Abstract
Meiosis is a dynamic process during which chromosomes undergo condensation, pairing, crossing-over and disjunction. Stringent regulation of the distribution and quantity of meiotic crossovers is critical for proper chromosome segregation in many organisms. In humans, aberrant crossover placement and the failure to faithfully segregate meiotic chromosomes often results in severe genetic disorders such as Down syndrome and Edwards syndrome. In most sexually reproducing organisms, crossovers are more evenly spaced than would be expected from a random distribution. This phenomenon, termed interference, was first reported in the early 20th century by Drosophila geneticists and has been subsequently observed in a vast range of organisms from yeasts to humans. Yet, many questions regarding the behavior and mechanism of interference remain poorly understood. In this review, we examine results new and old, from a wide range of organisms, to begin to understand the progress and remaining challenges to understanding the fundamental unanswered questions regarding genetic interference.
Keywords: Meiosis, recombination, crossover, Double-strand break, synaptonemal complex, chromosome Spo11, Rec8, Pch2.
BACKGROUND
Meiosis, a type of cell division, reduces the chromosomal complement by half to produce gametes that are essential for sexual reproduction. During meiotic prophase, chromosomes pair with their homologs and, in most organisms, undergo a physical exchange of DNA or an exchange of sequence information in a process called recombination [1]. Recombination is initiated by programmed double-strand breaks (DSBs) of chromosomes. During the repair of some DSBs, chromosome arms are exchanged generating crossovers (COs).
In most organisms, COs are not distributed randomly. Closely spaced COs are observed less frequently than would be expected from a random distribution. This phenomenon is known as crossover interference – though the more general term genetic interference may be more useful since there is growing evidence (described below) that other events can also interfere with one another. Alfred H. Sturtevant and Hermann J. Muller are typically given equal billing for the discovery of interference. Sturtevant clearly describes the phenomenon as early as 1913 [2]. Two years later, he coined the term “interference”, though in doing so he gives credit to Muller for suggesting the name and also for his influence in discovering the phenomenon [3]. For clarity, in this review ‘interference’ will refer to positive interference, which is the spacing of events that departs from a random distribution, as opposed to negative interference, which describes events that are more clustered than the null expectation. Although interference was originally observed almost a century ago and has subsequently been validated in numerous studies, fundamental questions regarding its underlying mechanisms still exist. The goal of this article is to outline unanswered questions about interference and also to review existing models.
The genomic distribution of COs is regulated in multiple ways. For example, they are distributed such that each chromosome typically receives at least one, which is known as ‘CO assurance’. A random distribution of COs among chromosomes predicts a class of chromosomes that have no COs, yet the observed number of chromosomes without a CO is quite small in most organisms [4, 5]. Growing evidence suggests that interference is the result of multiple levels of recombination regulation and CO assurance could be a result of the interference mechanism.
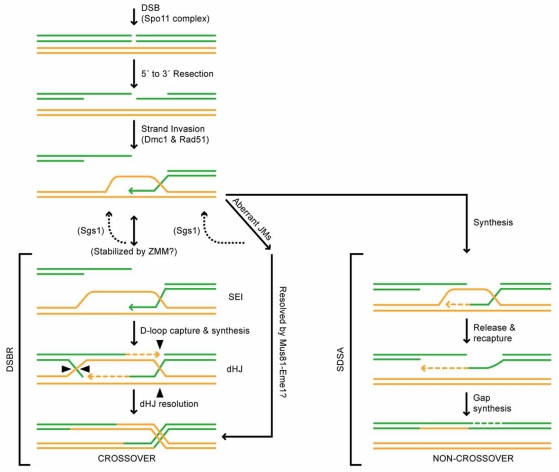
Meiotic DSBs are enzymatically catalyzed by a topoisomerase I-like protein called Spo11 that remains covalently attached to 5' ends of the break (Fig. 1). Following Spo11 removal and further end processing (resection), the breaks are left with single-stranded 3' tails. One of these tails can then invade a non-sister chromatid, which is known as strand invasion. Stabilized strand invasion intermediates are known as single-end invasion (SEI) intermediates. The free 3'-OH in the SEI structure is used as a substrate by DNA polymerase to extend the 3' tail and the size of the displaced DNA strand (D-loop). The DNA synthesis that occurs at this stage is primed by one chromatid but uses a non-sister chromatid as a template – therefore any polymorphisms that exist at this locus will be copied from the template chromatid to the invading chromatid. This transfer of parental information is called gene conversion (GC) [6, 7]. At this point the invading end can dissociate from the non-sister chromatid and re-associate with the other end of the break in a process called synthesis dependant strand annealing (SDSA) [8]. After additional DNA synthesis and ligation the break is repaired resulting in a non-crossover (NCO), potentially with associated GC if a polymorphism existed at the locus. It is important to note however that GC would result in heteroduplex DNA – i.e. that Watson and Crick strands of the converted chromatid would have non-complementary bases at the polymorphic site. The heteroduplex DNA can be recognized by the cell’s mismatch repair system and either repaired such that the original parental genotype is restored or such that the converted genotype is kept. Alternatively, the mismatch repair system can fail to recognize the mismatch. In this case, both genotypes in the heteroduplex will be propagated during the next mitotic division in a process called post-meiotic segregation (PMS). PMS results in two populations of cells, one that has experienced GC at the polymorphic site and the other that has not.
Fig. (1).
The DSBR and SDSA meiotic recombination models. Single strands of DNA are shown as either green (parent 1) or yellow (parent 2) rods. The Spo11 complex initiates programmed DSBs. DSBs are resected 5’ to 3’ to produce single ssDNA tails. ssDNA tails invade the homologous template which is aided by the ssDNA filament forming proteins Dmc1 and Rad51. At this stage, intermediates can undergo DSBR (left), which is thought to produce primarily COs or SDSA (right), which only produces NCOs. Also shown is a pathway (center) describing aberrant JMs, that are hypothesized to either be resolved back to the strand invasion stage by Sgs1 or resolved as COs by the Mus81-Eme1 heterodimer [52]. In the DSBR pathway, strand invasion complexes are stabilized (possibly by ZMM proteins) to form SEIs. Prior to stabilization, Sgs1 could wire SEIs back to the strand invasion stage [52, 89]. The displaced strand, called a D-loop is captured by the resected break of opposite homolog and subsequent DNA synthesis results in a dHJ intermediate. This intermediate is resolved as a CO upon appropriate resolution of the two HJs. in the SDSA pathway, the invading strand dissociates after a patch of DNA synthesis. This strand then re-anneals to the original parent, resulting in repair of the DSB and a patch of heteroduplex DNA. This pathway is always resolved as a NCO, but it can result in GC.
If SDSA does not occur, then as the D-loop is extended it can hybridize to the single-stranded 3' tail on the other side of the break in a process called second end capture (Fig. 1). Again, this structure can be acted on by DNA polymerase, which extends the second single-stranded 3' tail. As before, the priming and template DNA are from non-sister chromatids, therefore GC can occur during this stage. Following DNA synthesis and subsequent ligation, an intermediate called a double Holiday junction (dHJ) is formed. In principle, this structure can be resolved to produce either a crossover (CO) or NCO depending on how the individual junctions are cut to release the chromatids. However, evidence in Saccharomyces cerevisiae suggests that these intermediates are resolved predominantly as COs [4, 9, 10]. In either case, the resulting COs and NCOs can be associated with GC.
Early Observations
Sturtevant observed that in Drosophila melanogaster a three-point cross involving mutations on the X chromosome (yellow, white and miniature wings), a CO in the yellow-white interval occurred at a frequency of 1/69 gametes without the presence of a neighboring CO in white-miniature, but this frequency plummeted to 1/441 when there was a CO in the white-miniature interval (P = ~0.25) [3]. Sturtevant went on to define an “index of interference” as the expected probability of a double CO (the product of the two individual recombination frequencies) divided by the observed frequency of double COs. This marked the beginning of the debate as to the best way to measure interference, which is still being argued to this day [11]. With the index of interference and additional data Sturtevant decreed that “(interference) is less when the intervals are larger and vice versa [3].”
Data generated in Drosophila dominated advancements in interference research for the next 40 years [12-15]. Subsequent papers lacked empirical observations that changed the essential view of interference; instead they confirmed Sturtevant’s initial observations and focused on different ways to measure interference. Nonetheless, several key observations were made. Muller first proposed that interference does not act between different chromosomes, i.e. a CO on one chromosome does not affect the probability of a simultaneous CO in an interval on another chromosome [13]. Subsequent research by Weinstein established the reach of interference on the X chromosome. In this case, interference does not manifest when the distance between the two intervals being studied is greater than 46 cM [14]. Additionally, COs in intervals on the same chromosomal arm interfere more strongly than intervals on opposite arms that have approximately equivalent genetic distances [14]. This introduced the idea that interference does not cross the centromere. In 1932, Graubard observed that chromosome 2 carrying an inversion (max size 25 cM) did not affect interference values of intervals on that chromosome [15]. This result was the first to suggest that pre-existing chromosomal features are not the primary determinant of interference, which is widely believed to this day.
Model fungal organisms that form tetrads (fused meiotic products), allow for both the recovery of non-reciprocal recombination products and novel statistical approaches for interference analysis [16-18]. Non-Mendelian 6:2 segregation (the result of post-meiotic mitotic division of a 3:1 tetrad) at a single locus, indicative of a GC, was first observed in spore pigmentation mutants of Bombardia lunata [18]. These observations led researchers to question if GCs were born from the same mechanism that produces COs. If true, this would predict that they should exhibit interference. Using Neurospora crassa as a model system, Stadler tested this idea by seeing if GC events interfered with the probability of COs in an adjacent interval. He found that GCs did not interfere with COs and (errantly) concluded that GCs and COs arose from different precursors [19]. Subsequent research by Mortimer and Fogel demonstrated that GCs occur with or without flanking marker exchange establishing the idea of NCO and CO repair [20]. These data suggested that all recombination events have a common molecular basis (which we now know to be DSBs and strand invasion [6, 7]) and that initial events are distributed independently of one another, but that the occurrence of a CO at one site will subsequently influence nearby events to be resolved as NCOs, thus resulting in a CO distribution that displays interference [20].
UNANSWERED QUESTIONS
What Types of Events Interfere with One Another?
Interference is often referred to as ‘CO interference’, which does not capture the breadth of the phenomenon, since COs may not be the only recombination-related events to inhibit one another’s distribution. Determining what combinations of recombination events are subject to interference is key to understanding both when interference is imposed as well as the mechanism that mediates it.
DSB-DSB Interference
Interestingly, in many organisms, not even all COs are subject to interference (discussed below). However, those COs that do interfere could theoretically be a reflection of an inhibition of closely spaced DSBs. Whether or not, and to what degree DSBs interfere with one another remains an open question. Meiotic DSB mapping studies in S. cerevisiae have resulted in a detailed understanding of where DSBs are most likely to occur [21-24], but they are not ideal for addressing whether or not DSBs are subject to interference because DSB mapping is an amalgamation of thousands of independent meioses so even if the DSBs in individual meioses were subject to interference, it would likely be obscured by the layering of data. It may be relevant that the hottest DSB hotspots only have a break in ~10% of meioses [21] and periodicity in DSB hotspot distribution has never been reported. However, in S. cerevisiae, researchers have clearly shown competitive DSB inactivation whereby insertion of a strong DSB hotspot reduces the frequency of DSB formation and recombination in nearby regions [25, 26]. Ohta et al. have reported that insertion of an artificial DSB hotspot results in a parallel decrease of DSBs in a ~60 kb region around the insertion site [25]. This led to the proposal that strong DSBs sites could outcompete nearby sites for limiting factors essential for DSB formation. Another proposal is that structural features that influence DSB formation create domain boundaries that isolate regions from one another [25].
DSB distribution has also been addressed via mapping the physical position of structures called recombination nodules (RNs). RNs are proteinaceous structures that are associated with the axial elements of the synaptonemal complex (SC) from leptotene to pachytene and are thought to be locations where meiotic recombination reactions are occurring [27]. RNs are divided into two sub-classes: early nodules (ENs) and late nodules (LNs), which differ in respect to timing, size, shape and number. ENs, the smaller of the two, roughly correspond to Rad51/Dmc1 foci [28], while LNs, which most likely arise from a fraction of ENs, are thought to be the molecular machinery that execute COs and thus represent CO sites [27, 29]. In tomato, both ENs and LNs exhibit a distribution indicative of interference, but the strength of interference is much stronger among LNs [30, 31].
Analysis of the distribution of MSH4 foci in mouse meiocytes strongly supports the idea that DSBs interfere with one another. In zygotene, ~150 MSH4 foci are initially detected, which reduces to ~50 foci by late pachytene. Early MSH4 foci co-localize with RAD51/DMC1 and are thought to mark all DSBs in the early repair stages, while late MSH4 foci co-localize with MLH1 foci and are thought to mark CO sites only [32-34]. Early MSH4 foci exhibit a distribution indicative of interference, but do not exhibit as strong interference as MLH1 foci [31, 35, 36].
Taken together the above results argue that interference is the result of multiple layers of control [31, 32, 36]. DSBs may exhibit positive interference over short distances, but DSB-DSB competitive interaction would not be sufficient to explain CO interference over megabase (Mb) distances.
NCO-NCO and NCO-CO Interference
Another observation that suggests interference is not merely a reflection of an underrepresentation of closely spaced DSBs is that in S. cerevisiae, NCOs do not interfere with other NCOs. Mortimer and Fogel reported that GC events at ARG4 and THR1 (~19 kb apart) do not interfere with one another since alleles at the two loci co-converted at a frequency that is indistinguishable from independence predicted by their individual conversion frequencies. More recently, analysis of genome-wide recombination maps based on DNA tiling arrays reaffirmed that the distance between GCs not associated with COs does not differ significantly between experimental samples and randomized control data [37].
Whether or not COs interfere with NCOs (or vice versa) is a more controversial topic. The first results to address this issue came from S. cerevisiae when Mortimer and Fogel showed that a GC at HIS1 or ARG4 with exchange of flanking markers (CO) results in a decrease of genetic distance in an adjacent genetic interval, whereas a GC without exchange of flanking markers (NCO) actually promoted CO frequency in the adjacent interval (negative interference) [20]. The idea that NCOs do not interfere with COs was supported by Malkova et al. who showed that, at the met13 locus of S. cerevisiae, GCs without an accompanying CO did not exert positive interference on adjacent intervals, while GCs associated with a CO did [38]. However, they did not observe statistically significant negative interference between GCs not associated with CO at met13 and COs. Recently, Getz et al. showed that NCO GCs did not exert interference on adjacent intervals and the map distances in those intervals were actually increased indicative of negative interference, in support of the idea that NCOs do not positively interfere with COs [39]. However, most recently, a contradictory result showing that NCOs and COs interfere with one another was recently presented by Mancera et al. who, using the genome-tiling method, showed that inter-event distance between NCO and CO events were on average ~13kb larger than expected from a random distribution, representing statistically significant, albeit weak, positive interference [37]. While locus-by-locus studies have consistently shown negative or no interference between NCOs and COs, this genome-wide approach showed the opposite. It is difficult to reconcile these contradictory results, but because the Mancera data were generated using a genome-wide analysis the advantage appears to be with the idea that COs and NCOs exhibit positive interference. Nonetheless, it is interesting to note that models in which DSBs that are resolved as COs influence nearby DSBs to be resolved as NCOs predict negative interference between COs and NCOs. One reconciliatory possibility is that there exist both interfering NCOs and non-interfering NCOs (as in COs, see below) and the single locus studies mentioned above happened to measure only the latter class.
CO-CO Interference and Non-Interfering COs
In many organisms, there are at least two pathways for producing COs. Arabidopsis thaliana, humans, mouse and S. cerevisiae have one pathway constituting the majority of COs that is sensitive to interference and a secondary pathway that produces interference-insensitive (randomly distributed) COs [40]. In these organisms, primary pathway COs are characterized by the Msh4-Msh5 heterodimer while secondary pathway COs are dependent on the Mus81-Eme1 heterodimer [40]. In S. cerevisiae, msh4Δ or msh5Δ deletions have a ~60% reduction in COs and the remaining COs are interference insensitive [41-43], while mus81Δ or mms4Δ (eme1) deletions have a ~25% reduction in COs and the remaining COs are sensitive to interference [40, 41]. In Arabidopsis, an analogous situation exists where interference-sensitive COs mediated by the Msh4-Msh5 pathway make up ~80-85% of the total while interference insensitive COs mediated by the Mus81-Eme1 pathway make up ~15-20% of the total [44-47]. Not all organisms produce both interfering and non-interfering COs. Schizosaccharomyces pombe does not have CO interference and ~80-95% of its COs are dependent on Mus81-Eme1 [48, 49]. In contrast, Caenorhabditis elegans has ‘perfect’ interference (exactly one CO per bivalent) and all COs are dependent on Msh4-Msh5 [50]. D. melanogaster is not thought to produce interference-insensitive COs and is thought to exhibit absolute interference across distances < ~10 cM [51]. A comparison of interference patterns in different organisms is presented in Table 1.
Table 1.
CO Interference Comparisons Across Model Genetic Organisms. Haploid chromosome number (n) and presence or absence of CO interference is noted. Also shown are presence or absence of Msh4-Msh5 (interference-sensitive) and Mus81-Eme1 (interference-insensitive) mediated CO pathways
| Organism | N | Interference? | Msh4-Msh5 COs? | Mus81-Eme1 COs? | ~COs/meiosis |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae [24, 37, 40, 41, 43] | 16 | Yes | Yes | Yes | 90 |
| Schizosaccharomyces pombe [48, 89] | 3 | No | No | Yes | 38 |
| Neurospora crassa [75, 78, 90] | 7 | Yes | n.d. | n.d. | 20 |
| Aspergillus nidulans [91] | 8 | No | n.d. | n.d. | n.d. |
| Caenorhabditis elegans [92] | 6 | Yes | Yes | No | 6 |
| Arabidopsis thaliana* [44-47, 82, 93] | 5 | Yes | Yes | Yes | 10 |
| Lycopersicon esculentum [30] | 12 | Yes | n.d. | n.d. | 21 |
| Zea mays [94, 95] | 20 | Yes | n.d. | n.d. | 20 |
| Drosophila melanogaster [27, 51, 78] | 4 | Yes | No | No | 6 |
| Danio rerio*[96, 97] | 25 | Yes | n.d. | n.d. | 25-40 |
| Mus musculus*[31, 32, 35, 98-100] | 20 | Yes | Yes | Yes | 22-28 |
| Homo sapiens*[32, 34, 98, 101-106] | 23 | Yes | Yes | Likely | 50-70 |
n.d. = no data.
Reported differences between male and female COs/meiosis.
While it is known that both interfering and non-interfering COs can be produced in the same cell, the mechanistic difference between the two and specifically why one class exhibits interference and the other does not is unknown. One possibility is based on the work in Arabidopsis by Franklin et al. in which they found that, during leptotene, ~115 AtMUS81 foci form on chromosome axes and many of these co-localize with AtRAD51 and AtMSH4 foci, each of which form 80-100 foci during leptotene/early zygotene [47]. By early pachytene, the number of AtMUS81 foci drops precipitously to ~5. This suggests a ‘toolbox hypothesis’ where Mus81-Eme1 and Msh4-Msh5 are recruited to all DSBs and most are repaired via the Msh4-Msh5 pathway, while Mus81-Eme1 acts to resolve a subset that may consist primarily of aberrant joint molecules (JMs) as either COs or NCOs that could not be repaired using Msh4-Msh5. This could result in interfering and non-interfering COs (and perhaps non-interfering NCOs) if the Msh4-Msh5 pathway is subject to interference, but the smaller and randomly distributed population of aberrant JMs resolved as COs by Mus81-Eme1 acts later, after interference has already been established. In congruence with this idea, recent biochemical and genetic analysis of mus81 mutants in S. cerevisiae indicates that Mus81-Eme1 acts late in meiotic recombination and likely resolves aberrant JMs that cannot be resolved by the primary Msh4-Msh5 pathway [52, 53] (Fig. 1). Another related possibility is that primary CO reactions occur first and initiate an ‘interference signal’ (discussed below), while secondary COs take longer to process, as they must first be bypassed by the primary pathway. In this model, secondary COs do not produce an interference signal, and are resolved after the interference signal has been imposed.
An alternate hypothesis regarding the difference between interfering and non-interfering COs was introduced by Getz et al., who proposes two phases of COs. Early ‘pairing phase’ COs are non-interfering and late ‘disjunction phase’ COs, that are dependent on MSH4, exhibit positive interference [39]. In addition to being independent of MSH4, pairing phase COs are hypothesized to be less proficient at repairing mismatches. The hypothesized existence of MSH4-independent ‘pairing phase’ COs in S. cerevisiae is consistent with the lack of non-interfering (Mus81-dependent) COs in C. elegans and Drosophila, because these organisms do not use COs to pair their chromosomes [54-56]. This model is also compatible with the phenotype of ndj1 mutants, which have decreased interference and increased rates of nondisjunction [57, 58], which can be explained by increased pairing COs at the expense of disjunction phase COs. Since pairing phase COs are proposed to be interference insensitive it follows that they should be MUS81-dependent, which predicts that mus81 and eme1 mutants will be pairing defective. In yet another layer of complexity the toolbox and two-phase hypotheses are not mutually exclusive as there could be pairing phase non-interfering COs that have nothing to do with MUS81 as well as non-interfering disjunction phase COs produced by MUS81 that are reflective of MSH4-independent aberrant JM resolution.
What is the Timing of Events Leading to Interference?
Determining the timing of events that lead to interference is extremely challenging since diverse cellular processes likely play a role. Chromatin structure (e.g. nucleosome density, protein-DNA complexes, histone modifications etc.) and steric features of the chromosomes could influence recombination complex spacing. These features are not constant along chromosomes and are dynamic in both the mitotic and meiotic cell cycles. Meiotic chromosome condensation, which begins at the start of meiotic prophase and does not end until after recombination is complete, also likely influences interference [1, 29, 59]. However, pre-recombination chromosomal features are not the only important determinants. In many organisms, mutations in the meiosis-specific ZMM (ZIP, MSH, MER) recombination genes, which act after SEI formation, result in the abolition of interference. This strongly suggests that the assembly and distribution of recombination complexes is critical for the timing of the imposition of interference.
One attractive proposal is that interference is imposed during strand invasion when Msh4-Msh5 complexes stabilize CO-specific (SEI) recombination intermediates [60]. This idea is based on the observations that msh5 ndt80 mutants result in very low levels of JM accumulation along with absence of interference in S. cerevisiae [61], but spo16 ndt80 mutants, which are defective for synaptonemal complex (SC) extension, result in high JM accumulation but wild-type interference [60]. The ndt80 mutation was used in this case because it removes the late pachytene checkpoint and results in accumulation of recombination intermediates [60]. The spo16 mutant also offers important insight as to the latest interference could be acting. Because spo16 mutants are defective for SC extension and yet have wild-type interference it is likely that interference is fully implemented before late leptotene/early zygotene when the SC is formed [1]. Supporting this idea is the observation that SC initiation complexes exhibit a distribution indicative of interference [62]. Additional support for the idea that interference involves regulation of the strand invasion step comes from analysis of the tid1 mutant in S. cerevisiae [63]. Tid1 is an accessory factor that facilitates strand invasion and the tid1 mutant displays ~wild-type levels of COs yet interference is significantly weakened [63].
The timing of events leading to interference is different in Drosophila and C. elegans, in which pairing and synapsis occurs prior to the initiation of recombination. Thus, interference in these organisms is likely implemented after (though not dependent on) SC formation.
Do COs Influence Nearby DSBs to be Repaired as NCOs?
Analysis of the ZMM mutant phenotypes has led to the early decision model, in which the commitment of a DSB to be repaired as either a NCO or CO is made at, or prior to, stable SEI formation [10, 64]. How this decision is enforced is unknown. ZMM mutants are strongly CO defective, have abolished or greatly reduced interference, yet are not defective for NCO formation [42, 65, 66]. 2-D gel analysis of DNA intermediates has shown that the kinetics of DSB and NCO repair are normal in ZMM mutant backgrounds, but stable SEI production is inhibited. This observation strongly suggests that ZMM genes are not required for designation of DSB repair as either COs or NCOs and that this designation is made prior to SEI formation [10, 64]. NCO/CO designation is a key determinant of CO distribution and thus interference. It is important to determine if designation of a CO at one site subsequently results in NCO designation of nearby DSBs, which is an attractive, but as yet unproven, proposal. The other possibility is that all DSBs are designated as either COs or NCOs independently of one another in a distribution reflective of CO interference.
What is the Role of PCH2 in the Mediation of CO Interference?
Recent mutant analyses strongly implicate the AAA+ ATPase Pch2 as an important regulator of CO interference, however the pathway by which Pch2 mediates interference is unknown. Two independent studies in S. cerevisiae showed that pch2∆ mutants exhibit significantly weakened interference at several loci [67, 68]. Interestingly, both groups reported no significant changes in CO frequency on chromosome III (the shortest S. cerevisiae chromosome) indicating that the processes of CO formation and CO interference can be decoupled, at least on short chromosomes. However, the two studies presented incongruent results regarding CO frequency on other chromosomes in that Joshi et al. report no significant changes in CO frequency at any loci [67] while Zanders and Alani report significant increases in CO frequency on medium and large chromosomes [68].
pch2∆ mutants display elevated Zip3 foci (an early marker of CO designated sites), aberrantly diffuse Hop1 localization and increased SC length [67]. In light of these findings, the researchers propose a short-range interference model (discussed further below) in which Pch2 aids in the establishment of chromosomal domains in which only one CO can occur [67]. In addition to increased CO frequency, pch2∆ mutants display no increase in DSBs and exhibit elevated CO:NCO ratios at two GC loci, compared to wild-type [68]. Importantly, the researchers rule out excess COs as the explanation for weakened interference in pch2∆ since pch2∆; Spo11-hypomorph double mutants display ~74% of the COs of pch2∆ while maintaining the interference defect. Additionally, pch2∆; mms4 double mutants have significantly higher CO frequency than mms4 mutants, which one would not see if the interference defect and CO frequency increase was solely due to elevated secondary pathway (Mus81-Mms4 dependent) COs in pch2∆ [68]. To explain the pch2∆ phenotypes of increased CO frequency, increased CO;NCO ratio and weakened interference, the researchers propose that Pch2 acts to repress the CO designation at the CO/NCO bifurcation in the decision pathway [68]. While PCH2 very likely plays an important role in interference, this gene is a piece in the puzzle since a) the interference defect in pch2∆ is not present at all loci that display interference [67] b) pch2∆ mutants have residual positive interference [67, 68] and c) the interference defect in is most prominent in 50-100kb distances and can be mitigated at lower temperatures [67].
Do Meiosis-Specific Cohesins Mediate Interference?
In S. cerevisiae, the meiotic cohesin complex consists of Scc3, Smc1, Smc3 and Rec8, all which are also present in the mitotic cohesin complex except Rec8 [69, 70]. In addition to its role in sister chromatid cohesion, Rec8 has been implicated in a diverse set of meiotic processes including pairing, SC polymerization, recombination, and disjunction [70, 71]. Many of these functions have been shown to be separable from its role in sister chromatid cohesion [71]. In S. cerevisiae and S. pombe, Rec8 has a role in the binding of Spo11 to DSB sites in a region-specific manner [72, 73]. ChIP-chip reveals an interesting correlation between binding of Spo11 and cohesins. Spo11 has been shown to co-localize with Rec8 in early meiosis, and the frequency of co-localization decreases as meiosis progresses [73]. It has been proposed that Rec8 provides structural landmarks that dictate the proper distribution of Spo11 [73]. Spo11 could transfer from Rec8 binding sites to chromatin loops before initiating DSBs [73]. Since Rec8 has a role in dictating DSB distribution, a role in the establishment of the weak DSB-DSB interference discussed earlier is possible. The Rec8 binding landscape could serve to bias Spo11 distribution toward uniformity. However, a role for Rec8 in the mediation of interference has not yet been demonstrated, so additional work to elucidate this relationship will have to be conducted.
MODELS
The central unanswered question regarding interference is how it is achieved within the cell. The answer to this question has been elusive partly because traditional genetic screens designed to discover interference mutants are labor-intensive and problematic. Isolating the interference machinery is difficult because the relevant players likely have overlapping roles with other critical meiotic processes. Most mutations that affect interference also affect CO frequency and only a handful of mutants has been identified (all in S. cerevisiae) that exhibit an interference defect without a concurrent hypo- or hyper-CO phenotype [57, 63, 67]. Further complicating matters is that many of these mutants behave differently with regards to CO frequency in different studies and within studies at different loci [39, 57, 63, 67, 68, 74]. Additional complications arise from the fact that interference is not absolute in most species but instead reflects the reduced probability of an event in a population of events – making the implementation of efficient screens difficult. Models aimed at explaining the interference mechanism have been proposed, but there is no dominant paradigm as no empirical observations vastly favor one particular explanation. Several commonly referenced models are discussed below.
Mechanical Stress Model
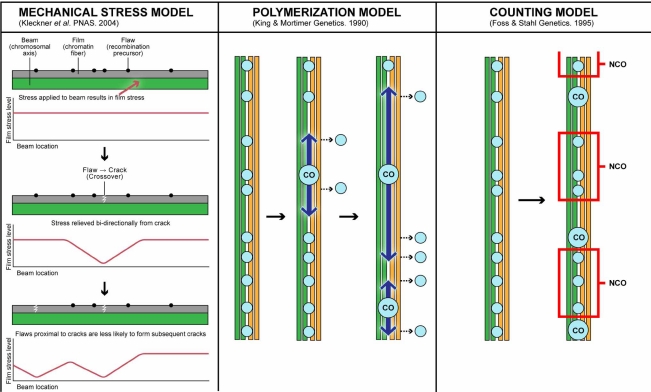
Because interference is maximized at close distances and decreases with increasing distance [3], interference models that rely on a mechanical explanation invoke a signal that spreads from CO sites. Muller’s original interference model posited that the stiffness of a chromosome would make it difficult to bend back on itself after a CO to form another in close proximity [13]. The modern mechanical stress model proposed by Kleckner et al., is based on the idea that in many physical systems, any increase or decrease in stress starts at a locus and propagates outward from that point (Fig. 2) [59]. Stress is generated as meiotic chromosomes compress and expand. COs result in a localized relief of this stress that spreads in both directions down the axis of the SC. This model is attractive because it posits a simple explanation that predicts many properties of interference including CO assurance and CO homeostasis (discussed below). As each chromosome will be under stress, the occurrence of a first event, CO assurance, is easily obtained. Each event defines a domain of inhibition resulting from stress relief spreading from a CO, which acts as an inhibitory signal. Chromosomal features such as centromeres could act as sinks that absorb the signal, which would explain how interference does not cross the centromere. Multiple events could occur on the same chromosome, and could take place in different locations in different nuclei, but would always have the tendency to be evenly spaced. A mathematical simulation of this model has been used to fit CO data from two species [59]. However, the stress model does not make easily testable predictions. Additionally, it is easy to imagine how DSBs would relieve tensile stress, but less so for COs, which would be necessary for this model to explain all aspects of interference. Furthermore, it is also difficult to account for how some COs could mediate stress relief while others don’t which would be necessary to explain the data in organisms that appear to have both interfering and non-interfering crossovers.
Fig. (2).
Interference Models. The left panel depicts the beam-film demonstration of the mechanical stress model proposed by Kleckner et al. [59]. The beam (chromosomal axis; green), film (chromatin fiber; grey), flaws (CO precursors; black dots). Diagrams depicting the stress level are shown under each beam in which the x axis represents beam position and stress level on the y. The center panel depicts the polymerization model proposed by King and Mortimer [75]. Chromatids are shown in green (parent 1) and yellow (parent 2). Small light blue circles represent recombination precursors and CO designates are shown as larger circles marked with ‘CO’. The interference polymer is shown as a large arrow emanating from CO sites, and CO precursors removed by the polymer are shown to the right accompanied with a dashed arrow. The right panel depicts the counting model proposed by Foss et al. [78]. Chromatids are shown in green (parent 1) and yellow (parent 2). Small light blue circles represent recombination precursors and CO designates are shown as larger circles marked with ‘CO’. In this diagram, m=3 and intervening NCOs between COs are outlined in a red box.
An additional layer to the mechanical stress model was put forward to explain the role of Pch2 in the mediation of short-range (< 100 kb) CO-interference [67]. Joshi et al. propose a one-CO module hypothesis in which Pch2 aids in the establishment of domains that tile chromosomes and incur one and only one CO per module [67]. Mechanical stress within each module promotes a single CO that that relieves stress within the module. Key features of one-CO modules, presumably mediated by Pch2, are a centrally located Zip3 focus along with Hop1 hyper-abundance that extends to the edges of the domain, establishing the reaches of short-range interference [67]. The hypothesized contents of the modules are based on the observation that pch2∆ mutants, along with weakened interference, display diffuse rather than domainal Hop1 staining and aberrant increased Zip3 foci [67]. The one-CO module model is capable of explaining interference across organisms with larger or smaller interference reach by varying the size of modules. For example, in C. elegans, which incurs exactly one CO per bivalent, each chromosome could be encompassed in entirety by one module. The one-CO module model does not explain interference over >100 kb distances in S. cerevisiae since in the pch2∆ mutant, where module establishment is presumed to be impaired, interference is unaffected in distances over 100 kb.
Polymerization Model
The polymerization model describes a situation where early recombination structures are distributed independently of one another and then have an equal chance per unit time of initiating a bi-directional polymerization event (Fig. 2) [75]. This polymer spreads from the site of initiation and has the ability to block additional early structures from binding to the bivalent. Sites of initiation are hypothesized to mature into LNs (COs) leading to chiasmata. This model is attractive because it explains interference and assurance while predicting a pattern in which interference is strongest nearest to initiation events with decreasing strength in a distance-dependant manner. A computerized simulation of the parameters described in the polymerization model was fit satisfactorily to CO data from Drosophila and S. cerevisiae [75].
Part of the rationale behind the polymerization model was that an optimal interference model should be useful in systems that differ by several orders of magnitude in genome size in bp. Physical distances measured in SC lengths rather than bp are much closer among species, thus the polymer is proposed to move down the axis of the SC. The idea that interference mediated over physical distance would be measured in SC length and that the interference signal is propagated along the SC axis is attractive because organisms of vastly different size genomes can be normalized by modifying the size of DNA loops that are associated with the SC axis. The main obstacle to the polymerization model is that the polymer itself has neither been identified nor observed. However, the search for a polymer as the signal may be a red herring as the signal could be a modification such as phosphorylation, methylation, acetylation or ubiquitination of a protein such as a histone or cohesin. SC polymerization itself has been proposed as an attractive mediator of interference [76, 77]. This would have demonstrated a clear role for the SC while being consistent with the observation that S. pombe lacks both SC and interference. However, the possibility that the SC is required for interference is extremely unlikely since in S. cerevisiae SC extension has been shown to be dispensable for interference [60] and in mouse, SC defective mutants have normal interference [31]. Lastly, in refutation of the idea that the SC is in any way required for interference, is that in Drosophila and C. elegans the SC seems to be complete before DSBs are formed.
Counting Model
The counting model is a mathematical construct in which COs are separated by a fixed number (m) of intervening NCO events (Fig. 2) [78]. It was proposed in part to reconcile the fact that, in terms of physical distance (bp of DNA or µm of SC), strength of interference varies by several orders of magnitude from organism to organism [78, 79]. The counting model can account for vast differences in genome size, as interference is dictated by genetic distance i.e. the initial density of precursors and the number of intervening events between COs. CO data from Drosophila and N. crassa fit the counting model predictions extremely well [78], but initially it was less successful at modeling CO distributions in S. cerevisiae and humans [79]. Additionally, m appeared to vary in the same organism between sexes and chromosomes [80]. Subsequently, a modified version of the counting model that allows a number of non-interfering COs (v) was suggested. This additional parameter allowed the counting model to satisfactorily fit CO data from S. cerevisiae [81], A. thaliana [45, 82], and humans [80] and in each of these systems, it has been shown that non-interfering COs exist [40, 44, 83].
Proposed biological equivalents for each parameter in the counting model include DSBs as precursors and NCOs as intervening events (m), however, the notion that DSBs are what is being ‘counted’ is not supported by subsequent observations. A prediction of the counting model when DSBs are ‘counted’ is that when the overall number of DSBs are reduced, COs and NCOs should reduce proportionally. It then follows that larger distances between COs should result. However, recent results failed to meet this expectation [84]. Cells appear to have a mechanism (called CO homeostasis) that ensures a certain number of COs per meiosis even if the pool of DSBs from which they arise is reduced. A series of spo11 hypomorphic mutants that produce DSBs in decreasing frequencies compared to wild-type do not show proportional reductions in COs [84]. Instead, these mutants maintain CO levels at the expense of NCOs, which are reduced proportionally to the reduction in DSBs. CO interference is maintained at wild-type levels in all spo11 hypomorphic backgrounds [84]. CO homeostasis is seen as a major obstacle to the counting model. However, if DSBs were not what is counted, but instead a factor that establishes DSB sites, the counting model could still be possible. Despite its clear predictions and modeling support, the counting model suffers from an absence of both in vivo evidence as well as a testable molecular mechanism for its execution.
Other Models
Interference models can be described in terms of point-process in which precursors i.e. ‘points’ are distributed according to a mathematical function and then COs are ‘processed’ from these points according to a second mathematical function [85]. The ‘hard-core’ model is a point-process variant where points are dispersed in a Poisson distribution with a minimum physical distance between any two [85]. At each point, chromatids have a 50% chance of being involved in a CO. The hard-core model is much like a situation where DSB-DSB interference is the driver of CO interference, which current data strongly suggests is not the case (discussed above). This model does not conform particularly well with CO data from Drosophila [85].
Another interesting proposal involves a ‘chiasma determining mechanism’ that moves along the bivalent [78, 86]. This mechanism is hypothesized to move along the bivalent at a constant rate occasionally firing and thus determining CO sites that mature into chiasmata. After firing, the mechanism requires time to recharge while still moving, resulting in CO interference [86].
CONCLUSIONS
How is Interference Imposed?
Interference modeling has traditionally been concerned with the synthesis of models capable of explaining CO interference across many organisms with vastly different genome sizes and recombination rates. Beyond this, accurate models must be able to reconcile additional complex properties of interference such as multi-level control, non-interfering COs and early decision. Even what falls under the purview of the term interference is currently in question. In addition to the strong likelihood that recombination events other than COs interfere, it is also possible that both CO homeostasis and CO assurance could be products of an over-arching CO control mechanism that results in the lack of closely spaced COs. While the above models can be altered to account for these complexities, the truth likely lies in combination of the ideas that have been proposed to explain interference.
Since a growing body of evidence supports the idea that interference is imposed at multiple levels, a comprehensive interference model will account for the presence of DSB-DSB interference, CO-CO interference and possibly CONCO interference. Although it remains unclear whether or not and to what degree DSBs interfere with one another, it is possible that DSB-DSB interference acts on a smaller scale and is mediated by multiple factors. First, the pattern of Spo11 distribution may be established by the meiosis-specific cohesin subunit Rec8. Rec8 has been shown to colocalize with Spo11 in S. cerevisiae, and rec8∆ mutants exhibit a drastic alteration in Spo11 distribution [73]. It has been speculated that Rec8 not only provides landmarks along the chromosomal axes that guide the distribution of Spo11, but it is also responsible for the transition of Spo11 from the axes to the loops, where breaks are subsequently formed [73] Rec8 could preferentially bind certain locations within the chromatin context of the chromosomal axis with intervening DNA loops, resulting in a minimum physical distance between subunits. This idea requires that only one DSB could be formed at each site of Rec8 localization, which would result in the minimization of closely spaced DSBs thus establishing a pattern of DSB-DSB interference on a small scale. Secondly, DSBs could recruit limiting break forming factors away from nearby sites [25].
In addition to small-scale DSB-DSB interference, we are intrigued by the possibility that CO-CO interference in S. cerevisiae and A. thaliana is mediated by an ‘interference signal’ that is initiated by the stabilization of SEIs by ZMM proteins. This hypothetical interference signal propagates either down the SC axial elements or the cohesin axis. A signal propagated down the axis rather than the DNA itself is attractive because it allows interference to act over very large distances of linear DNA by varying the DNA bp loop/axis ratio. One-CO modules can be incorporated as domains established on the chromosomal axes that create favorable conditions for signal propagation. Importantly, the SC itself is not required for interference in this model. Additionally, if we suppose that the amount of DNA traveled by the signal is dependent on the condensation of the chromosome, it could explain the ‘lack’ of interference in early pairing phase COs and ‘presence’ of interference in disjunction phase COs. COs could occur at various time points in the meiotic program and thus over a large range of states of chromosome condensation. The earlier a (interference-sensitive) CO is designated, the less it will interfere (and vice versa) due to the continual condensation of the chromosomes during the meiotic program. This idea predicts that if CO formation can be in some way be delayed, interference will strengthen, a hypothesis that can be most effectively tested in an organism (such as Drosophila), which has a single interference-sensitive CO pathway.
What is the nature of the interference signal? An intriguing and testable explanation is that a protein modification that is propagated from the majority of COs results in the inhibition of primary-pathway crossing over at sites where this modification spreads. The modified protein(s) could include axial elements, cohesins, or histones. This idea predicts that CO-CO interference can be largely eliminated by a mutation that blocks the spread of the inhibitory modification via the receiver or the modifier. Rec8 is a particularly interesting target since phosphorylation of Rec8 is important for many of its meiotic roles. Two rec8 mutants with mutations at multiple phosphorylation sites exhibit disrupted synapsis and have delayed production of mature recombinants [71]. Since Rec8 phosphorylation is required for recombination in S. cerevisiae, the protein modification could be a dephosphorylation of Rec8 that disables crossing over. A second possibility is de-methylation of H3K4me3, which has been shown to mark sites of meiotic recombination [87]. Thirdly, modification of axial-element protein Hop1, which is localized in discrete hyper-abundant domains on zygotene chromosomes, is an interesting possibility. pch2∆ mutants abolish domainal localization of Hop1 while concurrently weakening interference [67].
An alternate model for the interference signal is based on the recent observation that CO hotspots are significantly correlated with DNA methylation [88]. This group proposed that areas undergoing recombination could be secondarily methylated, which could result in the inhibition of further COs in that area [88]. Another possibility is that genomic regions that are methylated are preferential sites of recombination. These possibilities are not mutually exclusive and make testable predictions regarding local DNA methylation states prior to and after recombination has taken place.
Final Thoughts
As the initial report of interference reaches the century mark, some questions regarding how it works are in reach. These questions can be posed within the hypothesis that ZMM proteins stabilize CO intermediates at strand invasion (SEI), which are then committed to an interference-sensitive CO pathway. Does the stabilization of SEI complexes activate a spreading interference ‘signal’ or are CO-designated events evenly spaced to begin with? Do COs occur in two phases, one interfering and the other not? Does Rec8/Spo11 distribution play a role in interference? Does Mus81-Eme1 mediate the resolution of only aberrant recombination intermediates or does it also play a role in mediating a subset of traditional substrates? These questions and many more will have to be answered in order to solve the interference puzzle.
ACKNOWLEDGEMENTS
LEB and GPC thank NSF (MCB-0618691) and DOE (DE-FGO2-05ER15651) for financial support. We would also like to thank Drs. Corbin Jones, Jeff Sekelsky and Frank Stahl for critical comments. We also thank The University of North Carolina at Chapel Hill's Office of the Vice Chancellor for Research and Economic Development for providing support for Open Access publication.
ABBREVIATIONS
- CO
= Crossover
- dHJ
= Double Holliday Junction
- DSB
= Double-Strand Break
- DSBR
= Double-Strand Break Repair
- EN
= Early Nodule
- GC
= Gene Conversion
- HJ
= Holliday Junction
- JM
= Joint Molecule
- LN
= Late Nodule
- NCO
= Non-Crossover
- PMS
= Post-Meiotic Segregation
- RN
= Recombination Nodule
- SC
= Synaptonemal Complex
- SDSA
= Synthesis-Dependant Strand Annealing
- SEI
= Single-End Intermediate
- ssDNA
= Single-Stranded DNA
- ZMM
= ZIP, MSH, MER recombination genes
REFERENCES
- 1.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 2.Sturtevant AH. A Third Group of Linked Genes in Drosophila Ampelophila. Science. 1913;37:990–992. doi: 10.1126/science.37.965.990. [DOI] [PubMed] [Google Scholar]
- 3.Sturtevant AH. The behavior of chromosomes as studied through linkage. Z. Induct. Abstammungs-Vererbungsl. 1915;13:234–287. [Google Scholar]
- 4.Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 5.Jones GH. The control of chiasma distribution. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 6.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 7.Sun H, Treco D, Szostak JW. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 8.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 10.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 11.Stahl FW, Housworth EA. In: Methods for Analysis of Crossover Interference in Saccharomyces cerevisiae. in Meiosis volume 1. Housworth EA, editor. Vol. 1. New York: Springer; 2009. pp. 35–53. [DOI] [PubMed] [Google Scholar]
- 12.Muller HJ, Jacobs-Muller JM. The Standard Errors of Chromosome Distances and Coincidence. Genetics. 1925;10:509–524. doi: 10.1093/genetics/10.6.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller HJ. The Mechanism of Crossing Over. Am. Nat. 1916;50:193–434. [Google Scholar]
- 14.Weinstein A. Coincidence of Crossing over in Drosophila melanogaster (Ampelophila) Genetics. 1918;3:135–172. doi: 10.1093/genetics/3.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graubard MA. Inversion in Drosophila melanogaster. Genetics. 1932;17:81–105. doi: 10.1093/genetics/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogel S, Hurst DD. Meiotic gene conversion in yeast tetrads and the theory of recombination. Genetics. 1967;57:455–481. doi: 10.1093/genetics/57.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell MB. Aberrant recombination of pyridoxine mutants OF Neurospora. Proc. Natl. Acad. Sci. USA. 1955;41:215–220. doi: 10.1073/pnas.41.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zickler H. Genetische untersuchungen an einem heterothallischen askomuzeten (Bombardia lunata nov. spec.) Planta. 1934;22:574–613. [Google Scholar]
- 19.Stadler DR. The Relationship of Gene Conversion to Crossing over in Neurospora. Proc. Natl. Acad. Sci. USA. 1959;45:1625–1629. doi: 10.1073/pnas.45.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortimer RK, Fogel S. In: Genetical Interference and Gene Conversion. in Mechanisms in Recombination. Grell R.F, editor. New York: Plenum; 1974. pp. 263–275. [Google Scholar]
- 21.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 24.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohta K, Wu TC, Lichten M, Shibata T. Competitive inactivation of a double-strand DNA break site involves parallel suppression of meiosis-induced changes in chromatin configuration. Nucleic Acids Res. 1999;27:2175–2180. doi: 10.1093/nar/27.10.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter AT. Electron microscopy of meiosis in Drosophila melanogaster females: II. The recombination nodule--a recombination-associated structure at pachytene? Proc. Natl. Acad. Sci. USA. 1975;72:3186–3189. doi: 10.1073/pnas.72.8.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson LK, Offenberg HH, Verkuijlen WM, Heyting C. RecA-like proteins are components of early meiotic nodules in lily. Proc. Natl. Acad. Sci. USA. 1997;94:6868–6873. doi: 10.1073/pnas.94.13.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 30.Anderson LK, Hooker KD, Stack SM. The distribution of early recombination nodules on zygotene bivalents from plants. Genetics. 2001;159:1259–1269. doi: 10.1093/genetics/159.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA. 2006;103:9607–9612. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 33.Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 2000;14:1539–1547. doi: 10.1096/fj.14.11.1539. [DOI] [PubMed] [Google Scholar]
- 34.Neyton S, Lespinasse F, Moens PB, Paul R, Gaudray P, Paquis-Flucklinger V, Santucci-Darmanin S. Association between MSH4 (MutS homologue 4) and the DNA strand-exchange RAD51 and DMC1 proteins during mammalian meiosis. Mol. Hum. Reprod. 2004;10:917–924. doi: 10.1093/molehr/gah123. [DOI] [PubMed] [Google Scholar]
- 35.de Boer E, Dietrich AJ, Hoog C, Stam P, Heyting C. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J. Cell Sci. 2007;120:731–736. doi: 10.1242/jcs.003186. [DOI] [PubMed] [Google Scholar]
- 36.Mezard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: plants have their say. Trends Genet. 2007;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malkova A, Swanson J, German M, McCusker JH, Housworth EA, Stahl FW, Haber JE. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics. 2004;168:49–63. doi: 10.1534/genetics.104.027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Getz TJ, Banse SA, Young LS, Banse AV, Swanson J, Wang GM, Browne BL, Foss HM, Stahl FW. Reduced mismatch repair of heteroduplexes reveals "non"-interfering crossing over in wild-type Saccharomyces cerevisiae. Genetics. 2008;178:1251–1269. doi: 10.1534/genetics.106.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Argueso JL, Wanat J, Gemici Z, Alani E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics. 2004;168:1805–1816. doi: 10.1534/genetics.104.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. Global Analysis of the Meiotic Crossover Landscape. Dev. Cell. 2008;15:331–2. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de los Santos T, Loidl J, Larkin B, Hollingsworth NM. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics. 2001;159:1511–1525. doi: 10.1093/genetics/159.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins JD, Armstrong SJ, Franklin FC, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copenhaver GP, Housworth EA, Stahl FW. Crossover interference in Arabidopsis. Genetics. 2002;160:1631–1639. doi: 10.1093/genetics/160.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The Role of AtMUS81 in Interference-Insensitive Crossovers in A. thaliana. PLoS Genet. 2007;3:0001–0010. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higgins JD, Buckling EF, Franklin FC, Jones GH. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54:152–162. doi: 10.1111/j.1365-313X.2008.03403.x. [DOI] [PubMed] [Google Scholar]
- 48.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meneely PM, Farago AF, Kauffman TM. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics. 2002;162:1169–1177. doi: 10.1093/genetics/162.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinstein A. The Geometry and Mechanics of Crossing Over. Cold Spring Harb. Symp. Quant. Biol. 1958;23:177–196. doi: 10.1101/sqb.1958.023.01.022. [DOI] [PubMed] [Google Scholar]
- 52.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- 57.Chua PR, Roeder GS. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- 58.Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 59.Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover Assurance and Crossover Interference are Distinctly Regulated by the ZMM/SIC Proteins during Yeast Meiosis. Nat. Genet. 2008;40:299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 61.Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116:795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 63.Shinohara M, Sakai K, Shinohara A, Bishop DK. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 2003;163:1273–1286. doi: 10.1093/genetics/163.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 65.Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 66.Novak JE, Ross-Macdonald PB, Roeder GS. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 2001;158:1013–1025. doi: 10.1093/genetics/158.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joshi N, Barot A, Jamison C, Borner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanders S, Alani E. The pch2Delta mutation in baker's yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 2009;5:e1000571. doi: 10.1371/journal.pgen.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uhlmann F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 2003;13:R104–114. doi: 10.1016/s0960-9822(03)00039-3. [DOI] [PubMed] [Google Scholar]
- 70.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 71.Brar GA, Hochwagen A, Ee LS, Amon A. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol. Biol. Cell. 2009;20:1030–1047. doi: 10.1091/mbc.E08-06-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T, Shirahige K, Ohta K. Rec8 Guides Canonical Spo11 Distribution Along Yeast Meiotic Chromosomes. Mol. Biol. Cell. 2009;20:3064–76. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu HY, Burgess SM. Ndj1, a telomere-associated protein, promotes meiotic recombination in budding yeast. Mol. Cell. Biol. 2006;26:3683–3694. doi: 10.1128/MCB.26.10.3683-3694.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.King JS, Mortimer RS. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990;126:1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Egel R. Synaptonemal complex and crossing-over: structural support or interference? Heredity. 1978;41:233–237. doi: 10.1038/hdy.1978.92. [DOI] [PubMed] [Google Scholar]
- 77.Maguire MP. Crossover site determination and interference. J. Theor. Biol. 1988;134:565–570. doi: 10.1016/s0022-5193(88)80058-4. [DOI] [PubMed] [Google Scholar]
- 78.Foss E, Lande R, Stahl FW, Steinberg CM. Chiasma interference as a function of genetic distance. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foss EJ, Stahl FW. A test of a counting model for chiasma interference. Genetics. 1995;139:1201–1209. doi: 10.1093/genetics/139.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Housworth EA, Stahl FW. Crossover interference in humans. Am. J. Hum. Genet. 2003;73:188–197. doi: 10.1086/376610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stahl FW, Foss HM, Young LS, Borts RH, Abdullah MF, Copenhaver GP. Does crossover interference count in Saccharomyces cerevisiae? Genetics. 2004;168:35–48. doi: 10.1534/genetics.104.027789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lam SY, Horn SR, Radford SJ, Housworth EA, Stahl FW, Copenhaver GP. Crossover interference on nucleolus organizing region-bearing chromosomes in Arabidopsis. Genetics. 2005;170:807–812. doi: 10.1534/genetics.104.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McPeek MS, Speed TP. Modeling interference in genetic recombination. Genetics. 1995;139:1031–1044. doi: 10.1093/genetics/139.2.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fox DP. The control of chiasma distribution in the locust, Schistocerca gregaria (Forskal) Chromosoma. 1973;43:289–328. doi: 10.1007/BF00294277. [DOI] [PubMed] [Google Scholar]
- 86.Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination. Genome Res. 2009;19:581–589. doi: 10.1101/gr.086181.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez-Perez E, Colaiacovo MP. Distribution of meiotic recombination events: talking to your neighbors. Curr. Opin. Genet. Dev. 2009;19:105–112. doi: 10.1016/j.gde.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perkins DD. Crossing-over and interference in a multiply marked chromosome arm of Neurospora. Genetics. 1962;47:1253–1274. doi: 10.1093/genetics/47.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strickland WN. An analysis of interference in Aspergillus nidulans. Proc. R. Soc. Lond. B. Biol. Sci. 1958;149:82–101. doi: 10.1098/rspb.1958.0053. [DOI] [PubMed] [Google Scholar]
- 92.Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22:194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drouaud J, Mercier R, Chelysheva L, Berard A, Falque M, Martin O, Zanni V, Brunel D, Mezard C. Sex-Specific Crossover Distributions and Variations in Interference Level along Arabidopsis thaliana Chromosome 4. PLoS Genet. 2007;3:e106. doi: 10.1371/journal.pgen.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, Cande WZ. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J, Harper LC, Golubovskaya I, Wang CR, Weber D, Meeley RB, McElver J, Bowen B, Cande WZ, Schnable PS. Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics. 2007;176:1469–1482. doi: 10.1534/genetics.106.062604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kochakpour N, Moens PB. Sex-specific crossover patterns in Zebrafish (Danio rerio) Heredity. 2008;100:489–495. doi: 10.1038/sj.hdy.6801091. [DOI] [PubMed] [Google Scholar]
- 97.Moens PB. Zebrafish: chiasmata and interference. Genome. 2006;49:205–208. doi: 10.1139/g06-021. [DOI] [PubMed] [Google Scholar]
- 98.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koehler KE, Schrump SE, Cherry JP, Hassold TJ, Hunt PA. Near-human aneuploidy levels in female mice with homeologous chromosomes. Curr. Biol. 2006;16:R579–580. doi: 10.1016/j.cub.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 100.Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics. 2002;162:297–306. doi: 10.1093/genetics/162.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vallente RU, Cheng EY, Hassold TJ. The synaptonemal complex and meiotic recombination in humans: new approaches to old questions. Chromosoma. 2006;115:241–249. doi: 10.1007/s00412-006-0058-4. [DOI] [PubMed] [Google Scholar]
- 102.Tease C, Hartshorne G, Hulten M. Altered patterns of meiotic recombination in human fetal oocytes with asynapsis and/or synaptonemal complex fragmentation at pachytene. Reprod. Biomed. Online. 2006;13:88–95. doi: 10.1016/s1472-6483(10)62020-2. [DOI] [PubMed] [Google Scholar]
- 103.Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am. J. Hum. Genet. 2005;76:112–127. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barlow AL, Hulten MA. Crossing over analysis at pachytene in man. Eur. J. Hum. Genet. 1998;6:350–358. doi: 10.1038/sj.ejhg.5200200. [DOI] [PubMed] [Google Scholar]
- 105.Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 106.Anderson GW, Zhu Q, Metkowski J, Stack MJ, Gopinath S, Mariash CN. The Thrsp null mouse (Thrsp(tm1cnm)) and diet-induced obesity. Mol. Cell Endocrinol. 2009;302:99–107. doi: 10.1016/j.mce.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]