top panel shows the raw data while the bottom panel is a plot of the binding corrected for the dilution enthalpy (average dilution enthalpy = 232 cal mol−1). Data were fit to a one-site binding model. The results of a fit to one titration are shown in the box at the lower right corner. The average Kd of two independent experiments is 709 (± 47) nM.
(C) CD4 reporter assay. E4P4: CD4 enhancer and promoter; P131-265: core sequence of CD4 repressor; FL: CBFβ-SMMHC; d179-221: CBFβ-SMMHCd179-221; d134-236: CBFβ-SMMHCd134-236. Δ: statistically significant difference (P<0.05) between the top two conditions, which are P131-265 + Runx1 with either d179-221 or d134-236. *: statistically significant differences (P<0.05) between the top two conditions and the third one, which is P131-265 + Runx1 + FL.
(D) MCSFR reporter assay. pMCSFR: the luciferase reporter driven by human MCSFR promoter.
(E and F) Western blot analysis showing the expression of the transfected constructs in the reporter assays (C and D, respectively). The expression of CBFβ and full-length and variant CBFβ-SMMHC constructs was detected with a mouse monoclonal antibody (β141.2) specific for CBFβ.
The error bars in (C) and (D) represent one standard deviation.
We used isothermal titration calorimetry (ITC) to measure the binding affinity of CBFβ-SMMHCd134-236 with RUNX1 (Figure 1B). Fitting the ITC measurements for binding of CBFβ-SMMHCd134-236 to the runt domain of RUNX1 yielded a 1:1 stoichiometry (N=1.17) and a Kd of ~709 (± 47) nM. The stoichiometry is in agreement with our previous measurements of a similar HABD-deletion construct, CBFβ-SMMHCd179-221, as well as measurements of the wild type CBFβ, but differs from that of full-length CBFβ-SMMHC, which is 2:1 (Lukasik et al., 2002). The binding affinity data suggests a significant loss of binding affinity, since the Kd for CBFβ-SMMHCd134-236 binding to RUNX1 is ~100 fold weaker than full length CBFβ-SMMHC (7 nM), ~20 fold weaker than CBFβ-SMMHCd179-221 (34 nM), and even ~13 fold weaker than wild type CBFβ (54 nM).
CBFβ-SMMHC without HABD is less efficient than full length CBFβ-SMMHC for repressing RUNX1
We previously reported that CBFβ-SMMHC represses RUNX1 function in CD4 silencing and restores CD4 promoter transcriptional activity in a reporter assay (Zhao et al., 2007). We tested CBFβ-SMMHCd179-221 and CBFβ-SMMHCd134-236 in this reporter assay (Figure 1C). As we described before, in the presence of full length CBFβ-SMMHC (FL), CD4-CAT activity was significantly restored when compared to those with CD4 silencer (p131-265) alone or in the presence of RUNX1. CBFβ-SMMHCd179-221 (d179-221) and CBFβ-SMMHCd134-236 (d134-236) could partially restore CD4-CAT activity but were much less efficient than full length CBFβ-SMMHC (FL) (p<0.05). CBFβ-SMMHCd134-236 was the weakest, which correlated with its inability to bind RUNX1 efficiently.
We also performed reporter assay in which the expression of luciferase was driven by the macrophage colony-stimulating factor (M-CSF) receptor promoter (Rhoades et al., 1996). As can be seen in Figure 1D, full-length CBFβ-SMMHC repressed this reporter activity, which was activated synergistically by RUNX1 and CBFβ. On the other hand, both CBFβ-SMMHCd179-221 and CBFβ-SMMHCd134-236 variants partially restored the reporter activity.
Figure 1E and F are representative western blot data showing the expression levels of the transfected constructs in these two reporter assays, respectively.
Generation of knock-in mice expressing HABD-truncated CBFβ-SMMHC
We generated mouse ES cell lines expressing truncated CBFβ-SMMHC that lacks only the HABD (CBFβ-SMMHCd179-221). The knock-in strategy is shown in Figure 2A, and is similar to what we reported previously (Castilla et al., 1996) with the exception that the selection marker gene, neo, is flanked by two lox-P sites. A knock-in construct for the full-length CBFβ-SMMHC with a floxed neo cassette was also made as a control. Targeted ES cell clones were identified by Southern blot hybridization (Figure 2B). The expression of the fusion proteins from each targeted ES cell line was confirmed by western blot (Figure 2C). We produced chimeric mice using two independent Cbfb-MYH11d179-221 knockin ES cell clones (#220, #269) and one full-length Cbfb-MYH11 knock-in ES cell clone (#10), which served as a positive control. The chimeras from all three lines had successful germ-line transmission. The chimeras and F1 mice from the two independent Cbfb-MYH11d179-221 knock-in clones had identical phenotypes.
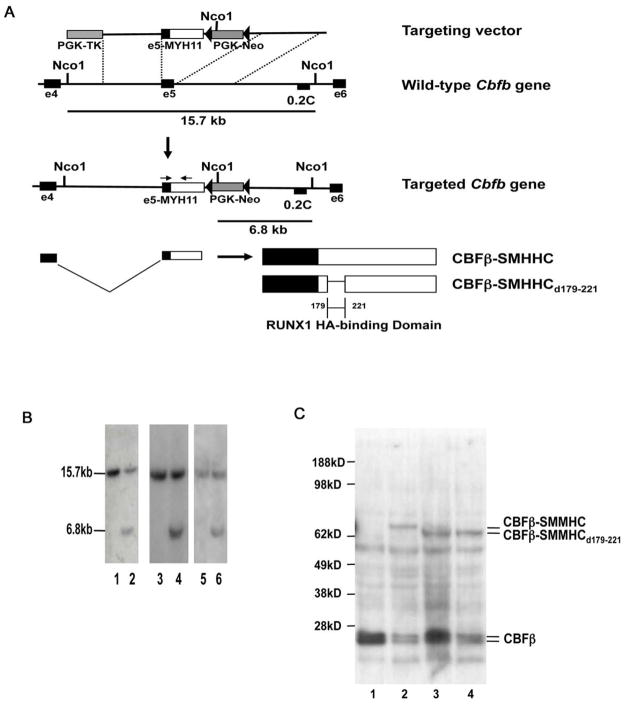
Figure 2. Generation of mouse ES cell lines with Cbfb-MYH11d179-221 and full length Cbfb-MYH11 knock-in constructs.
(A) Targeting strategy used to replace exon5 of Cbfb with the targeting constructs. Location of probe 0.2C and the sizes of NcoI fragments detected by 0.2C are indicated. The arrows indicate the locations of PCR primers used for genotyping. Filled triangles represent lox-P sites.
(B) Southern blot hybridization of NcoI-digested DNA from the parental ES cells (TC1; lanes 1, 3, and 5) and the targeted ES cell clones (lane 2: clone #10 for Cbfb-MYH11; lanes 4 and 6: clones #220 and #269 for Cbfb-MYH11d179-221) with probe 0.2C. The 15.7 kb band corresponds to the wild-type Cbfb allele and the 6.8 kb band corresponds to the knock-in allele.
(C) Western blot analysis of ES cells. Lane 1: the parental ES cell line TC-1; Lane 2: ES cell line #10 (knock-in of full-length Cbfb-MYH11), Lanes 3 and 4: ES cell lines #220 and #269 with knock-in of Cbfb-MYH11d179-221. The antibody used was the mouse monoclonal antibody (β141.2) specific for CBFβ. The calculated molecular weights for CBFβ-SMHHC, CBFβ-SMHHCd179-221, and CBFβ are 71 KD, 65 KD, and 22 KD, respectively.
Partial phenotypic rescue in embryos heterozygous for Cbfb-MYH11d179-221 knock-in
Two Cbfb+/MYH11d179-221 F1 mice were born alive, among 25 newborn mice observed (8%) (Figure 3A), suggesting decreased survival but significantly different from Cbfb+/MYH11 F1 mice, which never survived to birth (Castilla et al., 1996). Further studies showed that at E12.5, most Cbfb+/MYH11d179-221 embryos developed multiple hemorrhages in the central nervous system, as seen in Cbfb+/MYH11 embryos (Castilla et al., 1996). However, several Cbfb+/MYH11d179-221 embryos remained alive after E13.5 (Figure 3A), and one Cbfb+/MYH11d179-221 embryo had no hemorrhage at E16.5, while all Cbfb+/MYH11 embryos died at E13.5 (Castilla et al., 1996).
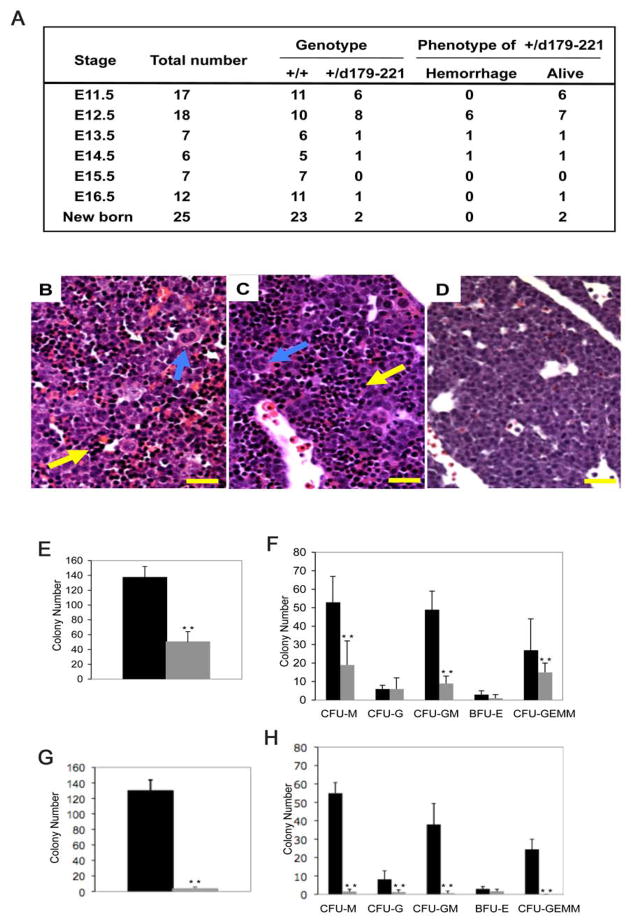
Figure 3. Partial phenotypic rescue in Cbfb+/MYH11d179-221 heterozygous embryos.
(A) Genotype and phenotype of embryos derived from crosses between Cbfb–MYH11 chimeras and normal females. +/+ are wild type littermates, +/d179-221 are embryos heterozygous for Cbfb-MYH11d179-221 knock-in. Viability was determined by embryo heart beats.
(B–D) Histologic sections of fetal livers from E12.5 embryos. (B) wildtype control, (C) Cbfb+/MYH11d179-221, (D) Cbfb+/MYH11. Sections were stained with H and E. Yellow arrows indicate hematopoietic cells in the livers. Blue arrows indicate megakaryocytes. Scale bars represent 100 μm.
(E–H) In vitro differentiation assay of fetal liver hematopoietic cells. Panels (E) and (F) compare colony numbers from embryos of wildtype (black bars) and Cbfb+/MYH11d179-221 (gray bars). Panels (G) and (H) compare colony numbers from embryos of wildtype (black bars) and Cbfb+/MYH11 (gray bars). Panels (E) and (G): total colony numbers. BFU-E: burst-forming unit-erythroid; CFU-E: colony-forming unit-erythroid; CFU-G: colony-forming unit-granulocyte; CFU-GM: colony-forming unit-granulocyte/macrophage; CFU-GEMM: colony-forming unit-granulocyte/erythroid/macrophage/megakaryocyte; CFU-M: colony-forming unit-macrophage. The error bars represent one standard deviation. **: P<0.001
The livers of the E12.5–13.5 Cbfb+/MYH11d179-221 embryos looked paler than those of the wildtype littermates but not as severe as in the Cbfb+/MYH11 embryos (data not shown). Histologically, hematopoietic progenitors were clearly visible in the liver sections of the Cbfb+/MYH11d179-221 embryos, in contrast to those in Cbfb+/MYH11 embryos, which contained no hematopoietic progenitors (Figure 3B–D). Megakaryocytes were observed in the liver of the Cbfb+/MYH11d179-221 embryos, even though in reduced numbers, contrary to Cbfb+/MYH11 fetal livers where there were no megakaryocytes (Figure 3B–D). In vitro colony-forming assay was then used to assess fetal liver hematopoiesis in the Cbfb+/MYH11d179-221 embryos. CFU-M, CFU-GM, CFU-GEMM, and total colony numbers were significantly reduced when compared to the wildtype littermates (P<0.001), while CFU-G and BFU-E numbers were not significantly changed (Figure 3E, F). On the other hand, as we reported before (Castilla et al., 1996), the number of colonies generated from Cbfb+/MYH11 embryos was about 0%–1% of the wild type (Figure 3G, H). Overall the data suggest that the hematopoietic defect was milder in the Cbfb+/MYH11d179-221 embryos than in Cbfb+/MYH11 embryos, and that the phenotype of hemorrhage and embryonic lethality was partially rescued by the HABD deletion.
Accelerated leukemogenesis in the Cbfb+/MYH11d179-221 mice
The partial phenotypic rescue in the Cbfb+/MYH11d179-221 embryos was consistent with reduced repression of RUNX1. We therefore predicted that the Cbfb+/MYH11d179-221 mice would have reduced frequency or prolonged latency in leukemogenesis. Unexpectedly, all Cbfb+/MYH11d179-221 chimeras (n = 22) died from leukemia by 19 weeks of age with no ENU treatment (Figure 4A), and the 2 surviving heterozygous F1 mice (Figure 3A) developed leukemia even faster, at 10 and 18 days after birth, respectively. As a comparison, Cbfb+/MYH11 chimeras (n = 17) did not develop leukemia spontaneously at similar ages (Figure 4A).
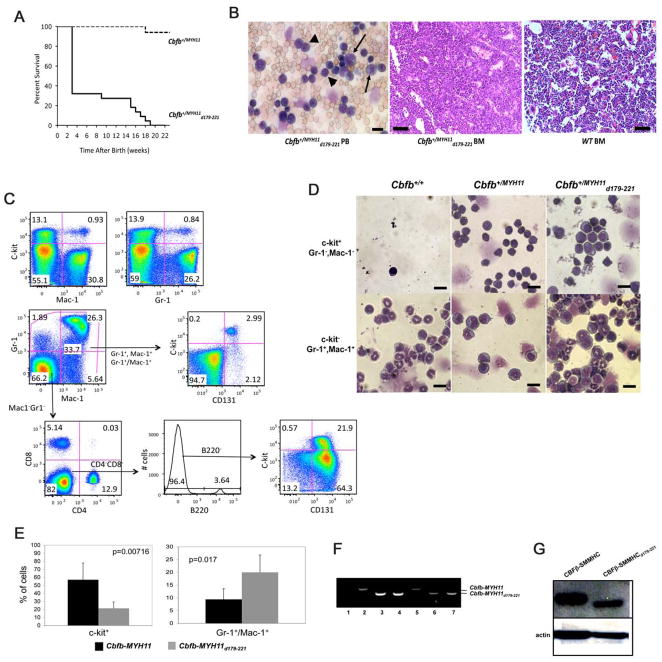
Figure 4. Acute myeloid leukemia development in Cbfb+/MYH11d179-221 and Cbfb+/MYH11 chimeras.
(A) Survival curves of Cbfb+/MYH11d179-221 and Cbfb+/MYH11 chimeras. Dotted line: the survival curve of Cbfb+/MYH11 chimeras (n=17); solid line: the survival curve of Cbfb+/MYH11d179-221 chimeras (n=22). The mice were not treated with any mutagens such as ENU.
(B) Morphology of leukemic cells in the Cbfb+/MYH11d179-221 mice. Left panel: wright-Giemsa stained peripheral blood smears showing poorly differentiated stem cell like cells (arrowheads) and myeloblasts (arrows). Middle and right panels: H&E stained bone marrow sections from mice of the indicated genotypes. Scale bar in the left panel represent 10 μm; those in the middle and right panels represent 50 μm.
(C) FACS analysis of peripheral blood cells in a Cbfb+/MYH11d179-221 mouse using antibodies against c-kit, Mac-1, GR-1, CD131, B220, CD4 and CD8.
(D) Correlation between cell morphology and surface marker expression for leukemic cells from the Cbfb+/MYH11d179-221 mice. FACS sorted c-kit+/GR1−/Mac1− and c-kit−/GR1+/Mac1+ populations from mice of the indicated genotypes were analyzed for morphologic features through Wright-Giemsa staining of cytospin preparations. Scale bars represent 10 μm.
(E) The percentages of leukemia cells expressing c-kit or Gr-1/Mac1 in the Cbfb+/MYH11 and Cbfb+/MYH11d179-221 mice (N = 5 for each genotype) as detected by FACS. The error bars represent one standard deviation.
(F and G) Expression of the knocked-in fusion genes in the leukemia cells. (F) RT-PCR with RNA samples from ES cells (lanes 1–4) and leukemic cells (lanes 5–7) with PCR primers flanking the Cbfb and MYH11 fusion junctions in Cbfb-MYH11 and Cbfb-MYH11d179-221. Lane 1: ES cell line TC1; Lane 2: ES cell clone #10 with knock-in of Cbfb-MYH11; Lanes 3 and 4: ES cell lines #220 and 269 with knock-in of Cbfb-MYH11d179-221; Lane 5: leukemia cells from a Cbfb+/MYH11 chimera; Lanes 6 and 7: leukemia cells from two Cbfb+/MYH11d179-221 chimeras. (G) Western blot analysis with protein samples from leukemic cells of the indicated genotype and the CBFβ-specific antibody (β141.2).
Histologically the leukemic cells in the Cbfb+/MYH11d179-221 chimeras were either poorly differentiated stem-like cells, or myeloblasts (Figure 4B). Correspondingly, by FACS, a significant population of the circulating leukemic cells was c-kit+, Mac-1−, and Gr-1− (stem- or progenitor-like cells), while another major cell population was c-kit−, Mac-1+, Gr-1+ (myeloblasts) (Figure 4C and D). Interestingly, almost all leukemic cells that were negative for Mac-1 and/or Gr-1 (either c-kit+ or c-kit−) were positive for CD131 (Csf2rb), which was also positive in almost all leukemic cells from mice expressing full-length Cbfb-MYH11 (Hyde et al., 2010). In addition, the myeloblast population (c-kit−, Mac-1+, GR-1+ ) in the Cbfb+/MYH11d179-221 chimeras was significantly larger than that in the Cbfb+/MYH11 chimeras (Figure 4E and data not shown). The leukemic cells mainly infiltrated bone marrow (Figure 4B) and spleen, as well as other organs such as liver and kidneys (Figure S1), similar to what was observed in the leukemic Cbfb+/MYH11 mice. PCR was performed to confirm the ES cell origin of the leukemic cells (Figure 4F). In addition, CBFβ-SMMHC protein products of expected sizes were detected in the leukemic cells (Figure 4G). Finally, sex-matched isogenic (C57BL/6 × 129/Sv-F1) recipients were transplanted with leukemic spleen cells (1 × 106 cells/mouse) from the Cbfb+/MYH11d179-221 chimeras. The transplanted mice developed the same type of AML as that of the donors 4 weeks after transplantation. Overall, the data demonstrated that deleting the HABD did not reduce the leukemogenic potential of Cbfb-MYH11. Instead, it led to acceleration of leukemogenesis in the Cbfb+/MYH11d179-221 knock-in mice.
Expression of HABD-deleted CBFβ-SMMHC in human CD34+ cells
Recently we established a human CD34+ culture system to characterize the effect of CBFβ-SMMHC on human hematopoietic stem and progenitor cells. We found that expressing full length CBFβ-SMMHC initially repressed progenitor activity in methylcellulose assays but eventually led to clonal expansion and the establishment of long-term cell lines (Wunderlich et al., 2006). To assess the effects of the HABD deletion, we introduced CBFβ-SMMHCd179-221 and CBFβ-SMMHCd134-226 into human CD34+ cells by retroviral transduction and analyzed the transduced cells as described previously (Wunderlich et al., 2006). Total colony numbers were significantly decreased initially in the cultures transduced by either CBFβ-SMMHC or the deletion mutants, as compared to vector alone (Figure 5A). This was associated with a significant decrease in the number of BFU-E colonies, but relatively increased proportion of GEMM and GM colonies (Figure 5B). These data showed that the effects of the deletion mutants on human progenitor cells were similar to that of CBFβ-SMMHC (Wunderlich et al., 2006). The transduced cells were then kept in long-term liquid cultures and were fed every 4th day. Beginning at week 4 the percentage of cells expressing the deletion mutants gradually increased in number, as did the cells expressing CBFβ-SMMHC (Wunderlich et al., 2006), and eventually grew robustly (data not shown). In contrast, those cells transduced with the empty vector grew slower over time and stopped expanding due to terminal differentiation by 10–12 weeks. Long-term cultures were established from the cells transduced with the deletion mutants as well as with CBFβ-SMMHC (Wunderlich et al., 2006). Cells at week 10 showed abnormal cellular morphology consistent with immature myelomonocytic cells at multiple stages of differentiation, similar to cells transduced by CBFβ-SMMHC (Figure 5C). These results demonstrate that CBFβ-SMMHCd179-221 and CBFβ-SMMHCd134-226, like full length CBFβ-SMMHC (Wunderlich et al., 2006), are able to promote the expansion of human CD34+ progenitor cells in vitro. Figure 5D shows a representative western blot that the fusion proteins were expressed at similar levels in the CD34+ cell lines.
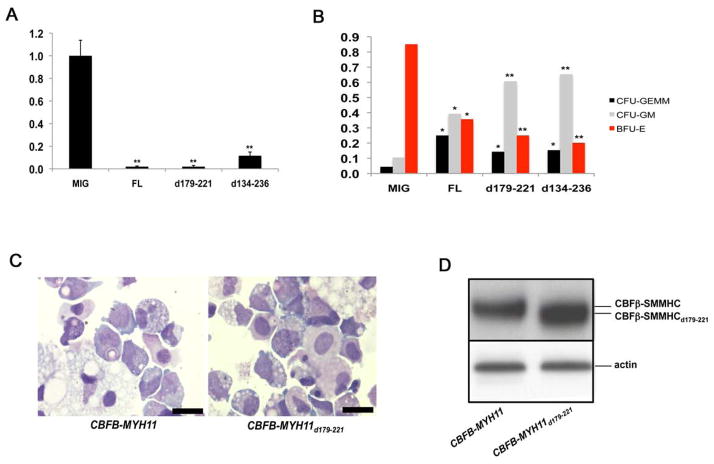
Figure 5. Growth and differentiation defects of human CD34+ cells transduced with CBFB-MYH11 variants.
(A) and (B) In vitro differentiation of human CD34+ cells. Human CD34+ cells were transduced with empty retroviral vector (MIG), and vectors expressing CBFB-MYH11 (FL), CBFB-MYH11d179-221 (d179-221), or CBFB-MYH11d134-236 (d134-236). The transduced cells were sorted by FACS and two thousand GFP+ cells were plated in serum-free methylcellulose cultures. (A) Total colony numbers scored after 14 days. The total colony number from MIG-transduced cells was set at 1, and the total colony numbers from the other two transduced cell populations were calculated relative to that of MIG. The data shown are averages of three independent experiments. (B) Frequencies of BFU-E, CFU-GM, and CFU-GEMM colonies from transduced human CD34+ cells. The data shown are averages of two independent experiments. *: p<0.01 between clones transduced with MIG and those with CBFB-MYH11 constructs; **: p<0.001 between clones transduced with MIG and those with CBFB-MYH11 constructs. The error bars represent one standard deviation.
(C) Cell morphology from long-term cultures of human CD34+ cells transduced with CBFB-MYH11 or CBFB-MYH11d179-221. Shown are cytospin preparations of non-adherent cells at week 10, stained with Wright-Giemsa stain. Scale bars represent 10 μm.
(D) Western blot analysis with protein samples from transduced human CD34+ cells and the CBFβ-specific antibody (β141.2).
Up-regulation of MN1 in cells expressing CBFβ-SMMHCd179-221
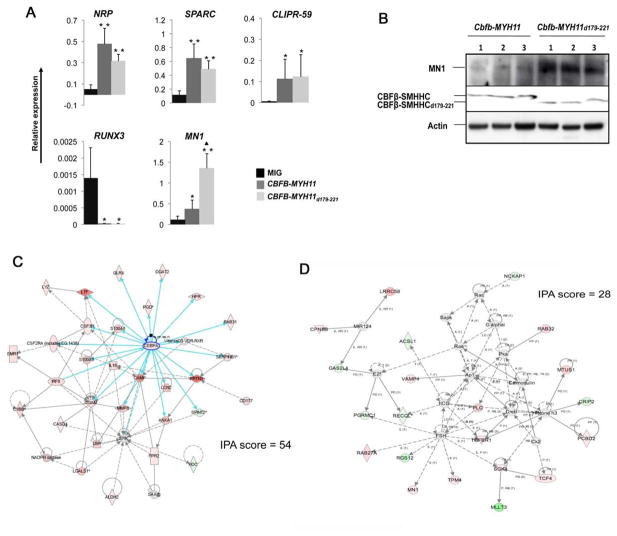
We performed qRT-PCR to assess the expression level of 5 genes previously shown to be altered in CBFβ-SMMHC-transformed human patient samples. We found similar expression alterations for all 5 genes in human CD34+ cells expressing CBFβ-SMMHCd179-221 when compared to normal human CD34+ cells (Figure 6A). Specifically, NRP1, SPARC, and CLIPR-59 were all up-regulated, while RUNX3 was down-regulated in CBFβ-SMMHCd179-221 expressing cells. The expression changes for these four genes were not significantly different from those in CBFβ-SMMHC expressing cells. Interestingly, MN1 was significantly increased in CBFβ-SMMHCd179-221 compared to both full-length CBFβ-SMMHC and the control (Figure 6A).
Figure 6. Similar gene expression changes induced by CBFB-MYH11 and CBFB-MYH11d179-221.
(A) qRT-PCR analysis of RNA samples from human CD34+ cell transduced with MIG, CBFB-MYH11, and CBFB-MYH11d179-221. Error bars represent SDs of 2 to 6 samples. *: P<0.05 between MIG and CBFB-MYH11 or CBFB-MYH11d179-221; **: P<0.01 between MIG and CBFB-MYH11 or CBFB-MYH11d179-221; ▲: P<0.05 between CBFB-MYH11 and CBFB-MYH11d179-221. The error bars represent one standard deviation.
(B) Western blot detection of MN1 expression in leukemia cells from Cbfb+/MYH11 and Cbfb+/MYH11d179-221 mice.
(C) Overexpression of myeloid genes in the Cebpa network from analysis of microarray data by Ingenuity Pathway Analysis (IPA). The blue arrows highlights connections directly from Cebpa. Pink colored genes are those that upregulated. More detailed information about the genes in this network is available in Table S1.
(D) The MN1 network identified by IPA analysis of microarray data. More detailed information about the genes in this network is available in Table S2.
We have shown previously that MN1 is overexpressed in many human inv(16) AML cells and that MN1 cooperates with CBFB-MYH11 for leukemogenesis in mouse models (Carella et al., 2007). Higher expression level of MN1 observed in human CD34+ cells expressing CBFβ-SMMHCd179-221 may explain why CBFβ-SMMHCd179-221 could induce leukemia more efficiently in the knockin mice. We therefore determined the expression level of MN1 in leukemia cells from the CBFβ-SMMHCd179-221 and CBFβ-SMMHC chimeras. The results showed that MN1 was expressed at higher levels in the CBFβ-SMMHCd179-221 mice than in the CBFβ-SMMHC mice (Figure 6B).
We also performed microarray analysis of gene expression changes in the leukemic cells isolated from spleens of Cbfb+/MYH11d179-221 and Cbfb+/MYH11 chimeras with AML. We detected significant changes in gene expression between leukemia cells of the two genotypes in several pathways using Ingenuity Pathway Analysis (IPA, data not shown). Notably, the network with the highest score (IPA score = 54) was centered on Cebpa, with most of the genes in this network upregulated in the Cbfb+/MYH11d179-221 mice, including Cebpa itself (Figure 6C; more detailed information about the genes in this network is available in Table S1). As predicted based on the function of Cebpa, many genes in this network are related to functions in more mature myeloid cells. These results suggest that leukemia cells in the CBFβ-SMMHCd179-221 mice were more differentiated along the myeloid lineage, consistent with FACS and morphological findings (Figure 4B–E).
MN1 did not pass the cut off IPA score, probably because its RNA level was too low. Therefore it was not included in the initial IPA analysis. In order to determine gene expression networks affected by MN1, we added MN1 to the dataset manually with an arbitrary assignment of 2-fold upregulation (since we know through other approaches described above that it was upregulated in the leukemia cells). The IPA analysis assigned MN1 to a pathway that includes Tgfbeta1, Tcf4 (both were upregulated), and Mllt3 (down-regulated) (Figure 6D; more detailed information about the genes in this network is available in Table S2). This network has a score of 28 (#6 on the score list) and the top functions assigned to the network are cellular assembly and organization, endocrine system disorders, and tissue morphology.
RUNX1 phosphorylation in cells expressing CBFβ-SMMHCd179-221
Phosphorylation of RUNX1 at serine residues 249, 276, and 303 has been demonstrated to play important roles for its degradation, transactivation, and functions in cell cycle (Aikawa et al., 2006; Biggs et al., 2006; Wee et al., 2008; Zhang et al., 2008). HIPK2 phosphorylates RUNX1 at Ser249 and Ser276 in a CBFβ-dependent manner, which in turn leads to phosphorylation of p300 (Aikawa et al., 2006; Wee et al., 2008). Interestingly CBFβ-SMMHC disrupts the phosphorylation of both RUNX1 and p300, which have been proposed as a potential mechanism for leukemogenesis by CBFβ-SMMHC (Wee et al., 2008). In addition, cyclin-dependent kinase phosphorylation of RUNX1 at 3 sites (including Ser303) has been shown to regulate RUNX1 degradation (Biggs et al., 2006), increase RUNX1 transactivation potency and stimulate cell proliferation (Zhang et al., 2008). We therefore decided to investigate the effect of CBFβ-SMMHCd179-221 on RUNX1 phosphorylation.
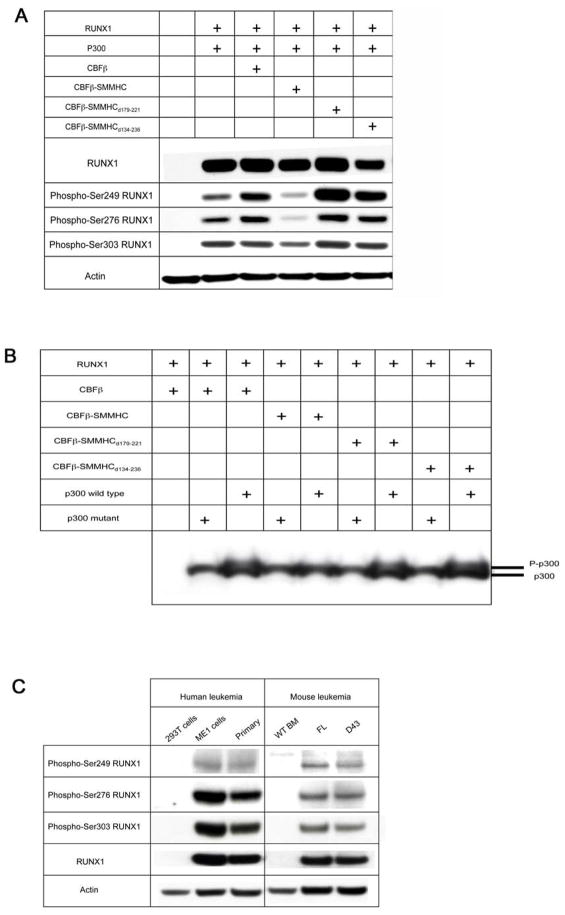
As shown in Figure 7A and B, we confirmed that CBFβ-SMMHC repressed RUNX1 and p300 phosphorylation, when the proteins were overexpressed in transient transfection assays. However, both the type I fusion, CBFβ-SMMHCd134-236, and CBFβ-SMMHCd179-221 were fully capable of promoting RUNX1 and p300 phosphorylation in this assay, similar to wildtype CBFβ. Furthermore, when mouse leukemia cells that express CBFβ-SMMHC or CBFβ-SMMHC d179-221 were examined, their RUNX1 phosphorylation levels seemed to be similar to each other (Figure 7C). This was confirmed in human leukemia cells with inv(16). As shown in Figure 7C, RUNX1 was phosphorylated at all three serine residues in a primary leukemia sample (Liu et al., 1996) as well as in the cell line ME1 (Yanagisawa et al., 1991). These results suggest that RUNX1 was phosphorylated in the presence of CBFβ-SMMHC after leukemic transformation even though CBFβ-SMMHC reduced RUNX1 phosphorylation in transient cell culture assays.
Figure 7. Runx1 and p300 phosphorylation.
(A) RUNX1 phosphorylation by CBFβ and deletion variants of CBFβ-SMMHC. CBFβ, CBFβ-SMMHC, or deletion variants of CBFβ-SMMHC fusion proteins were co-expressed in 293T cells with RUNX1 and wild-type p300. Western blot analyses were performed using the indicated antibodies.
(B) p300 phosphorylation by CBFβ and deletion variants of CBFβ-SMMHC fusion proteins. Wild-type p300 or mutant p300 (p300 ΔSTP1,2,3) were transfected with RUNX1 and CBFβ, CBFβ-SMMHC, or the CBFβ-SMMHC deletion constructs. Immunoblotting with p300 antibody was performed with lysates from transiently transfected 293T cells.
(C) RUNX1 phosphorylation in human and mouse leukemia samples. ME1: a cell line derived from an inv(16)+ AML case (Yanagisawa et al., 1991), primary: primary leukemia cells from bone marrow of an inv(16)+ AML case (Liu et al., 1996). Both human cases contain the type A CBFB-MYH11 fusion. FL: full length CBFβ-SMMHC, D43: CBFβ-SMMHC d179-221, WT BM: wildtype whole bone marrow cells after erythrocyte lysis. Protein lysates were isolated from human and mouse leukemic cells and western blot analyses were performed using the indicated antibodies.
DISCUSSION
CBFβ-SMMHC dominantly represses RUNX1 function, generates defects in definitive hematopoiesis (Castilla et al., 1996), and predisposes mice to leukemia with cooperating gene mutations (Castilla et al., 1999; Castilla et al., 2004). In comparable mouse models the leukemia fusion gene RUNX1-ETO (also known as AML1-ETO) functions very similarly: it dominantly suppresses RUNX1 function, blocks hematopoiesis, and requires additional ‘hits’ for leukemogenesis (Yergeau et al., 1997; Yuan et al., 2001). Therefore the hypothesis of RUNX1 inhibition as a common leukemogenic pathway has been proposed for CBF-related leukemias (Speck and Gilliland, 2002).
Previously we identified a RUNX1 high-affinity binding domain, HABD, at a proximal region of SMMHC in the CBFβ-SMMHC fusion protein (Lukasik et al., 2002). The HABD has been considered as one of the most important domains for dominant repression of RUNX1, since the domain makes it possible for CBFβ-SMMHC to outcompete CBFβ for RUNX1 binding.
Clinically the fusion junctions between CBFB and MYH11 are heterogeneous, 10 different fusion junctions have been reported (van Dongen et al., 1999). All other CBFB-MYH11 fusion types contain the HABD except for the type I fusion. Earlier reports suggested that type I fusion is rare and tends to be associated with therapy related AML (t-AML) or myelodysplastic syndrome (t-MDS) (Dissing et al., 1998; Grardel et al., 2002; van der Reijden et al., 1995; Yamamoto et al., 2006). But a more recent publication indicated that type I fusion is potentially more frequent and can be associated with de novo AML (Monma et al., 2007). It is noticeable that most commonly used RT-PCR primers for diagnosis of CBFB-MYH11 would not be able to detect the type I fusion, so it may have been under-diagnosed, especially in cytogenetically negative cases (Van der Reijden et al., 2001; van Dongen et al., 1999). The existence of the type I fusion suggests that the HABD or high-efficient RUNX1 repression is not always required for leukemogenesis.
To understand the importance of HABD for leukemogenesis, we generated mice expressing a knock-in Cbfb-MYH11 fusion gene with the HABD removed (Cbfb+/MYH11d179-221). In vitro and in vivo analyses indicated that the encoded CBFβ-SMMHCd179-221 was less efficient in binding and repressing RUNX1, as expected. Unexpectedly, leukemia development was accelerated in the Cbfb+/MYH11d179-221 knock-in mice, so that all the chimeras and F1 heterozygotes developed leukemia shortly after birth without ENU treatments. These findings cast doubts on the model that strong, dominant repression of RUNX1 is a key step in leukemogenesis.
Human CD34+ cells expressing the CBFβ-SMMHC variants behaved essentially the same as those expressing the full-length. This data is consistent with our conclusion that RUNX1 dominant inhibition may not be a critical step for leukemogenesis by CBFβ-SMMHC. However, the CBFβ-SMMHC variants did not accelerate the phenotype development in the human CD34+ cells, as compared to the accelerated leukemogenesis in the knockin mouse model. This difference is likely related to the fact that transgenic expression from retroviral vectors was used in the human CD34+ cells, while knockin technology was used in the mouse model. Both alleles of CBFB are intact in the human CD34+ cells; on the other hand, one Cbfb allele in the mouse genome is replaced by the Cbfb-MYH11 fusion gene. The endpoint of the human CD34+ cell assay system is also different from that in the mouse model, being clonal expansion rather than leukemia. All these factors could have contributed to the observed differences between these two systems.
The mechanism for the accelerated leukemogenesis in the Cbfb+/MYH11d179-221 mice is not clear. It is possible that the deletion released an anti-leukemia effect or provided a leukemia-promoting effect, such as upregulation of a cooperating gene. One potential candidate is MN1, which was up-regulated in leukemia cells in the Cbfb+/MYH11d179-221 mice, as well as in human CD34+ cells expressing CBFβ-SMMHCd179-221. MN1 is an important cooperating gene for inv(16) leukemogenesis, as has been demonstrated by its specific upregulation in human inv(16) leukemia and its ability to accelerate leukemogenesis in our mouse Cbfb-MYH11 knock-in model (Carella et al., 2007). It is therefore plausible that CBFβ-SMMHCd179-221 further up-regulates MN1, which in turn cooperates with CBFβ-SMMHCd179-221 for leukemogenesis.
CBFβ-SMMHCd179-221 may up-regulate MN1 through multiple mechanisms. First, it appears that both full-length and d179-221 forms of CBFβ-SMMHC are able to up-regulate MN1 at the level of transcription (Figure 6A). Secondly, at the protein level, MN1 is recruited by p300 to act as a co-activator (van Wely et al., 2003). It is likely that MN1 protein level/functionality is enhanced in cells expressing CBFβ-SMMHCd179-221 since there is more p300 phosphorylation in these cells than in those expressing full-length CBFβ-SMMHC (Figure 7B). Thirdly, hematopoietic blockage is less severe in the CBFβ-SMMHCd179-221 mice, resulting in a larger population of myeloid progenitors in the bone marrow (Figure 4E), which express the highest level of MN1 (Grosveld, 2007). In summary, we believe the combined effect of a larger population of cells expressing high baseline level of MN1, more efficient p300 recruitment, and the transcriptional up-regulation contributes to higher expression level of MN1 in cells with CBFβ-SMMHCd179-221.
The finding that the type I fusion and CBFβ-SMMHCd179-221 are fully capable of promoting RUNX1 and p300 phosphorylation, in contrast to CBFβ-SMMHC, is interesting. However, because this activity is similar to wildtype CBFβ, it raises the question as how this would contribute to the enhanced leukemogenic activity.
A likely explanation is that RUNX1 phosphorylation is a modifying or cooperating step, which contributes to leukemogenesis only in the presence of CBFβ-SMMHC proteins. More RUNX1 and p300 phosphorylation in mice expressing CBFβ-SMMHCd179-221 is consistent with partial rescue of hematopoietic blockage in these mice. It is therefore not surprising that this activity is similar to wildtype CBFβ, since CBFβ supports rather than blocks RUNX1 function in hematopoiesis.
The implications of normal Runx1 phosphorylation and hematopoietic rescue in the CBFβ-SMMHCd179-221 mice are at least two fold. First, normal Runx function may be required for leukemogenesis. We have observed that reduction of RUNX1 activity by introducing a Runx1 dominant negative allele to the Cbfb+/MYH11 mice delayed leukemia development (LZ and PPL, unpublished observations). It was also shown recently that Runx2 cooperated with Cbfβ-SMMHC for leukemogenesis (Kuo et al., 2009). Secondly, reduced blockage of hematopoiesis as a result of partial RUNX1 inhibition may have led to the expansion of a “leukemia-prone” cell population, which provides more “target” cells for leukemogenesis. We speculate that HSCs were the target cells in the Cbfb+/MYH11 mice while both HSCs and myeloid progenitors could be the target cells in the Cbfb+/MYH11d179-221 mice. The fact that more leukemia cells in the Cbfb+/MYH11d179-221 mice express myeloid markers (Figure 4E) suggests that leukemia may have initiated in myeloid progenitors in these mice.
Our finding that partial inhibition of RUNX1 may be more leukemogenic than complete RUNX1 inhibition is similar to what happened with PU.1, a hematopoietic transcription factor downstream of RUNX1. Mice with homo- or heterozygous deletion of Pu.1 do not develop leukemia; on the other hand, mice carrying hypomorphic alleles of Pu.1 with reduced expression (20% of normal) developed AML rapidly and efficiently (Rosenbauer et al., 2004).
Moreover, a variant of the AML1-ETO fusion protein, AML1-ETO9a that contains C-terminal truncation, was found to be a much more potent inducer of leukemia than the full-length AML1-ETO in mouse models. It has been hypothesized that the deleted region inhibits the leukemogenic potential of AML1-ETO (Peterson et al., 2007). Interestingly AML1-ETOtr, a C-terminally truncated protein similar to AML1-ETO9a, lost the ability to inhibit cell cycle progression of myeloid cells, which may contribute to its enhanced leukemogenic potential (Yan et al., 2004). This AML1-ETO variant is thus similar to the CBFβ-SMMHC179-221 variant reported here, in that they are both less potent in repressing RUNX1/CBFB function yet are more potent in leukemogenesis.
Recent results (Kwok et al., 2009; Park et al., 2009; Roudaia et al., 2009) show that while very modest effects on the heterodimerization of CBFβ with AML1-ETO have no effect on leukemogenesis, substantial loss of binding results in a protein that is incapable of causing leukemia, likely a reflection of the relatively high concentration of CBFβ in cells. This point is echoed by recent studies on recurrent mutations in RUNX1 in patients with AML subtype M0 and familial platelet disorder with predisposition to AML. The mutations abolish DNA binding and transactivation by RUNX1 in vitro (Michaud et al., 2002; Osato et al., 1999) and cause hematopoietic defects and embryonic lethality in mice (Matheny et al., 2007), suggesting that they are mostly loss-of function mutations. However, it was recently shown that these mutations also altered differentiation and increased serial replating ability (Cammenga et al., 2007), indicating that RUNX1 has DNA-binding independent activities that play a role in leukemogenesis. As the primary role of CBFβ is to stabilize RUNX1’s interaction with DNA, these findings provide further argument for CBFB/RUNX repression independent mechanisms in leukemogenesis.
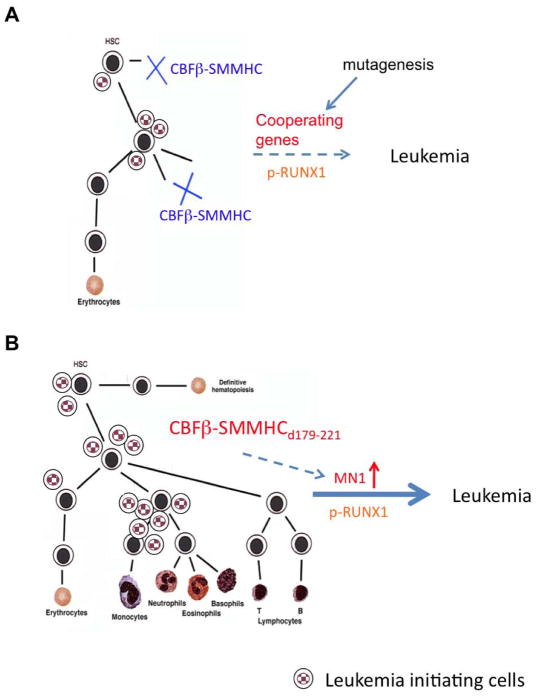
Our current model for leukemogenesis in the Cbfb+/MYH11d179-221 mice is illustrated in Figure 8. More severe blockage of hematopoiesis and RUNX1 inhibition by Cbfb-MYH11 results in a smaller leukemia target cell pool, where additional mutations are needed to introduce cooperating genes and to restore RUNX1 phosphorylation. On the other hand, a larger population of leukemia target cells is available in the Cbfb+/MYH11d179-221 mice, and leukemogenesis is further accelerated by upregulation of MN1 and retention of RUNX1 phosphorylation.
Figure 8.
Working model for leukemogenesis in Cbfb+/MYH11d179-221 mice.
In summary, loss of the HABD from CBFβ-SMMHC unexpectedly potentiated its leukemogenic activity, raising questions to the proposed dominant-negative mechanism of leukemogenesis. Partial reduction of key transcription factors such as RUNX1 and PU.1 may be a common mechanism for leukemogenesis. Moreover, CBFβ-SMMHC may contribute to leukemogenesis through pathways other than RUNX1 repression. Recent work from our group has provided evidence for such RUNX1-repression independent pathways (Hyde et al., 2010). However, it still needs to be determined whether the leukemogeneic activities of CBFβ-SMMHC depend on its interaction with RUNX1, even for those that do not seem to require RUNX1-repression. Such studies will provide important mechanistic insight guiding the design and development of small molecule inhibitors targeting the CBFβ/AML1 interaction for leukemia treatments (Gorczynski et al., 2007).
EXPERIMENTAL PROCEDURES
ES Cell Targeting and Mouse Experiments
The targeting constructs were assembled in the pPNT vector, which included a thymidine kinase gene as positive selection. A 3.5 kb KpnI–StuI mouse genomic clone (strain 129/Sv), including mouse Cbfb intron 4 and the first 25 bp of mouse exon 5, was ligated to the StuI site of a 1.5 kb StuI–NotI human CBFB–MYH11 cDNA clone or a mutant CBFB–MYH11 cDNA with HABD deletion between aa179 to 221. The PGK neo was ligated downstream of BGHpA. A 4.7 kb mouse genomic clone of Cbfb intron 5 (strain 129/Sv) was then ligated to the 3′ end of the PGK neo fragment. The constructs were linearized at an NotI site and transfected into the TC 1 ES cells (from mouse strain 129/SvEv). Screening of the targeted ES cell clones by Southern blot analysis was performed as described (Castilla et al., 1996; Kuo et al., 2006). ES cells were injected into C57BL/6-derived blastocysts. Chimeric mice were crossed with C57BL/6 females for further embryo analysis. The mouse studies were approved by the NHGRI Animal Care and Use Committee, and all experiments on mice were performed in accordance with relevant NIH and national guidelines and regulations.
Western Blot Analysis
Protein isolation, gel eletrophoresis, blot transfer, and detection with monoclonal antibody β141.2 and actin (Millipore) were performed as described previously (Kundu et al., 2002).
Histological Sections
E12.5 embryos and adult tissues (such as liver, kidney and spleen) were fixed in 10% formalin and the sections were stained with either Wright–Giemsa or hematoxylin and eosin (H and E) stains (American HistoLabs, Inc., Gaithersburg, MD). The histopathology slides were examined and photographed as described (Castilla et al., 1999).
In Vitro Differentiation Experiments for fetal liver
Fetal liver cells from E11.5 embryos were isolated and cultured in methylcellulose medium M3434 (Stemcell Technologies, Vancouver, Canada). Colonies were scored at day 7 as described before (Castilla et al., 1996).
Flow cytometry analysis
Peripheral blood and leukemic spleen cells were stained with PE-TexRed-B220, PE-Ter119, PE-Cy7-GR1, APC-Cy7-C-kit, PerCp-Cy5.5-Mac1, Pacific Blue-CD4, APC-CD8 (BD Biosciences Pharmingen, CA) for flow cytometry analysis. Appropriate isotype controls were used in each experiment. LSRII (BD Biosciences, CA) was used to acquire data, which were analyzed using Flowjo 9.0.1 (Tree Star, Inc., Ashland, OR).
RNA isolation and RT-PCR
Total RNA was isolated from ES cells and leukemic cells using RNA STAT-60™ (TEL-TEST, INC.). The presence of the Cbfb–MYH11 transcript was identified using a forward primer that is in exon 3 of the mouse Cbfb gene (E3: 5′-CAA ACA CCT AGC CGG GAA TA-3′) and a reverse primer in the human MYH11 cDNA (MYH11: 5′-CTT CCA AGC TCT TGG CTT TCT TC-3′). The quality of the reverse-transcribed cDNA was confirmed by PCR using primer mC3/4.1F and a reverse primer in exon 6 of the mouse Cbfb gene (mC6.1R: 5′-GAACCAGGACTAGGGTCTTGC-3′). In all instances, PCR conditions were the same as those for genotype analysis described below.
Genotype Analysis
The presence of the knock-in alleles was assessed by PCR using DNA isolated from tail snips, PB, yolk sacs, ES cells or whole embryos. We amplified 50 ng of template DNA by PCR using neo-specific primer (neo forward 5′-AGAGGCTATTCGGCTATGA CTG-3′, and neo reverse 5′TTCGTCCAGATCATCCTGATC-3′). The PCR cycle conditions were 94°C for 2 min, 30 cycles of amplification (30 s each at 94°C, 60°C, and 72°C), and a final 5 min extension step at 72°C. The genotypings were confirmed in a parallel PCR reaction with a forward primer in exon 5 of the mouse Cbfb gene (E5: 5′-CAG GAA GAT GCA TTA GCA CAA-3′) and the reverse primer MYH11.
Human CD34+ cell assays
Human CD34+ umbilical cord blood cells were obtained from the Translational Research Development and Support Laboratory of Cincinnati Children’s Hospital under an approved IRB protocol. Retroviral production, transduction, cell culture, CFU assays, and qRT-PCR were performed as previously described (Wunderlich et al., 2006).
Microarray analysis
Spleen cells were isolated from knock-in mice with AML, including 2 adult Cbfb+/MYH11 chimeras at 2 months after ENU treatment and 2 Cbfb+/MYH11d179-221 chimeras at 3 weeks after birth. For each sample we performed two independent experiments with Affymetrix GeneChip 430. All intensity values were scaled to an average value of 150 per GeneChip according to the method of global scaling, or normalization, provided in the Affymetrix Microarray Suite software. The normalized results were then analyzed using Genesifter (VizX Labs, Seattle, WA).
Expression and Purification of CBFβ-SMMHCd134-236
CBFb-MYH11d134-236 was cloned in the pHis-parrallel vector(Sheffield et al., 1999). CBFβ-SMMHCd134-236 was expressed in Rosetta (DE3) cells after induction with IPTG and purified with a Ni-NTA column (Qiagen, Valencia CA). The 6xHis-tag was cleaved with the AcTEV protease (Invitrogen, Carlsbad CA). Further purification of the protein was accomplished using ion-exchange chromatography on a Q-Sepharose (GE Healthcare, Piscataway NJ).
Isothermal titration Calorimetry (ITC)
ITC measurements were carried out at 30°C on a VP-ITC MicroCalorimeter system (MicroCal Inc., Northampton MA). Protein samples were dialyzed against 12.5 mM KPi (pH 6.5), 150 mM NaCl, 2.5 mM MgCl2, and 1 mM DTT, centrifuged to remove precipitates, and degassed for 15 min prior to use. 15 μM RUNX1 Runt Domain was titrated with 200 μM CBFβ-SMMHCd134-236. Dilution enthalpies were determined through protein into buffer experiments, and were subtracted from the initial experiments. Data was analyzed using Origin 7.0 (Origin Lab, Northampton MA).
In vitro reporter assay
CAT reporter assay was performed as described before (Zhao et al., 2007). Briefly, the CAT reporter constructs were transfected into the CD4- Jurkat cell clone (D1.1, from American Type Culture Collection, Manassas, VA) alone or with RUNX1, CBFB, CBFB-MYH11, CBFB-MYH11d179-221, and CBFB-MYH11d134-236 cDNA constructs by electroporation (BTX electroporation system, Harvard Apparatus, Holliston, MA). CAT activity was measured following manufacture’s protocol (CAT ELISA, Roche Diagnostics, Basel, Switzerland). A luciferase vector was cotransfected to standardize transfection efficiency. The transfection was performed for 3 times with similar results.
For the MCSFR reporter assay, 293T cells were seeded into 6 well plates (5×105 cells/well) one day before transfection. 1.2 μg of pMCSFR-luc (Rhoades et al., 1996), and 1 μg of RUNX1, CBFB, CBFB-MYH11, CBFB-MYH11d179-221, and CBFB-MYH11d134-236 were transfected using Lipofectamin 2000 (Invitrogen). The experiments were performed three times.
Runx1 and p300 phosphorylation
Typically, 105 293 cells were plated in 6-well plate and cultured overnight before transiently transfected with expression constructs for RUNX1 (0.1 μg), Cbfβ or each Cbfβ-SMMHC truncated mutant (0.3 μg), and with p300(0.5 μg), using the FuGENE 6 reagent (Roche Diagnostics, Indianapolis, IN) according to manufacturer’s instructions. The expression constructs for pEF-neo-RUNX1, pEF-bos-CBFβ, pEF-bos-Full length CBFβ-SMMHC, CBFβ-SMMHC d179-221, and CBFβ-SMMHCd134-236, were described previously (Huang et al., 2001). pcDEF-FLAG-p300 and pcDEF-FLAG-p300ΔSTP1,2,3 were also described previously (Wee et al., 2008).
Lysates from transiently transfected 293T cells were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). After probing with appropriate antibodies, protein bands were detected using the enhanced chemiluminescence detection system (GE Healthcare). The following primary antibodies were used in the study. Rabbit anti-AML1 (Active Motif, Carlsbad, CA), Phospho-AML1 Ser249 (Cell signaling, MA), RUNX1/AML1 phospho S276 antibody ab55291 (Abcam, MA), RUNX1/AML1 phospho S303 antibody ab55308 (Abcam, MA), and anti-p300 antibody (N15) (Santa Cruz Biotechnology, CA). Secondary horseradish peroxidase-linked goat anti–rabbit IgG or anti–mouse antibodies used in Western blotting were obtained from GE Healthcare (Little Chalfont, UK).
Accession Number
The microarray data have been deposited in the Nation Center for Biotechnology Information's Gene Expression Omnibus database with the accession number of GSE21155.
HIGHLIGHTS
A variant CBFβ-SMMHC in AML patients missing the HABD binds RUNX1 weakly
Hematopoiesis is partially rescued in mice expressing a HABD-deleted CBFβ-SMMHC
Leukemogenesis is accelerated in mice expressing a HABD-deleted CBFβ-SMMHC
The variant CBFβ-SMMHC increases MN1 expression and retains RUNX1 phosphorylation
SIGNIFICANCE
CBFβ-SMMHC is a common mutation in human AML and is causally related to leukemogenesis in a knock-in mouse model. Dominant repression of RUNX1 has been considered as the main function of CBFβ-SMMHC; however, its importance in leukemogenesis is unclear. One of the 10 CBFB-MYH11 fusions detected in AML patients also lacks HABD and does not bind or repress RUNX1 efficiently; suggesting that dominant RUNX1 inhibition is not critical for leukemogenesis by CBFB-MYH11. We generated knock-in mice expressing a deletion mutant of CBFβ-SMMHC with reduced RUNX1 binding and inhibition. The contrasting phenotype of partial hematopoiesis rescue and accelerated leukemogenesis suggests that the ability of CBFβ-SMMHC to induce leukemia does not correlate with its ability to repress RUNX1.
Supplementary Material
Acknowledgments
We thank Martha Kirby and Stacie Anderson for FACS analysis, Bert A. van der Reijden for the human leukemia sample, Chenwei Wang for statistical analysis of microarray data, Abdel Elkahloun for microarray hybridization, Kirin Brewery for the cytokine TPO, and Amgen for FLT3L, SCF, and IL-6. This work was supported by the Intramural Research Programs of National Human Genome Research Institute, NIH, NIH grant CA118319, U.S.P.H.S. Grant Number MO1 RR 08084, and General Clinical Research Centers Program, National Center for Research Resources, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol Cell Biol. 2006;26:7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammenga J, Niebuhr B, Horn S, Bergholz U, Putz G, Buchholz F, Lohler J, Stocking C. RUNX1 DNA-binding mutants, associated with minimally differentiated acute myelogenous leukemia, disrupt myeloid differentiation. Cancer Res. 2007;67:537–545. doi: 10.1158/0008-5472.CAN-06-1903. [DOI] [PubMed] [Google Scholar]
- Carella C, Bonten J, Sirma S, Kranenburg TA, Terranova S, Klein-Geltink R, Shurtleff S, Downing JR, Zwarthoff EC, Liu PP, Grosveld GC. MN1 overexpression is an important step in the development of inv(16) AML. Leukemia. 2007;21:1679–1690. doi: 10.1038/sj.leu.2404778. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, Owens J, Eckhaus M, Bodine D, Liu PP. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Perrat P, Martinez NJ, Landrette SF, Keys R, Oikemus S, Flanegan J, Heilman S, Garrett L, Dutra A, et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4924–4929. doi: 10.1073/pnas.0400930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, Marin-Padilla M, Collins FS, Wynshaw-Boris A, Liu PP. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- Dissing M, Le Beau MM, Pedersen-Bjergaard J. Inversion of chromosome 16 and uncommon rearrangements of the CBFB and MYH11 genes in therapy-related acute myeloid leukemia: rare events related to DNA-topoisomerase II inhibitors? J Clin Oncol. 1998;16:1890–1896. doi: 10.1200/JCO.1998.16.5.1890. [DOI] [PubMed] [Google Scholar]
- Gorczynski MJ, Grembecka J, Zhou Y, Kong Y, Roudaia L, Douvas MG, Newman M, Bielnicka I, Baber G, Corpora T, et al. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chem Biol. 2007;14:1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Grardel N, Roumier C, Soenen V, Lai JL, Plantier I, Gheveart C, Cosson A, Fenaux P, Preudhomme C. Acute myeloblastic leukemia (AML) with inv (16)(p13;q22) and the rare I type CBFbeta-MYH11 transcript: report of two new cases. Leukemia. 2002;16:150–151. doi: 10.1038/sj.leu.2402332. [DOI] [PubMed] [Google Scholar]
- Grosveld GC. MN1, a novel player in human AML. Blood Cells Mol Dis. 2007;39:336–339. doi: 10.1016/j.bcmd.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. Embo J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde RK, Kamikubo Y, Anderson S, Kirby M, Alemu L, Zhao L, Liu PP. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood. 2010;115:1433–1443. doi: 10.1182/blood-2009-06-227413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Chen A, Anderson S, Kirby M, Xu L, Castilla LH, Bodine D, Liu PP. Role of Cbfb in hematopoiesis and perturbations resulting from expression of the leukemogenic fusion gene Cbfb-MYH11. Blood. 2002;100:2449–2456. doi: 10.1182/blood-2002-04-1064. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, Le Beau MM, Kogan SC, Castilla LH. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9:57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Qiu J, Dong S, So CW. Transforming activity of AML1-ETO is independent of CBFbeta and ETO interaction but requires formation of homo-oligomeric complexes. Proc Natl Acad Sci U S A. 2009;106:2853–2858. doi: 10.1073/pnas.0810558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Tarl:e SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia [see comments] [published erratum appears in Blood 1997 Mar 1;89(5):1842] Blood. 1995;85:2289–2302. [PubMed] [Google Scholar]
- Liu PP, Wijmenga C, Hajra A, Blake TB, Kelley CA, Adelstein RS, Bagg A, Rector J, Cotelingam J, Willman CL, Collins FS. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chromosomes Cancer. 1996;16:77–87. doi: 10.1002/(SICI)1098-2264(199606)16:2<77::AID-GCC1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Lukasik SM, Zhang L, Corpora T, Tomanicek S, Li Y, Kundu M, Hartman K, Liu PP, Laue TM, Biltonen RL, et al. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol. 2002;9:674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- Matheny CJ, Speck ME, Cushing PR, Zhou Y, Corpora T, Regan M, Newman M, Roudaia L, Speck CL, Gu TL, et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007;26:1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud J, Wu F, Osato M, Cottles GM, Yanagida M, Asou N, Shigesada K, Ito Y, Benson KF, Raskind WH, et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- Monma F, Nishii K, Shiga J, Sugahara H, Lorenzo Ft, Watanabe Y, Kawakami K, Hosokai N, Yamamori S, Katayama N, Shiku H. Detection of the CBFB/MYH11 fusion gene in de novo acute myeloid leukemia (AML): a single-institution study of 224 Japanese AML patients. Leuk Res. 2007;31:471–476. doi: 10.1016/j.leukres.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y. Biallelic and Heterozygous Point Mutations in the Runt Domain of the AML1/PEBP2alphaB Gene Associated With Myeloblastic Leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- Park S, Speck NA, Bushweller JH. The role of CBFbeta in AML1-ETO’s activity. Blood. 2009;114:2849–2850. doi: 10.1182/blood-2009-07-231233. [DOI] [PubMed] [Google Scholar]
- Peterson LF, Boyapati A, Ahn EY, Biggs JR, Okumura AJ, Lo MC, Yan M, Zhang DE. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades KL, Hetherington CJ, Rowley JD, Hiebert SW, Nucifora G, Tenen DG, Zhang DE. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc Natl Acad Sci U S A. 1996;93:11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S, Lee CT, Kaur P, Williams O, Bushweller JH, Speck NA. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Van der Reijden BA, de Wit L, van der Poel S, Luiten EB, Lafage-Pochitaloff M, Dastugue N, Gabert J, Lowenberg B, Jansen JH. Identification of a novel CBFB-MYH11 transcript: implications for RT-PCR diagnosis. Hematol J. 2001;2:206–209. doi: 10.1038/sj.thj.6200103. [DOI] [PubMed] [Google Scholar]
- van der Reijden BA, Lombardo M, Dauwerse HG, Giles RH, Muhlematter D, Bellomo MJ, Wessels HW, Beverstock GC, van Ommen GJ, Hagemeijer A, et al. RT-PCR diagnosis of patients with acute nonlymphocytic leukemia and inv(16)(p13q22) and identification of new alternative splicing in CBFB-MYH11 transcripts. Blood. 1995;86:277–282. [PubMed] [Google Scholar]
- van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, Gottardi E, Rambaldi A, Dotti G, Griesinger F, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- van Wely KH, Molijn AC, Buijs A, Meester-Smoor MA, Aarnoudse AJ, Hellemons A, den Besten P, Grosveld GC, Zwarthoff EC. The MN1 oncoprotein synergizes with coactivators RAC3 and p300 in RAR-RXR-mediated transcription. Oncogene. 2003;22:699–709. doi: 10.1038/sj.onc.1206124. [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. Embo J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee HJ, Voon DC, Bae SC, Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: implications for leukemogenesis. Blood. 2008;112:3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Krejci O, Wei J, Mulloy JC. Human CD34+ cells expressing the inv(16) fusion protein exhibit a myelomonocytic phenotype with greatly enhanced proliferative ability. Blood. 2006;108:1690–1697. doi: 10.1182/blood-2005-12-012773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Nishikawa S, Minagawa K, Yakushijin K, Okamura A, Matsui T. Therapy-related myelodysplastic syndrome with inv(16)(p13q22) and I type CBFbeta/MYH11 after autologous transplantation: undetectable fusion transcript in pretransplant progenitor cells. Leuk Res. 2006;30:354–361. doi: 10.1016/j.leukres.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, Hines R, Akashi K, Zhang DE. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci U S A. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa K, Horiuchi T, Fujita S. Establishment and characterization of a new human leukemia cell line derived from M4E0. Blood. 1991;78:451–457. [PubMed] [Google Scholar]
- Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, Marin-Padilla M, Tenen DG, Speck NA, Zhang DE. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, Burel SA, Lagasse E, Weissman IL, Akashi K, Zhang DE. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fried FB, Guo H, Friedman AD. Cyclin-dependent kinase phosphorylation of RUNX1/AML1 on 3 sites increases transactivation potency and stimulates cell proliferation. Blood. 2008;111:1193–1200. doi: 10.1182/blood-2007-08-109702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Cannons JL, Anderson S, Kirby M, Xu L, Castilla LH, Schwartzberg PL, Bosselut R, Liu PP. CBFB-MYH11 hinders early T-cell development and induces massive cell death in the thymus. Blood. 2007;109:3432–3440. doi: 10.1182/blood-2006-10-051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.