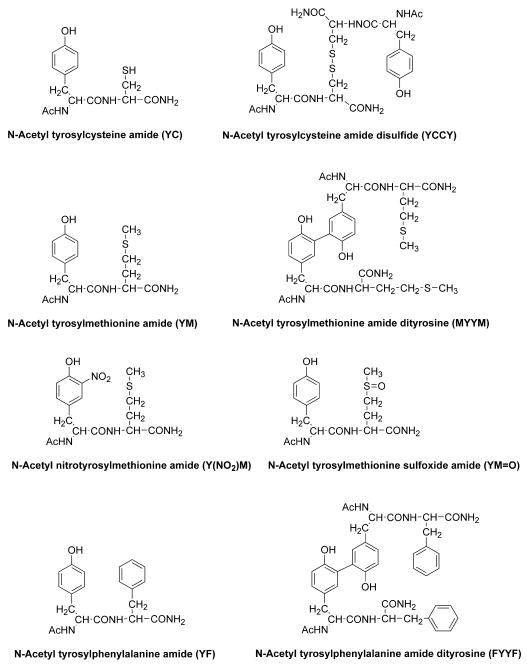
Abstract
Recent reports suggest that intramolecular electron-transfer reactions can profoundly affect the site and specificity of tyrosyl nitration and oxidation in peptides and proteins. Here we investigated the effects of methionine on tyrosyl nitration and oxidation induced by myeloperoxidase (MPO), H2O2 and NO2- and peroxynitrite (ONOO-) or ONOO- and bicarbonate (HCO3-) in model peptides, tyrosylmethionine (YM), tyrosylphenylalanine (YF) and tyrosine. Nitration and oxidation products of these peptides were analysed by HPLC with UV/Vis and fluorescence detection, and mass spectrometry; radical intermediates were identified by electron paramagnetic resonance (EPR)-spin-trapping. We have previously shown (Zhang et al., J. Biol. Chem. (2005) 280, 40684-40698) that oxidation and nitration of tyrosyl residue was inhibited in tyrosylcysteine(YC)-type peptides as compared to free tyrosine. Here we show that methionine, another sulfur-containing amino acid, does not inhibit nitration and oxidation of a neighboring tyrosine residue in the presence of ONOO- (or ONOOCO2-) or MPO/H2O2/NO2- system. Nitration of tyrosyl residue in YM was actually stimulated under the conditions of in situ generation of ONOO- (formed by reaction of superoxide with nitric oxide during SIN-1 decomposition), as compared to YF, YC and tyrosine. The dramatic variations in tyrosyl nitration profiles caused by methionine and cysteine residues have been attributed to differences in the direction of intramolecular electron transfer mechanism in these peptides. Further confirmation of HPLC data analysis was obtained by steady-state radiolysis and photolysis experiments. Potential implications of the intramolecular electron-transfer mechanism in mediating selective nitration of protein tyrosyl groups are discussed.
Keywords: Nitrotyrosine, reactive nitrogen species, electron-transfer, spin-trapping, EPR, radiolysis
Introduction
Emerging research strongly supports the role of pro-inflammatory, reactive oxygen/nitrogen species (ROS/RNS) in cardiovascular, pulmonary, and neurodegenerative diseases [1-9]. Post-translational nitrative modifications of proteins and lipids have been identified as biomarkers for RNS in diseases [9-12]. These post-translational modifications include nitration of tyrosyl and lipid moieties. Two major pathways are responsible for protein and lipid nitration. These include heme peroxidases (e.g., myeloperoxidase, eosinophil peroxidase) using nitrite and H2O2 as substrates and/or peroxynitrite and bicarbonate [13-19]. In addition, the interaction between peroxynitrite and metal ions (including heme proteins) may act as a biologically relevant source of nitrating species [20].
Factors influencing nitration of tyrosyl residues in protein are not fully known. Several studies have shown that tyrosine nitration is a selective process that is controlled by various microenvironmental factors (hydrophobicity, CO2 levels, membrane oxygen concentration, acidic environment and amino acid sequence) [21-29]. Although no specific amino acid sequence criteria exist for predicting tyrosine nitration or lack thereof, it has been suggested that protein tyrosyl nitration is (i) enhanced when a tyrosyl moiety is situated closer to a negatively–charged amino acid (i.e., glutamate or aspartate) and (ii) decreased when the tyrosyl residue is present in the vicinity of a cysteinyl or methionine residue [12; 29; 30]. Detailed quantitative analyses of the effects of methionine residue on tyrosyl nitration are, however, lacking. Understanding the biophysical/biochemical mechanisms that determine the motif for nitration-sensitive tyrosine residues is clearly pathophysiologically significant [31].
Methionine, along with cysteine and tryptophan, represents one of the most easily oxidized natural amino acids [32]. It is known that methionine can be directly oxidized by peroxynitrite via a radical and non-radical mechanism, leading to the formation of ethylene and methionine sulfoxide as a biologically relevant product [33-36]. Early on, it was suggested that the presence of a methionine residue adjacent to tyrosine in peptides and proteins will result in decreased tyrosyl nitration [12; 28; 30] due to methionine's ability to react with peroxynitrite and radicals derived from it [37-43]. However, recent proteomic studies have yielded conflicting results [44-47]. In some cases, the neighboring methionine residue inhibited tyrosine nitration [44; 45]; whereas, in other studies, nitrotyrosine formation was enhanced in the proximity of a methionine residue [46; 47].
To understand how a neighboring methionine residue can influence tyrosyl nitration, we used a model system consisting of simple dipeptides (e.g., tyrosylmethionine (YM), YF, etc. (Fig. 1)) and monitored their nitration/oxidation products in the presence of myeloperoxidase (MPO), H2O2 and NO2- or ONOO- and HCO3-. We have also monitored the products of one-electron oxidation of the peptides by Br2•- or CO3•- using a combination of radiolysis and photolysis techniques. Results indicate that, contrary to the other sulfur-containing amino acid cysteine, methionine did not inhibit tyrosine nitration in the dipeptide. In case of SIN-1 (slow in situ generators of ONOO-), we actually observed stimulation of tyrosine nitration/oxidation in YM peptide as compared to YF, YC or tyrosine. This is attributed to a possible intramolecular electron-transfer between the methionine sulfide radical cation and the tyrosyl residue resulting in increased oxidation and nitration of tyrosine. The influence of intramolecular electron transfer mechanism in protein nitration reactions is also discussed.
Figure 1. Structures of peptides and their oxidation and nitration products.
Experimental
The following chemicals and enzymes were purchased from different sources as indicated: Tyrosine, hydrogen peroxide, sodium nitrite, sodium bicarbonate, 3-nitrotyrosine, and D,L-methionine were from Sigma; myeloperoxidase was from CalBiochem. Rink amide methylbenzhydrylamine (MBHA) resin and Fmoc protected amino acids were from Calbiochem-Novabiochem (LaJolla). Di-iso-propylcarbodiimide (DIC), 1-hydroxybezotriazole (HOBt), triisopropylsilane (TIS), methylmethane-thiosulfonate (MMTS), N-methyl-pyrrolidione (NMP) and piperidine were purchased from Fisher Scientific. Peroxynitrite was synthesized in-house as reported previously [48].
Peptide synthesis and purification
The structures of the peptides used in this study are shown in Figure 1. Peptides were chemically synthesized using the standard Fmoc solid phase peptide based synthetic procedure on an Advanced Chemtech Model 90 synthesizer (Louisville, Ky) [29]. Rink Amide MBHA resin (loading 0.72 mmol/g) was used as a solid support. Fmoc-protected amino acids were coupled as HOBt-esters. All amino acids were double-coupled using HOBt/DIC. The following steps were performed in the reaction vessel for each double-coupling: deprotection of the Fmoc group with 20% piperidine in NMP for 30 min (twice), three NMP washes, two dichloromethane (DCM) washes, first coupling for 1 h with a five-fold excess of Fmoc amino acid in 0.5 M HOBt and 0.5 M DIC, second coupling using a fresh addition of the same reagent for 1 h, followed by three NMP washes, and two DCM washes. Final acetylation was performed using an acetic anhydride/HOBt/DIC mixture for 30 min (twice). The resin was washed twice with DCM, three times with methanol, and then dried at room temperature under vacuum prior to cleavage. The peptide was deprotected and cleaved from the resin with 90% TFA containing TIS for three hours at room temperature. The resin was removed by filtration, washed with TFA, and the combined TFA filtrates were evaporated to dryness under a stream of dry N2 gas. The oily residue was washed three times with cold ether and the dry crude peptide was dissolved in acetonitrile/H2O (1:1) and lyophilized. The crude peptides were purified by a semi-preparative reverse phase-HPLC on a RP-C18 (10 × 250 mm) column using a CH3CN/water gradient (5% to 25% CH3CN over 60 minutes) containing 0.1% TFA at a flow rate of 3 ml/min with UV absorption detection at 280 nm. The identity of YM peptide was verified by its UV absorption spectrum and ESI-MS (M+H+: 356)
Synthesis of nitrated YM
Nitrated YM peptide (Y(NO2)M) was prepared as follows: The YM peptide (1 mM) was mixed with MPO/H2O2 and NO2− in a phosphate buffer (100 mM, pH 7.4) containing DTPA (100 μM) for 20 min. The reaction was terminated by adding catalase (100 U/ml). The resulting Y(NO2)M peptide was purified using a semi-preparative HPLC. Nitrated peptides show characteristic UV-Vis absorption spectra. Upon adding NaOH, the absorption peak at 350 nm was shifted to 430 nm. The structure of Y(NO2)M was verified by LC-MS spectrometry on an Agilent 1100 series LC-MS (M+H+; 401). An extinction coefficient of 4100 M-1 cm-1 (pH > 11) at 430 nm was used for quantitation of Y(NO2)M.
Synthesis of YM sulfoxide
The peptide YM (1 mM) was mixed with H2O2 (2 mM) in phosphate buffer (100 mM, pH 7.4) containing DTPA (1 mM) at room temperature for 12 hours. The resulting YM sulfoxide was purified by HPLC and its identity was further verified by ESI-MS (M+H+; 372).
Synthesis of carbonato tetrammine cobalt(III) perchlorate
Carbonato tetrammine cobalt(III) perchlorate [Co(NH3)4CO3]ClO4 was synthesized in two steps, as described elsewhere: (1) synthesis of [Co(NH3)4CO3]Cl [49] and (2) exchange of chloride anion with perchlorate anion [50].
Nitration and oxidation of YM peptide induced by MPO/H2O2/NO2-
Typically, peptides (250 μM) were incubated with NaNO2 (500 μM), H2O2 (100 μM) and MPO (30 nM) in a phosphate buffer (100 mM, pH 7.4) containing DTPA (100 μM) at room temperature for 30 min. Reactions were terminated by adding catalase (200 U/ml) enzyme and directly analyzed by HPLC. Repeated injections in 24 h showed no significant oxidation of YM under these experimental conditions.
Nitration and oxidation of YM peptide induced by peroxynitrte
YM (250 μM) was rapidly mixed with with peroxynitrite (20 - 500 μM) in a phosphate buffer (100 mM, pH 7.4) containing DTPA (100 μM) and incubated at room temperature for 10 min, and analyzed by HPLC. In control experiments peroxynitrite was first added to the buffer followed by the addition of YM after 10 min.
One-electron oxidation of peptides by dibromine radical anion
The peptides (100 μM) were incubated with KBr (100 mM) in phosphate buffer (50 mM, pH 7.4) saturated with N2O. Dibromine radical anion (Br2•-) was generated in situ by radiolysis of solutions contained in HPLC vials using X-rays (dose rate of 3 Gy/min) [51]. Under these conditions Br2•- was the major species responsible for peptide consumption. After irradiation, samples were analyzed by HPLC.
One-electron oxidation of peptides by carbonate radical anion
The peptides (100 μM) were incubated with phosphate buffer (50 mM, pH 7.4) in aqueous solution containing 4 mM [Co(NH3)4CO3]ClO4. The carbonate radical anion (CO3•-) was generated in situ by irradiation of the samples placed in quartz cuvettes with UV light [50; 51]. Photolysis was carried out using a 300 W Xe high pressure compact arc lamp (Eimac Model VIX 300 UV). The light was passed through a water filter prior to sample irradiation. Under those conditions CO3•- was the major species responsible for peptide consumption. After photolysis the samples have been analyzed by HPLC.
HPLC analyses of nitration and oxidation products
Typically, a 20 μl of sample was injected into a HPLC system (HP1100) equipped with C-18 column (250 × 2.0 mm) equilibrated with a 5% CH3CN in 0.1% TFA. Products were separated by a linear increase of CH3CN concentration to 25% in 60 min at a flow rate of 0.2 ml/min. The elution was monitored using the on-line UV-vis and fluorescence detectors. YM, dityrosyl YM and nitrated YM were eluted at 9.5, 17 and 18 min, respectively.
Nitration of tyrosine with or without methionine by MPO/H2O2/NaNO2 or peroxynitrite was performed under the same conditions and the products analyzed as reported previously [29].
EPR spin-trapping experiments
Incubations consisting of a peptide or tyrosine (1 mM), MPO (150 nM), and DBNBS (20 mM) in a phosphate buffer (100 mM, pH 7.4) containing DTPA (100 μM) were rapidly mixed with H2O2 (1 mM). Samples were subsequently transferred to a 100 μl capillary tube and ESR spectra were taken within 30 s after starting the reaction in a Bruker EMX spectrometer. Typical spectrometer parameters were: scan range, 100 G; field set, 3505 G; time constant, 0.64 ms; scan time, 10 s; modulation amplitude, 2.0 G; modulation frequency, 100 kHz; receiver gain, 5 × 105; and microwave power, 20 mW. The spectra shown were the average of 30 scans.
Results
MPO/H2O2/NaNO2-dependent oxidation and nitration of tyrosylmethionine
The effects of a methionine residue on tyrosine nitration and oxidation were determined by measuring the MPO/H2O2/NaNO2-dependent nitration/oxidation of YM peptides (Fig. 2).
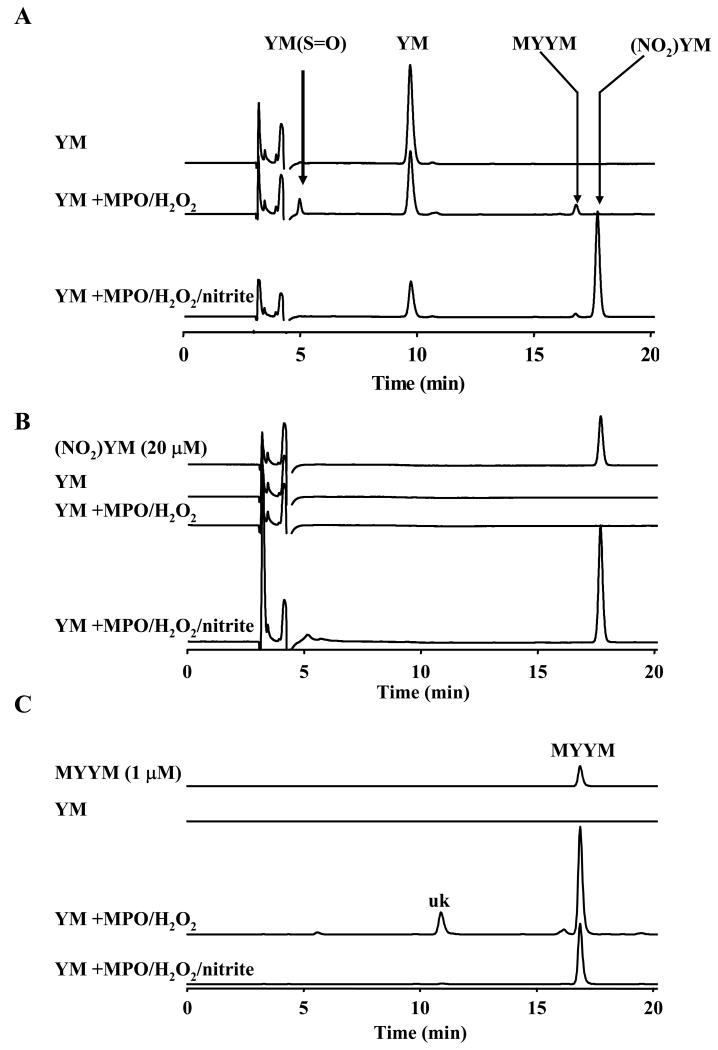
Figure 2. HPLC analyses of products formed from tyrosylmethionine in MPO/H2O2/NO2- system.
YM (250 μM) was incubated with MPO (30 nM), H2O2 (100 μM) and NaNO2 (500 μM) for 30 minutes at room temperature. The reaction was terminated by adding catalase and the resulting mixture was analyzed by HPLC using different detection conditions. (A) YM and (NO2)YM were detected as major components at 9.5 and 18 min using the HPLC/UV detection at 280 nm. (B) Major peak due to (NO2)YM was detected at 18 min using a detection wavelength of 350 nm. (C) The dityrosyl product, MetTyrTyrMet, was detected as a major product using the HPLC-fluorescence mode (λmax = 290 nm, λmin = 410 nm). In addition, there was an unidentified peak (uk).
HPLC with UV-Vis absorption detection at 280 nm for tyrosyl residues and at 350 nm for nitro tyrosyl residues were employed. The HPLC/fluorescence (λexc = 290 nm, λemi = 410 nm) was used for detecting the dityrosyl product of YM (Fig. 2A-C). HPLC product analyses of incubations containing YM (250 μM), MPO (30 nM), H2O2 (100 μM), DTPA (100 μM) and NaNO2 (500 μM) in a phosphate buffer (100 mM, pH 7.4) for 30 min at room temperature showed a significant depletion of YM peptide and formation of a new major product (retention time, 18 min). Based on its UV absorption maximum (λ = 350 nm) and its pH-dependent changes, this new product was assigned to a methionyl nitrotyrosine ((NO2)YM). Additional evidence in favor of this structural assignment was obtained from the ESI-MS analysis of this product (M+H+; 401). Another minor product (retention time, 17 min) was also detected by fluorescence that was assigned to a dityrosyl product (Fig. 2A and C). This structural assignment was further confirmed by the ESI/MS analysis (M+H+; 709). Incubations of YM (250 μM) and MPO (30 nM), H2O2 (100 μM) in the absence of NaNO2 also yielded the same dityrosyl product (Fig. 2A). In this case, an additional product was detected (retention time, 5 min) which was assigned to an YM sulfoxide structure on the basis of MS analysis. From these results, we conclude that only the tyrosyl residue but not the methionine residue undergoes nitration and oxidation induced in the MPO/H2O2/NO2-/YM system. This is in stark contrast to oxidation of YC peptides in MPO/H2O2/NO2-which yielded a corresponding disulfide (i.e., YCCY) as a major product with little or no nitrated product formation [29].
As MPO/H2O2/NaNO2-dependent nitration/oxidation of YM peptide yielded exclusively tyrosyl nitration and oxidation products, we compared the product profiles with another dipeptide, tyrosinephenylalanine (YF). Incubation of YF with MPO/H2O2/NaNO2− resulted in the depletion of YF at nearly the same rate as that of YM (Fig. 3A and B). Incubation of YF or YM with hydrogen peroxide alone showed no significant depletion of these peptides, which indicated that direct oxidation of YM or YF peptide by H2O2 is negligible. As with YM, incubation of YF with MPO/H2O2 also resulted in its oxidation. However, the extent of depletion of YM peptide in both systems is the same as that of YF, suggesting that the rate of oxidation of both dipeptides by MPO is similar. Under similar conditions, the dityrosine yields of YM (i.e., MYYM) induced by MPO/H2O2 or MPO/H2O2/NaNO2 were significantly higher than those of YF (FYYF) (Fig. 3C). Nitration of YM by MPO/H2O2/NaNO2 was slightly increased as compared to nitration of YF (Fig. 3D). These results clearly demonstrate that tyrosine nitration and oxidation induced by MPO/H2O2/NO2- system was not inhibited by a neighboring methionine residue. On the contrary, the methionyl residue enhanced tyrosyl nitration in YM, as compared to YF peptide.
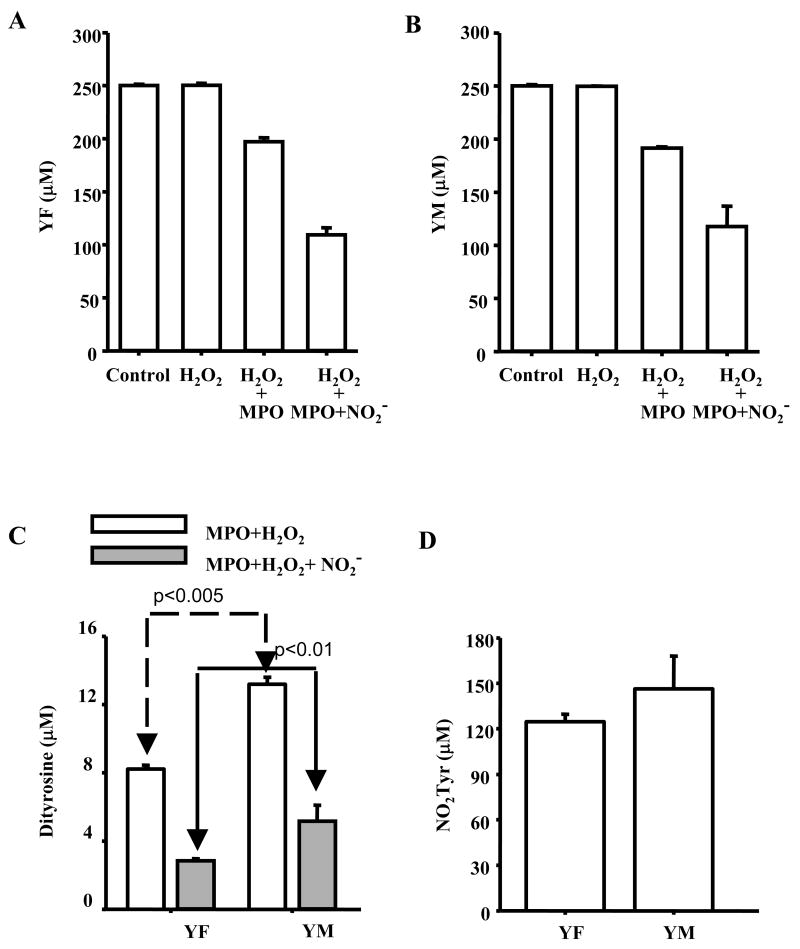
Figure 3. Substrate depletion and product formation from oxidation/nitration of dipeptides in in MPO/H2O2/NO2- system.
(A) The dipeptide, YF (250 μM), was incubated with MPO (30 nM), H2O2 (100 μM) and NaNO2 (500 μM) in a phosphate buffer (100 mM, pH 7.4) containing DTPA (100 μM) for 30 min at room temperature. The reaction was terminated by adding catalase (200 U/ml) and reaction mixtures analyzed by HPLC using conditions described in Fig. 2. Bar graphs show the concentrations of YF under various incubating conditions. (B) Same as (A) except that YF was substituted by YM. (C) The concentrations of dipeptides obtained from YF and YM during MPO/H2O2 and MPO/H2O2/NO2- oxidation. The incubation conditions are as indicated in (A) and (B). (D) The concentrations of nitrotyrosines obtained from YF and YM during MPO/H2O2/NO2- mediated nitration. Experimental conditions are as in (A) and (B).
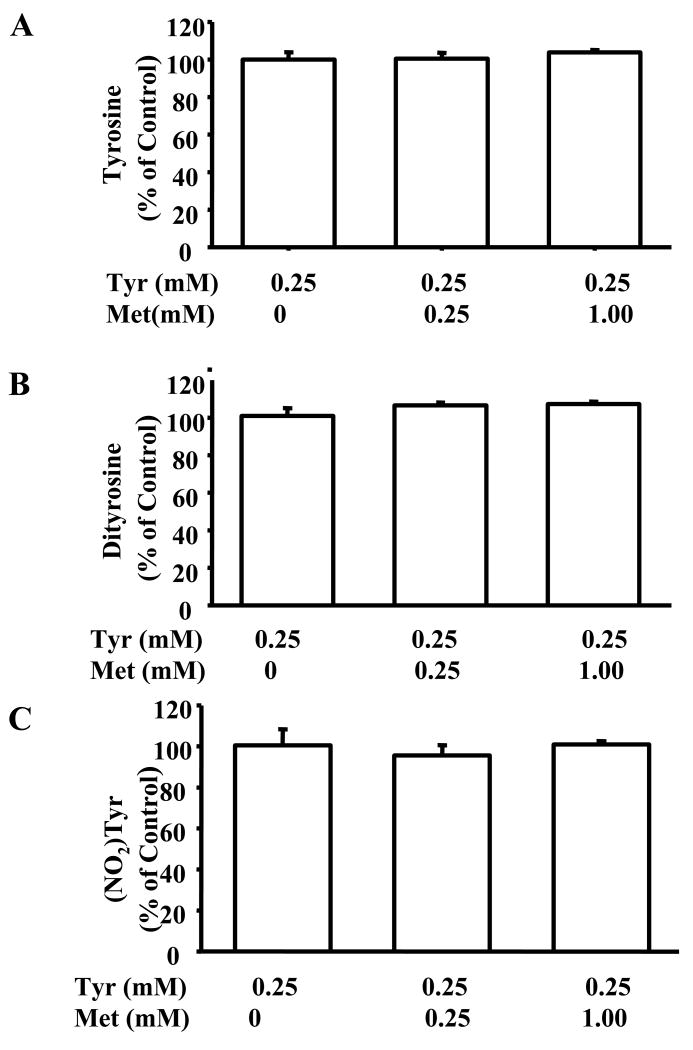
To further understand the effect of methionine on tyrosyl oxidation/nitration by MPO/H2O2/NO2-, we incubated tyrosine (250 μM) with MPO, H2O2 and various amounts of methionine (0, 0.25 and 1 mM) and observed that oxidation of tyrosine to dityrosine by MPO/H2O2 was not affected by methionine (Fig 4A). This indicates that methionine did not react with the tyrosyl radicals formed in this system (Fig. 4A). In addition, nitration of tyrosine by MPO/H2O2/NaNO2 was also not affected by methionine (Fig. 4C), suggesting that the nitrogen-dioxide radical (•NO2) formed in this system reacts with tyrosine more rapidly than with methionine. Collectively, these results show that the exogenously-added methionine does not affect tyrosyl nitration or oxidation in MPO/H2O2/NO2- system.
Figure 4. The effect of methionine on tyrosine oxidation/nitration in MPO/H2O2/NO2- system.
(A) Incubation mixtures contained Tyr (250 μM), MPO (30 nM), H2O2 (100 μM), DTPA (100 μM) and varying amounts of Met for 30 min at room temperature. Graphs show Tyr concentrations measured after the incubation period. (B) Incubation conditions are as in (A). Graphs show dityrosine levels measured after 30 min of incubation. (C) Same as in (A) except that nitrotyrosine was measured following the incubation period.
Peroxynitrite-dependent oxidation and nitration of tyrosylmethionine peptide
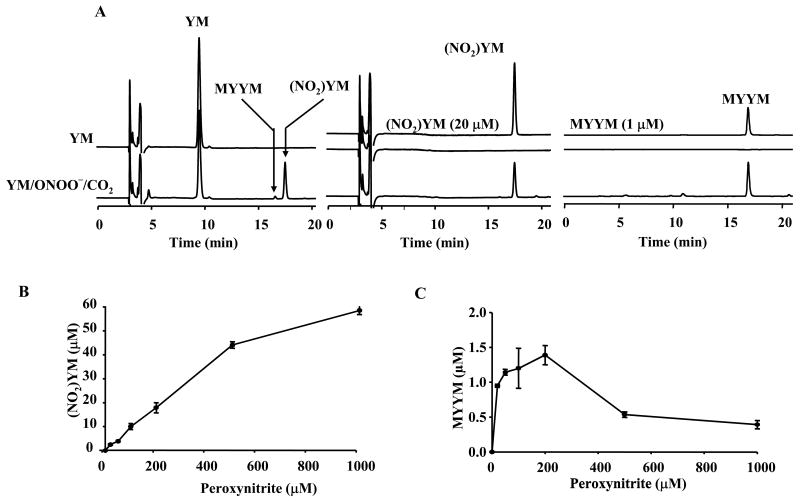
Figure 5A (left) shows the HPLC traces obtained from an incubation mixture containing YM (250 μM), DTPA (100 μM), peroxynitrite (100 μM), and bicarbonate (25 mM) in a phosphate buffer (100 mM, pH 7.4). HPLC traces were recorded 10 min after the addition of peroxynitrite. Predominant products were (NO2)YM (eluting at 18 min with absorption at 280 and 350 nm) and MYYM (the dityrosyl product detected using fluorescence paameters: λexc = 290 nm, λemi = 410 nm) (Fig. 5A, middle and right). Incubation of YM (250 μM) with different amounts of peroxynitrite showed a dose-dependent increase in formation of the nitrated product (Fig. 5B) and the corresponding dityrosyl product (MYYM) (Fig. 5C).
Figure 5. HPLC ananlysis of nitration and oxidation products formed from tyrosylmethionine and peroxynitrite and bicarbonate.
(A) YM (250 μM) was incubated with peroxynitrite (100 μM) in a phosphate buffer (pH 7.4, 100 mM) containing DTPA (100 μM) for 10 min and analyzed by HPLC. (bottom trace) Incubations contained YM alone and (top trace) Incubations contained YM, peroxynitrite, and bicarbonate (25 mM). (B) Dose-dependent formation of nitrated YM under incubation conditions shown in (A). (C) Dose-dependent formation of dityrosine formed from YM.
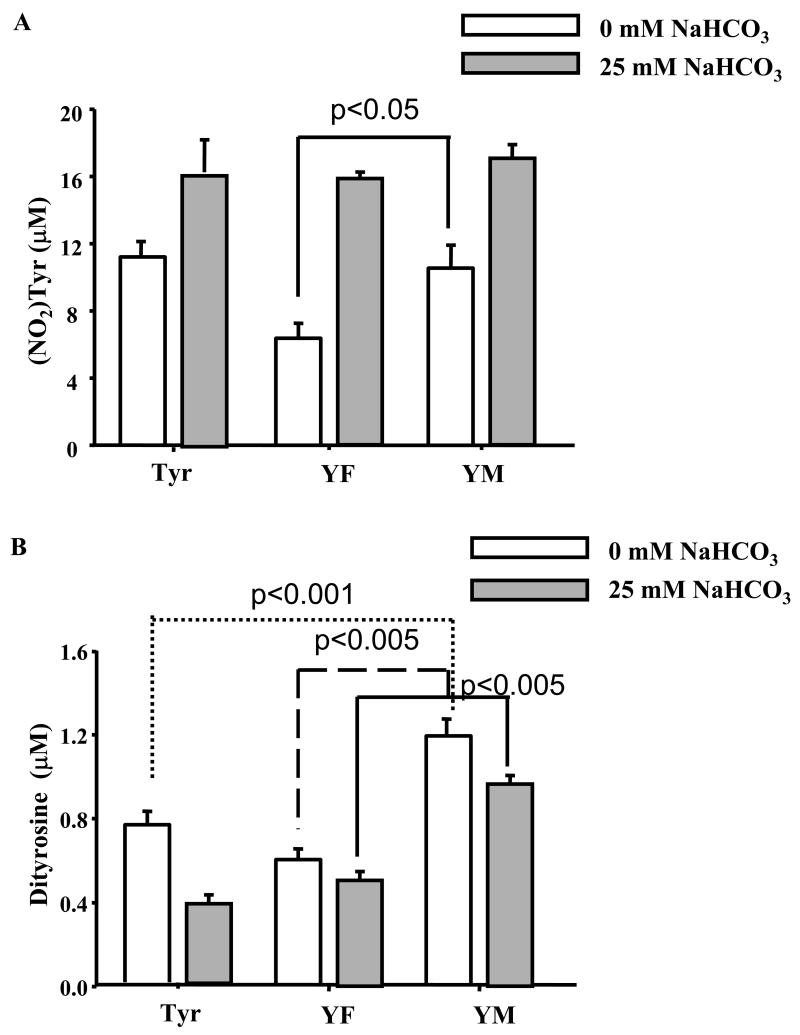
As with MPO/H2O2/NO2- system, we compared the yields of nitration/oxidation products of YM peptide with those obtained from YF peptide and with tyrosine treated with peroxynitrite (Fig. 6). Oxidation of YM (250 μM) and Y (250 μM) with 100 μM peroxynitrite yielded 10.5±1.2 μM of (NO2)YM and 11.1±0.8 μM of Y(NO2) and treatment of FY with peroxynitrite yielded 6.3±0.8 μM of (NO2)YF (Fig. 6A). Under these conditions, higher yields of MYYM (1.19±0.08 μM) were obtained as compared to FYYF (0.61±0.05 μM) and YY (0.78±0.06 μM). As it is well-known that bicarbonate can enhance the nitration yields of tyrosine induced by peroxynitrite through intermediate formation of carbonate radical and nitrogen dioxide radical [19; 21; 27; 32; 52], we investigated the effects of bicarbonate on nitration and oxidation products of YM. Incubation of YM (250 μM) with 100 μM peroxynitrite in the presence of 25 mM bicarbonate yielded 16.8±0.7 μM of (NO2)YM similar to those formed from YF and tyrosine (Fig. 6A). However, under the same conditions, the amount of dityrosyl product formed from YM (MYYM) was significantly higher than the dityrosyl products (YY and FYYF) formed from oxidation of Y or YF peptide (Fig. 6B).
Figure 6. Relative formation of nitrotyrosine and dityrosine from different dipeptides in peroxynitrite/bicarbonate system.
Dipeptides (250 μM) were incubated with peroxynitrite (100 μM) in a phosphate buffer (pH 7.4, 100 mM) containing DTPA (100 μM) for 5 min and reaction contents analyzed by HPLC. (A) nitrated product of Y, YF, and YM, and (B) dityrosyl product of Tyr, YF, YM.
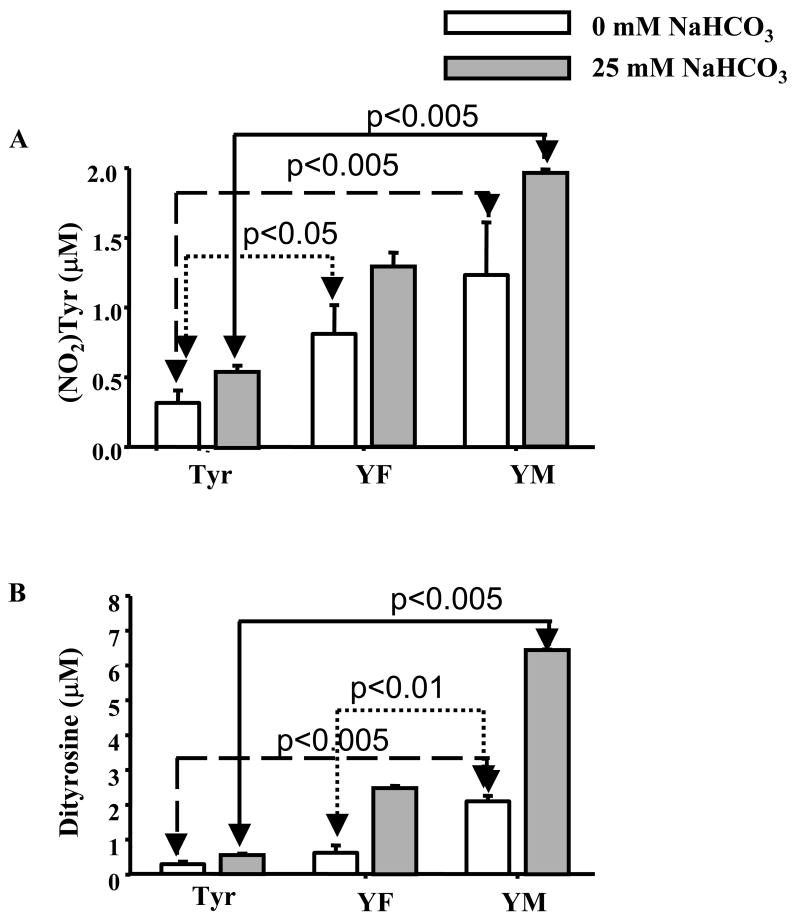
To better understand the effects of methionine on peroxynitrite-induced tyrosine nitration under physiologically-relevant conditions, we used SIN-1 that simultaneously generated equal amounts of •NO and superoxide. SIN-1 has been widely used to simulate the in vivo system, in which peroxynitrite is formed by reaction of •NO with O2•-. Incubation of YM (250 μM) with SIN-1 (250 μM) yielded the corresponding nitration product, (NO2)YM, in a much higher yield as compared to that formed from tyrosine or YF (Fig. 7A). There was a 4-fold and 7-fold increase in the formation of (NO2)YM and MYYM as compared to Y(NO2) and YY (Fig. 7B). Similar results were obtained in the presence of bicarbonate. In agreement with literature reports, the nitration efficiency of the tyrosyl residues was much lower in the case of SIN-1 (Fig. 7A and B) than in the case of bolus peroxynitrite (Fig. 6) or slowly infused (5 μM/min) peroxynitrite (data not shown). From these studies, we conclude that a neighboring methionine residue actually enhances the nitration and oxidation of the adjacent tyrosyl group in the presence of in situ generated peroxynitrite with or without bicarbonate.
Figure 7. SIN-1 induced nitrotyrosine and dityrosine formation from dipeptides.
Dipeptides (250 μM) were incubated with SIN-1 (250 μM) in a phosphate buffer (pH 7.4, 100 mM) containing DTPA (100 μM) for 10 h at room temperature and solutions analyzed by HPLC. (A) Relative levels of nitrated products of tyrosine and (B) dityrosyl products of Tyr, YF, and YM.
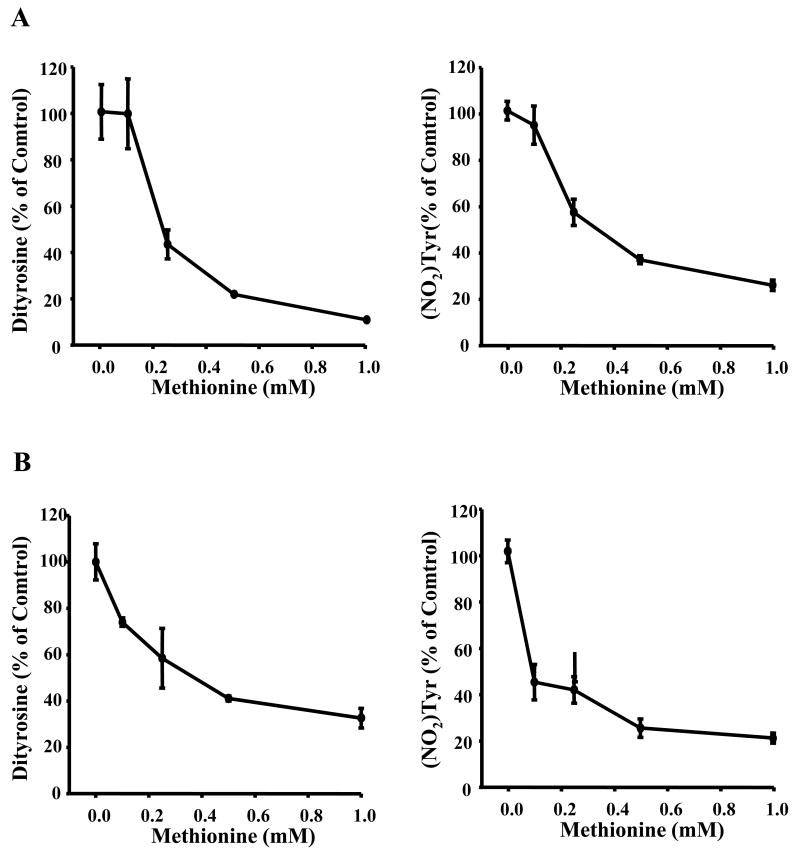
Previous data suggest that peroxynitrite is scavenged by methionine [38]. We investigated the effects of free methionine on tyrosine nitration and oxidation induced by peroxynitrtite (Fig. 8A-D). In agreement with literature data, addition of methionine dose-dependently inhibited tyrosine oxidation and nitration induced by peroxynitrite (Fig. 8C and D). Methionine also inhibited the nitration and oxidation of tyrosine in the presence of bicarbonate (Fig. 8A and B). Thus, these results clearly indicate the differential effects of free methionine and the peptidyl methionyl residue on tyrosyl nitration. Whereas exogenously added methionine inhibited peroxynitrite-mediated nitration of tyrosine, the methionyl residue in YM dipeptide did not inhibit nitration of adjacent tyrosine but in fact stimulated nitration of adjacent tyrosyl group as compared to YF. This is a potentially new finding.
Figure 8. The effects of methionine on tyrosine nitration/oxidation induced by peroxynitrite and peroxynitrite/bicarbonate.
(A) Tyr (250 μM) and Met were mixed with peroxynitrite (100 μM) in the presence and absence of bicarbonate (25 mM) in a phosphate buffer (pH 7.4, 100 mM), incubated for 5 minutes and analyzed by HPLC. (A) Dityrosine (Y-Y) and nitrotyrosine (NO2Y) were measured in incubations containing peroxynitrite, tyrosine, and methionine at concentrations as indicated. (B) Same as (A) but without bicarbonate.
One-electron oxidation of YM and other peptides
To investigate the mechanism of the nitration/oxidation of tyrosyl residue in YM peptide, we performed steady-state radiolysis and photolysis techniques to generate short-lived oxidizing radicals. We used dibromine radical anion (Br2•-) as a one-electron oxidant (Figures 1S-4S) [51]. In the case of YM peptide (Figure 1S) it is likely that the methionine residue is the primary site of attack, as the rate constant of the reaction of methionine with Br2•- (k = 2 × 109 M-1s-1) is two orders of magnitude higher than the reaction of Br2•- with tyrosine (k = 2 × 107 M-1s-1). Although the primary site of reaction is a methionyl residue, the final product of the reaction is dityrosyl tetrapeptide (MYYM) (Fig. 1S), indicating a fast intramolecular electron transfer from tyrosyl residue to the one-electron oxidized methionine moiety. MYYM peptide as the major product was also detected in the presence of other one-electron oxidants generated radiolytically, including N3• and (SCN)2•- or the carbonate radical (CO3•-) generated by photolysis of the [Co(NH3)4CO3]+ complex (Figure 2S). Those radicals are expected to oxidize mostly the tyrosyl residue (with N3•) or both residues to a similar extent (with (SCN)2•- and CO3•-); however, in all cases the corresponding dityrosyl peptide was the major product, again supporting the role of intramolecular electron transfer from tyrosine to the one-electron oxidized methionine. The dityrosine-type peptide has been observed as a major product also in the case of tyrosine and YF peptide, when oxidized by Br2•- (Figure 3S and 5S), N3•, (SCN)2•- and CO3•- (Figure 4S and 6S). On the other hand, the one electron oxidation of YC peptide leads exclusively to a disulfide product, regardless of the oxidant used (Figure 7S and 8S). With Br2•- (Figure 7S) it is likely that the primary site of attack in YC peptide is the cysteinyl residue, as the rate constant of the reaction cysteine with Br2•- is an order of magnitude higher (k = 1.8 × 108 M-1s-1) than of the reaction of tyrosine with Br2•- (see above). Thus the disulfide product could be expected in the absence of an intramolecular electron transfer mechanism. On the other hand, in the case of azidyl radical (N3•), the primary site of attack is expected to be the tyrosyl residue, as the rate constant of the reaction of N3• with cysteine (k = 1.4 × 107 M-1s-1) is an order of magnitude lower than of the reaction with tyrosine (k = 1.0 × 108 M-1s-1). As the final product is also a disulfide, the occurrence of an intramolecular electron transfer from the cysteinyl moiety to the tyrosyl radical is plausible. This conclusion is in agreement with the observed protective effect of the cysteinyl residue on tyrosyl nitration in YC-type peptides [29].
EPR spin-trapping of radical intermediates
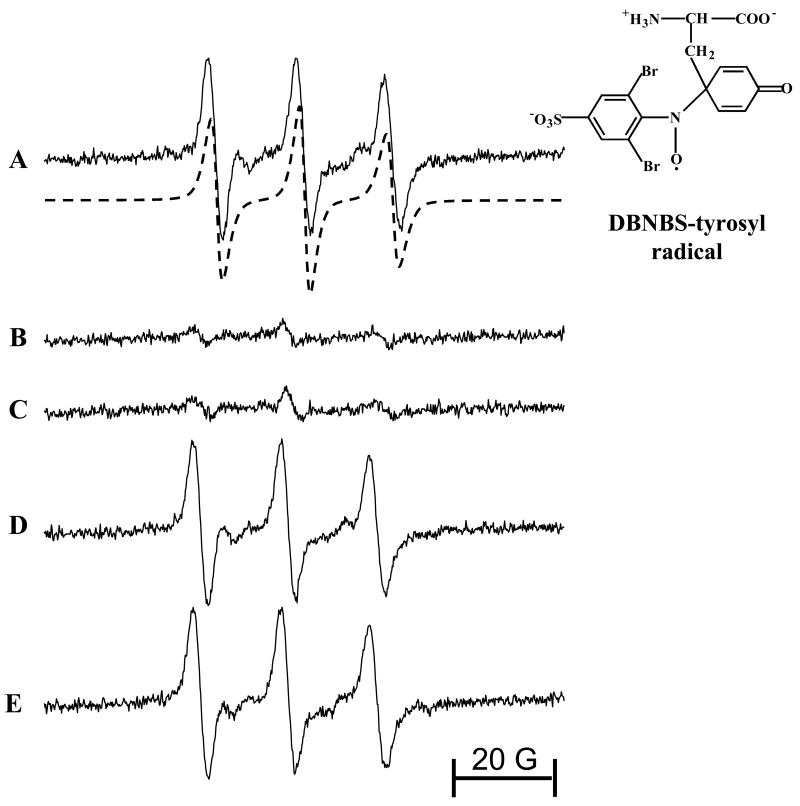
To further understand the paradoxical differences between the exogenously-added free methionine and the endogenously present methionyl residue on tyrosyl nitration, we used the EPR spin-trapping to detect radical intermediates formed in MPO/H2O2/NO2- and peroxynitrite/bicarbonate systems. Incubations containing tyrosine and MPO/H2O2 in the presence of DBNBS yielded a three-line EPR spectrum (Fig. 9A). This spectrum was assigned to a DBNBS-tyrosyl adduct (αN=13.6 G) [53]. In the absence of MPO, the DBNBS adduct formation was minimal (Fig. 9B). Addition of methionine (1 and 5 mM) had no effect on the DBNBS-tyrosyl adduct formation (Fig. 9D and E), suggesting that methionine did not effectively inhibit the tyrosyl radical formation. The DBNBS adduct was also minimal in incubations containing methionine (1 mM), MPO (150 nM), DBNBS (25 mM), H2O2 (1 mM) and DTPA (100 μM) (Fig. 9C). This is consistent with the report that methionine is a poor substrate for MPO/H2O2 [54]. These results are in agreement with the HPLC results shown in Figure 4.
Figure 9. The effects of methionine on DBNBS-tyrosyl radical adduct formation by MPO/H2O2.
(A) Incubations contained Tyr (1 mM), MPO (150 nM), H2O2 (1 mM), DTPA (100 μM), and DBNBS spin trap (20 mM) in a phosphate buffer (pH 7.4, 100 mM). (B) Same as (A) but without tyrosine. (C) Same as (A) but without MPO. (D) Same as (A) but containing Met (1 mM), and (E) Same as (A) but containing 5 mM of Met. EPR spectra were obtained immediately after the addition of MPO. EPR spectral conditions are described under Materials and Methods.
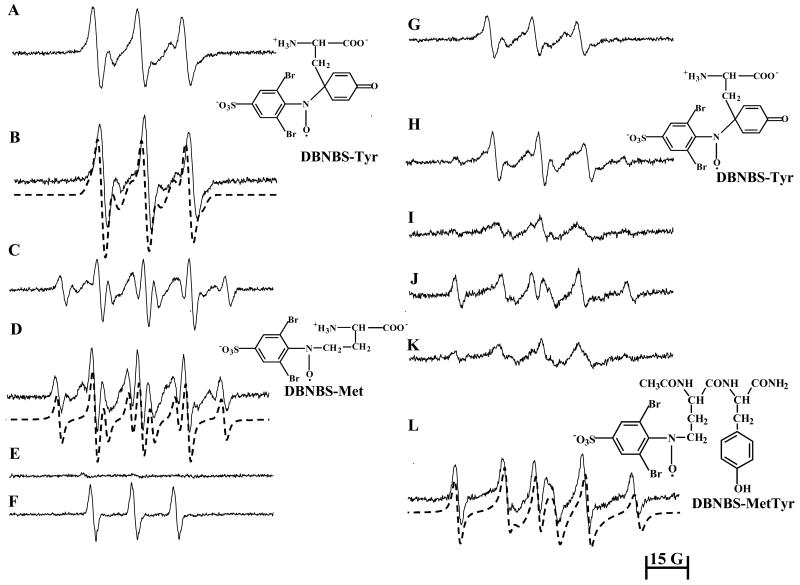
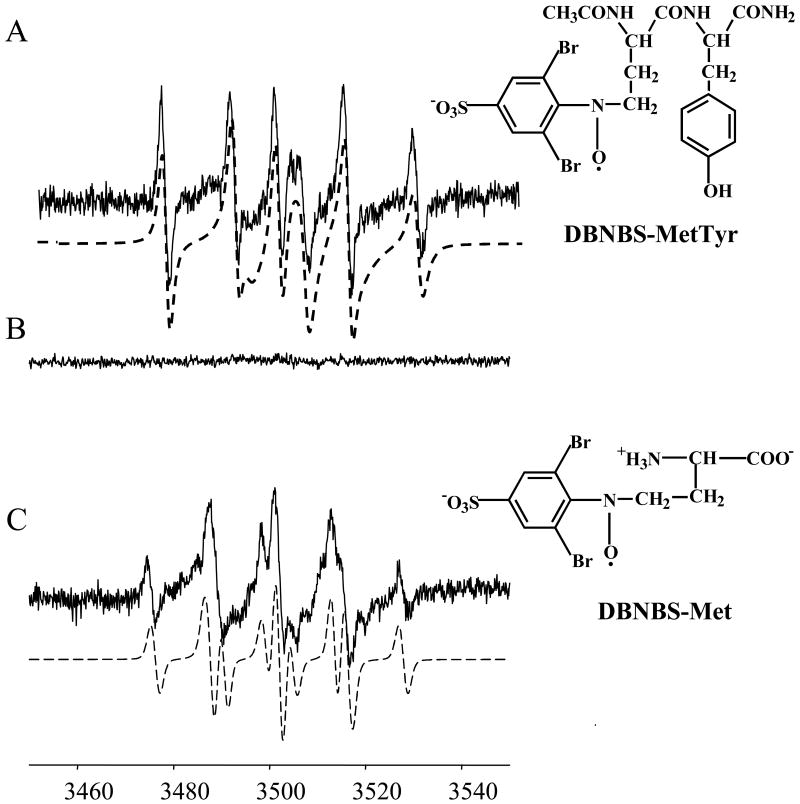
Figure 10A shows the EPR spectrum obtained from incubations containing tyrosine (2 mM), DTPA (100 μM), DBNBS (25 mM) and peroxynitrite (2.5 mM) which was attributed to the DBNBS-tyrosyl adduct. The intensity of the EPR signal was enhanced in the presence of bicarbonate (Fig. 10B). The computer simulation shows the presence of the two radical adduct species (at C-3 and C-5 positions of the tyrosyl group) (αN=13.6 G and αN=13 G, αH=6 G), as previously reported [53]. Unlike the MPO/H2O2 oxidizing system, peroxynitrite or peroxynitrite/bicarbonate-mediated oxidation of methionine yielded a DBNBS-methionine radical adduct (DBNBS-Met) (αN=14 G, αH(2)=11 G) (Fig. 10C). On the basis of published results [55], this spectrum was assigned to the DBNBS- CH2S(CH2)2CH(NH2)COOH and/or the DBNBS-(CH2)2CH(NH2)COOH adduct [55-57]. Bicarbonate enhanced this DBNBS-methionine adduct signal (Fig. 10D). Decomposed peroxynitrite did not stimulate any adduct formation (Fig. 10E). In the absence of either tyrosine or methionine, the active peroxynitrite induced a three-line nitroxide signal in the presence of DBNBS alone (Fig. 10F) [53]. Addition of methionine (at various concentrations) inhibited the DBNBS-tyrosyl adduct formation. Clearly, the EPR results suggest that methionine oxidation by peroxynitrite or peroxynitrite/bicarbonate inhibits tyrosyl radical formation (Figs. 10G and H) as compared to results (obtained in the absence of methionine) shown in Figures 10A and B. Incubation mixtures containing the YM peptide, DBNBS, DTPA, and peroxynitrite stimulated the formation of the DBNBS-MY adduct (DBNBS-MetTyr), the spectral intensity of which was enhanced with 25 mM of bicarbonate (Figs. 10I-10L). Based on the computer simulations (shown in dotted lines in Fig. 10L), the spectrum was assigned to the DBNBS-MetTyr, formed from trapping of a radical derived from the methionine residue in YM. The structure was further confirmed by identifying the pronase-induced degradation of the primary adduct. Figure 11 shows the effect of pronase on the DBNBS-MetTyr adduct formation. Upon adding the enzyme, pronase, to incubations containing tyrosylmethionine, DBNBS, and peroxynitrite (after the formation of the DBNBS-MetTyr adduct), the EPR spectrum (Fig. 11A) changed to that of the DBNBS-Met adduct resulting from a peptidic cleavage of the DBNBS-MetTyr (Fig. 11C).
Figure 10. EPR spectra of DBNBS-derived spin adducts formed from oxidation of tyrosine and tyrosylmethionine by peroxynitrite.
(A) Incubations contained Tyr (2 mM), DBNBS (20 mM), DTPA (100 μM) and peroxynitrite (2.5 mM) in a phosphate buffer (pH 7.4, 100 mM). (B) Same as (A) but in the presence of bicarbonate (25 mM). Dotted spectrum represents a computer simulation using the parameters described in the text. (C) Same as (A) but in the presence of Met (2 mM). (D) Same as (C) but containing bicarbonate (25 mM). Dotted EPR spectrum is a computer simulation using the parameters as indicated in the text. (E) Incubations contained Tyr (2 mM), Met (2.5 mM), DBNBS (25 mM), DTPA (100 μM), and decomposed peroxynitrite (2.5 mM) in a phosphate buffer (pH 7.4; 100 mM). (F) Peroxynitrite (2.5 mM) was incubated with DBNBS (20 mM) in a phosphate buffer. (G) Same as (A) but in the presence of Met (2 mM). (H).Same as (B) but in the presence of Met (2.5 mM). (I) Same as (A) but tyrosine was replaced by the dipeptide, tyrosylmethionine (YM) (0.5 mM). (J) Same as (I) but with bicarbonate (25 mM). (K) Incubations contained YM (1 mM) and peroxynitrite (2.5 mM). Other conditions are the same as in (A). (L) Same as (K) but in the presence of bicarbonate (25 mM). Note that the possibility of an alternate chemical structure, DBNBS-CH2S(CH2)2CH(NH2)COOH is not ruled out in spectrum (E).
Figure 11. The effect of pronase on DBNBS-derived spin adduct formed from oxidation of tyrosylmethionine by peroxynitrite.
(A) Incubations contained YM (1 mM), DBNBS (20 mM), DTPA (100 μM), and peroxynitrite (2.5 mM) in a phosphate buffer (pH 7.4, 100 mM). (B) Same as (A) but in the absence of decomposed peroxynitrite. (C) Same as (A) but the enzyme, pronase (1 mg/ml) was added to the incubation mixtures 5 min after the addition of peroxynitrite. Note that the possibility of an alternate chemical structure, DBNBS-CH2S(CH2)2CH(NH2)COOH is not ruled out in spectrum (C).
Although the yield of both nitration and oxidation of YM (forming Y(NO2)M and MYYM) was increased by MPO/H2O2/NO2− and peroxynitrite/bicarbonate, we were unable to detect the corresponding tyrosyl radical formed from YM (i.e., •YM). At this time we do not have an explanation for the lack of detection of the DBNBS-tyrosylmethionine adduct (formed from trapping of •YM by DBNBS) by EPR. However, we compared the effects of DBNBS on the nitration and oxidation products formed from Y, YF, YM, and YC(MTSL modified). DBNBS effectively inhibited the dimeric products (YY, FYYF, CYYC) formed from oxidation of the parent tyrosines by MPO/H2O2 (data not shown). Concomitantly, the corresponding DBNBS-tyrosyl adducts were detected by EPR [29]. In contrast, DBNBS had little or no effect on MYYM formation in MPO/H2O2 system (data not shown), indicating that DBNBS either does not effectively trap the tyrosylmethionine radical or the resulting spin adduct is not stable enough to be detected by EPR. However, the failure to trap •YM by DBNBS remains enigmatic. Additional computational analysis of the stability of tyrosyl radical in YM peptide is, therefore, warranted.
Discussion
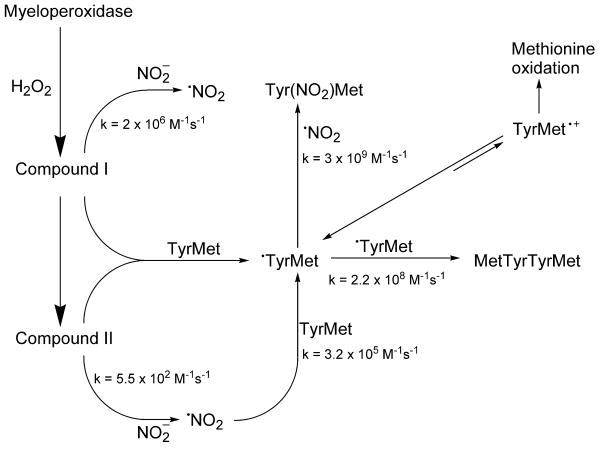
Results from this study show that the presence of a methionine residue adjacent to a tyrosyl group does not inhibit tyrosyl nitration and oxidation catalysed by MPO/H2O2/NO2-. In contrast, the methionine residue next to a tyrosyl group enhances its nitration and oxidation reactions induced by in situ-generated peroxynitrite and peroxynitrite/bicarbonate systems. This is tentatively attributed to an intramolecular radical transfer from methionine sulfide radical cation to the adjacent tyrosyl group. The possibility of an electron transfer from tyrosyl residue to one-electron oxidized methionine moiety has been confirmed by the steady-state radiolysis and photolysis experiments, showing the formation of the corresponding dityrosyl product in the presence of different one-electron oxidants, including Br2•-, N3•, (SCN)2•- and CO3•. In contrast, no dityrosyl product has been detected after one-electron oxidation of the YC peptide; instead, only the disulfide product was formed exclusively. We have used the EPR spin-trapping and HPLC/mass spectrometry techniques to identify intermediates and products, respectively. Schemes 1 and 2 summarize the interactions between tyrosylmethionine and nitrating and oxidizing species generated in MPO/H2O2/NO2- and peroxynitrite/bicarbonate systems. Rate constants shown in these schemes were obtained from the previous publications [54-65].
Scheme 1. Oxidative and nitrative pathways of TyrMet in MPO/H2O2/nitrite system.
In the presence of H2O2, myeloperoxidase (MPO) forms higher oxidants, compounds I and II which oxidizes NO2- to •NO2. Both compounds I and II and •NO2 will oxidize tyrosine to a tyrosyl radical which reacts with •NO2 to form nitrotyrosine. The rate constants shown for •TyrMet represent the reactivity of free tyrosyl radical.
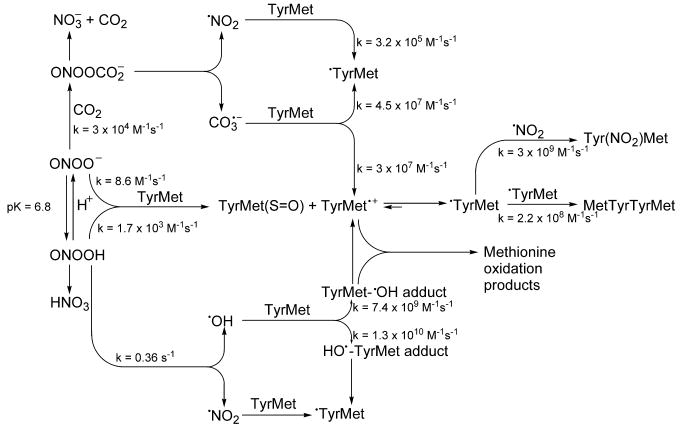
Scheme 2. Oxidative and nitrative pathways of TyrMet by peroxynitrite and bicarbonate.
Peroxynitrite anion exists in equilibrium with peroxynitrous acid (pKa = 6.9). The peroxynitrite anion reacts with CO2 (which, in solution, is in equilibrium with bicarbonate anion) to form a transient intermediate, ONOOCO2-, which homolytically decomposes to give nitrogen-di-oxide (•NO2), and the carbonate anion radical (CO3•-). ONOOH homolytically decomposes to •NO2 and •OH. Nitration of tyrosine occurs following a rapid recombination of •NO2 and tyrosyl radical. The rate constants shown for TyrMet represent the reactivity of free amino acids (tyrosine and methionine).
Reactions between peroxynitrite/bicarbonate, methionine, and methionine-containing peptides
Pioneering works by Pryor at al. and others have shown that peroxynitrite can oxidize methionine by both one-electron and two-electron pathways, but the 2-electron oxidation, which results in the formation of methionine sulfoxide predominates [40; 42]. The methionine sulfide radical cation was proposed as an intermediate during peroxynitrite-induced one-electron oxidation of methionine [40]. Bicarbonate stimulated peroxynitrite-induced one-electron oxidation of methionine [40]. This was attributed to the formation of an intermediate, ONOOCO2-, which decomposed to form a potent one-electron oxidant and nitrating species (i.e., CO3•- and •NO2). The carbon dioxide was shown to stimulate peroxynitrite-mediated nitration of tyrosine residues and inhibit oxidation of methionine residues of glutamine synthetase [39; 66]. The present data show that there is negligible formation of the tyrosyl methionine sulfoxide during oxidation of YM by both peroxynitrite or peroxynitrite and bicarbonate. The major products induced by peroxynitrite were the nitration and oxidation products of YM (e.g., Y(NO2)M and MYYM) which were increased in the presence of bicarbonate. Previously, it has been shown that the product profiles of peroxynitrite or peroxynitrite and bicarbonate mediated oxidation of methionine are different from those obtained from methionine-containing peptides or proteins [67]. The presence of a methionine residue stimulates oxidation and nitration of the neighboring tyrosine, suggesting that the one-electron oxidized form of methionine (i.e., methionine sulfide radical cation) can be “repaired” by tyrosine [64; 65], resulting in its oxidation and/or nitration. This situation is different for other oxidants such as the hypochlorous acid (HOCl) generated by MPO/H2O2/Cl-. Methionine residues inhibit tyrosine chlorination by HOCl in proteins due to stoichiometric scavenging of HOCl by methionine [68; 69]. Mutation of methionine to another amino acid (e.g., alanine) that does not react with HOCl enhanced the yield of chlorotyrosine [68].
Influence of intramolecular electron-transfer on biological nitration and nitrosation
Intracellular nitrosative and nitrative modifications are complex and not always predictable. Obtaining a deeper understanding of the structural motif and sequence specificity that facilitate nitrative and nitrosative posttranslational modification is key to mapping functional regions of proteins that mediate redox signaling [70; 71]. Although tyrosine nitration and cysteinyl thiol nitrosation are induced by NO and oxygen or NO-derived oxidants, this process appears to be selective as not all tyrosine residues or cysteine residues undergo nitration or nitrosation. Previously we proposed that intramolecular electron transfer reactions between tyrosyl radical and cysteine residues in peptides may have a profound effect on the selective nitration of tyrosine residues and oxidation/nitrosation of cysteinyl residues [29]. Analysis of peptide sequences of S-nitrosylated proteins revealed that S-nitrosation occurs predominantly when cysteinyl residues are present adjacent to or closer to a tyrosine residue. The proposed mechanism involved a rapid intramolecular electron transfer between the tyrosyl radical and cysteinyl thiol forming the thiyl radical that reacts with •NO to form the corresponding S-nitrosated cysteine [29]. In the case of tyrosylmethionine peptide, the electron transfer to the methionine sulfide radical cation from tyrosine will be favored. Pulse radiolysis studies performed nearly 20 years ago indicate that tyrosine is a better “radical sink” for methionine radical cation than is methionine as an “radical sink” for tyrosyl radicals (EoTyr•/Tyr= +0.94 V [72]; EoMet+•/Met= +1.48 V [73] vs. NHE). Thus, the occurrence of a rapid and efficient intramolecular electron transfer to MetS•+ from Tyr-OH should induce tyrosyl radical formation. The lack of the inhibitory action and in some cases the stimulatory action of neighboring methionine on the oxidation and nitration of tyrosine residues is attributed to this electron transfer mechanism, although efforts to trap and detect the corresponding tyrosyl radical by DBNBS have been unsuccessful.
Biological Implications
It was postulated that the presence of a redox-active amino acid (cysteine, methionine) residue adjacent to a tyrosyl residue in proteins could inhibit its ability to undergo nitration [30]. As discussed earlier, inhibition of tyrosyl nitration by neighboring cysteinyl residues is attributed to a rapid electron transfer to the tyrosyl radical from the cysteinyl thiols, and not due to direct scavenging of reactive nitrogen species (•NO2 and/or ONOO-) by cysteinyl thiols. Tyrosyl nitration appears to be a selective process and not all tyrosyl residues in a protein are nitration-sensitive. From looking at the peptide sequences of nitrated tyrosine (Table 1), it appears that tyrosyl nitration occurs more frequently when tyrosine residues are juxtaposed to a methionine residue and that the presence of methionine does not inhibit nitration of adjacent tyrosines. The proposed electron transfer mechanism is one of the predictive tools. As reported previously [30], the presence of a positively charged amino acid nearer to tyrosine residues might be another structural motif that facilitate nitration mechanism. However, the use of proteomics methods to identify structural sequences in posttranslational nitrative modifications has recently been questioned [74]. Posttranslational nitrative modifications of alpha-synuclein tyrosine were linked to the onset and progression of neurodegenerative synucleinopathy lesions [8]. The presynaptic protein alpha-synuclein has four tyrosines and four methionines and has no cysteinyl residues. Previously we reported that nitration and oxidation of alpha-synuclein tyrosine were inhibited in the presence of added methionine [75]. Preliminary results suggest that a tyrosyl residue (Tyr-125) was nitrated that is adjacent to a methionine residue (Met-127) during peroxynitrite-induced modification of alpha-synuclein (data not shown). Clearly, the role of methionine on protein tyrosyl nitration deserves further investigation.
Table 1. Sequences of the peptides that have a neighboring methionine adjacent to nitrotyrosine.
| Proteins | Sequence |
|---|---|
| Eukaryotic translation initiation factor 2 | MY2SGAGPVLASPAPTTSPIPGYAFKPPPRPDFGTTGR |
| Nuclear factor 1A, isoform 1 | MY2SPLCLTQDEFHPFIEALLPHVR. |
| Pyruvate kinase | ITLDNAY148MEKCDENILWLDYK |
| Actinin | AIMTY241VSSFYHAFSGAQKAETAANR |
| Vacuolar protein sorting 16, isoform 1 | MDCY4TANWNPLGDSAFYR. |
| T-complex associated-testisexpressed 1-like (Tcte1l) | MEGY4QRPCDEVGFNADEAHNIVK. |
| Interleukin 1 family member 6 | KDIMDLY96NQPEPVKSFLFYHSQSGR |
| Hypothetical fimbral chaperone | TRIKMFY139RPAQHLK |
Summary
The presence of a methionine residue adjacent to a tyrosyl moiety in peptides does not inhibit nitration and oxidation of tyrosine by MPO/H2O2/NO2-. The methionine residue actually stimulated peroxynitrite and peroxynitrite/bicarbonate-mediated nitration and oxidation of tyrosine in TyrMet peptide. An intramolecular electron-transfer to the methionine sulfide radical cation from the tyrosyl residue has been proposed to account for enhanced tyrosyl nitration and oxidation.
Supplementary Material
Acknowledgments
This research was supported by a grant from National Institutes of Health Grant HL63119.
- YM
N-acetyl tyrosylmethionine amide
- MYYM
bis-N-acetyl tyrosylmethionine amide
- Y(NO2)M
N-acetyl nitrotyrosylmethionine
- YF
N-acetyl tyrosylphenylalanine amide
- DBNBS
3,5-dibromo-4-nitrosobenzenesulfonic acid
- DMPO
5,5′–dimethyl-1-pyrroline N-oxide
- DTPA
diethylenetriaminepentaacetic acid
- EPR
electron paramagnetic resonance
- MMTS
methylmethanethiosulfonate
- MPO
myeloperoxidase
- •NO2
nitrogen-di-oxide radical
- NO2Tyr
3-nitrotyrosine
- Tyr
tyrosine
- YC
N-acetyl tyrosylcysteine amide
- YCysCysY
disulfide formed from YC
- FYYF
bis-N-acetyl tyrosylphenylalanine amide
- YM=O
N-acetyl tyrosylmethionine sulfoxide amide
- Recommended IUPAC nomenclature: NaHCO3
sodium hydrogen carbonate
- ONOO-
oxoperoxonitrate (1-)
- ONOOH
hydrogen oxoperoxonitrate
- •NO2
dioxidonitrogen(•)
- CO3•-
trioxidocarbonate(•1-)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kooy NW, Lewis SJ, Royall JA, Ye YZ, Kelly DR, Beckman JS. Crit Care Med. 1997;25:812–819. doi: 10.1097/00003246-199705000-00017. [DOI] [PubMed] [Google Scholar]
- 2.Turko IV, Murad F. Pharmacol Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 3.Baldus S, Eiserich JP, Brennan ML, Jackson RM, Alexander CB, Freeman BA. Free Radic Biol Med. 2002;33:1010. doi: 10.1016/s0891-5849(02)00993-0. [DOI] [PubMed] [Google Scholar]
- 4.Wu W, Chen Y, Hazen SL. J Biol Chem. 1999;274:25933–25944. doi: 10.1074/jbc.274.36.25933. [DOI] [PubMed] [Google Scholar]
- 5.Leeuwenburgh C, Hardy MM, Hazen SL, Wagner P, Oh-ishi S, Steinbrecher UP, Heinecke JW. J Biol Chem. 1997;272:1433–1436. doi: 10.1074/jbc.272.3.1433. [DOI] [PubMed] [Google Scholar]
- 6.Pavlick KP, Laroux FS, Fuseler J, Wolf RE, Gray L, Hoffman J, Grisham MB. Free Radic Biol Med. 2002;33:311–322. doi: 10.1016/s0891-5849(02)00853-5. [DOI] [PubMed] [Google Scholar]
- 7.Haddad IY, Pataki G, Hu P, Galliani C, Beckman JS, Matalon S. J Clin Invest. 1994;94:2407–2413. doi: 10.1172/JCI117607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paxinou E, Chen Q, Weisse M, Giasson BI, Norris EH, Rueter SM, Trojanowski JQ, Lee VM, Ischiropoulos H. J Neurosci. 2001;21:8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schopfer FJ, Baker PR, Freeman BA. Trends Biochem Sci. 2003;28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Rubbo H, Radi R, Trujillo M, Telleri R, Kalyanaraman B, Barnes S, Kirk M, Freeman BA. J Biol Chem. 1994;269:26066–26075. [PubMed] [Google Scholar]
- 11.Lanone S, Manivet P, Callebert J, Launay JM, Payen D, Aubier M, Boczkowski J, Mebazaa A. Biochem J. 2002;366:399–404. doi: 10.1042/BJ20020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenacre SA, Ischiropoulos H. Free Radic Res. 2001;34:541–581. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 13.Brennan ML, Wu W, Fu X, Shen Z, Song W, Frost H, Vadseth C, Narine L, Lenkiewicz E, Borchers MT, Lusis AJ, Lee JJ, Lee NA, bu-Soud HM, Ischiropoulos H, Hazen SL. J Biol Chem. 2002;277:17415–17427. doi: 10.1074/jbc.M112400200. [DOI] [PubMed] [Google Scholar]
- 14.Beckman JS. Chem Res Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- 15.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Arch Biochem Biophys. 1992;298:431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 16.Pfeiffer S, Lass A, Schmidt K, Mayer B. FASEB J. 2001;15:2355–2364. doi: 10.1096/fj.01-0295com. [DOI] [PubMed] [Google Scholar]
- 17.Jourd'heuil D, Jourd'heuil FL, Kutchukian PS, Musah RA, Wink DA, Grisham MB. J Biol Chem. 2001;276:28799–28805. doi: 10.1074/jbc.M102341200. [DOI] [PubMed] [Google Scholar]
- 18.MacPherson JC, Comhair SA, Erzurum SC, Klein DF, Lipscomb MF, Kavuru MS, Samoszuk MK, Hazen SL. J Immunol. 2001;166:5763–5772. doi: 10.4049/jimmunol.166.9.5763. [DOI] [PubMed] [Google Scholar]
- 19.Pryor WA, Squadrito GL. Am J Physiol. 1995;268:L699–L722. doi: 10.1152/ajplung.1995.268.5.L699. [DOI] [PubMed] [Google Scholar]
- 20.Sampson JB, Rosen H, Beckman JS. Methods Enzymol. 1996;269:210–218. doi: 10.1016/s0076-6879(96)69023-5. [DOI] [PubMed] [Google Scholar]
- 21.Khairutdinov RF, Coddington JW, Hurst JK. Biochemistry. 2000;39:14238–14249. doi: 10.1021/bi001270x. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Miller MJ, Joshi MS, Thomas DD, Lancaster JR., Jr Proc Natl Acad Sci U S A. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marla SS, Lee J, Groves JT. Proc Natl Acad Sci U S A. 1997;94:14243–14248. doi: 10.1073/pnas.94.26.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denicola A, Batthyany C, Lissi E, Freeman BA, Rubbo H, Radi R. J Biol Chem. 2002;277:932–936. doi: 10.1074/jbc.M106589200. [DOI] [PubMed] [Google Scholar]
- 25.Denicola A, Souza JM, Radi R. Proc Natl Acad Sci U S A. 1998;95:3566–3571. doi: 10.1073/pnas.95.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ischiropoulos H. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 27.Lymar SV, Hurst JK. J Am Chem Soc. 1995;117:8867–8868. [Google Scholar]
- 28.Zhang H, Joseph J, Feix J, Hogg N, Kalyanaraman B. Biochemistry. 2001;40:7675–7686. doi: 10.1021/bi002958c. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Xu Y, Joseph J, Kalyanaraman B. J Biol Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]
- 30.Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Arch Biochem Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 31.Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Am J Physiol Lung Cell Mol Physiol. 2004;287:L262–L268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- 32.Augusto O, Bonini MG, Amanso AM, Linares E, Santos CC, De Menezes SL. Free Radic Biol Med. 2002;32:841–859. doi: 10.1016/s0891-5849(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 33.Schöneich C. Biochim Biophys Acta. 2005;1703:111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Stadtman ER, Levine RL. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 35.Vogt W. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 36.Schöneich C, Sharov VS. Free Radic Biol Med. 2006;41:1507–1520. doi: 10.1016/j.freeradbiomed.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Alvarez B, Rubbo H, Kirk M, Barnes S, Freeman BA, Radi R. Chem Res Toxicol. 1996;9:390–396. doi: 10.1021/tx950133b. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez B, Radi R. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 39.Stadtman ER, Moskovitz J, Levine RL. Antioxid Redox Signal. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- 40.Pryor WA, Jin X, Squadrito GL. Proc Natl Acad Sci U S A. 1994;91:11173–11177. doi: 10.1073/pnas.91.23.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez B, Ferrer-Sueta G, Freeman BA, Radi R. J Biol Chem. 1999;274:842–848. doi: 10.1074/jbc.274.2.842. [DOI] [PubMed] [Google Scholar]
- 42.Perrin D, Koppenol WH. Arch Biochem Biophys. 2000;377:266–272. doi: 10.1006/abbi.2000.1787. [DOI] [PubMed] [Google Scholar]
- 43.Tien M, Berlett BS, Levine RL, Chock PB, Stadtman ER. Proc Natl Acad Sci U S A. 1999;96:7809–7814. doi: 10.1073/pnas.96.14.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haqqani AS, Kelly JF, Birnboim HC. J Biol Chem. 2002;277:3614–3621. doi: 10.1074/jbc.M105730200. [DOI] [PubMed] [Google Scholar]
- 45.Tedeschi G, Cappelletti G, Negri A, Pagliato L, Maggioni MG, Maci R, Ronchi S. Proteomics. 2005;5:2422–2432. doi: 10.1002/pmic.200401208. [DOI] [PubMed] [Google Scholar]
- 46.Kanski J, Hong SJ, Schöneich C. J Biol Chem. 2005;280:24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 47.Sacksteder CA, Qian WJ, Knyushko TV, Wang H, Chin MH, Lacan G, Melega WP, Camp DG, Smith RD, Smith DJ, Squier TC, Bigelow DJ. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 48.Papee HM, Petriconi GL. Nature. 1964;204:142–144. [Google Scholar]
- 49.Lamb AB, Damon EB. J Am Chem Soc. 1937;59:383–390. [Google Scholar]
- 50.Cope VW, Chen SN, Hoffman MZ. J Am Chem Soc. 1973;95:3116–3121. [Google Scholar]
- 51.Neta P, Huie RE, Ross AB. J Phys Chem Ref Data. 1988;17:1027–1284. [Google Scholar]
- 52.Meli R, Nauser T, Latal P, Koppenol WH. J Biol Inorg Chem. 2002;7:31–36. doi: 10.1007/s007750100262. [DOI] [PubMed] [Google Scholar]
- 53.Gunther MR, Tschirret-Guth RA, Lardinois OM, Ortiz de Montellano PR. Chem Res Toxicol. 2003;16:652–660. doi: 10.1021/tx0256580. [DOI] [PubMed] [Google Scholar]
- 54.Tien M. Arch Biochem Biophys. 1999;367:61–66. doi: 10.1006/abbi.1999.1226. [DOI] [PubMed] [Google Scholar]
- 55.Nakao LS, Iwai LK, Kalil J, Augusto O. FEBS Lett. 2003;547:87–91. doi: 10.1016/s0014-5793(03)00674-4. [DOI] [PubMed] [Google Scholar]
- 56.Davies MJ, Gilbert BC, Norman ROC. J Chem Soc Perkin Trans. 1983:731–738. [Google Scholar]
- 57.Pietraforte D, Minetti M. Biochem J. 1997;321(Pt 3):743–750. doi: 10.1042/bj3210743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kettle AJ, Winterbourn CC. Biochemistry. 2001;40:10204–10212. doi: 10.1021/bi010940b. [DOI] [PubMed] [Google Scholar]
- 59.Burner U, Furtmuller PG, Kettle AJ, Koppenol WH, Obinger C. J Biol Chem. 2000;275:20597–20601. doi: 10.1074/jbc.M000181200. [DOI] [PubMed] [Google Scholar]
- 60.Marquez LA, Dunford HB. J Biol Chem. 1995;270:30434–30440. doi: 10.1074/jbc.270.51.30434. [DOI] [PubMed] [Google Scholar]
- 61.Prütz WA, Mönig H, Butler J, Land EJ. Arch Biochem Biophys. 1985;243:125–134. doi: 10.1016/0003-9861(85)90780-5. [DOI] [PubMed] [Google Scholar]
- 62.Chen SN, Hoffman MZ. Radiat Res. 1975;62:18–27. [PubMed] [Google Scholar]
- 63.Prütz WA, Butler J, Land EJ, Swallow AJ. Int J Radiat Biol. 1989;55:539–556. doi: 10.1080/09553008914550591. [DOI] [PubMed] [Google Scholar]
- 64.Bobrowski K, Wierzchowski KL, Holcman J, Ciurak M. Int J Radiat Biol. 1992;62:507–516. doi: 10.1080/09553009214552431. [DOI] [PubMed] [Google Scholar]
- 65.Bobrowski K, Wierzchowski KL, Holcman J, Ciurak M. Int J Radiat Biol. 1990;57:919–932. doi: 10.1080/09553009014551041. [DOI] [PubMed] [Google Scholar]
- 66.Berlett BS, Levine RL, Stadtman ER. Proc Natl Acad Sci U S A. 1998;95:2784–2789. doi: 10.1073/pnas.95.6.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen JL, Miller BL, Zhang X, Hug GL, Schöneich C. J Am Chem Soc. 1997;119:4749–4757. [Google Scholar]
- 68.Shao B, Oda MN, Bergt C, Fu X, Green PS, Brot N, Oram JF, Heinecke JW. J Biol Chem. 2006;281:9001–9004. doi: 10.1074/jbc.C600011200. [DOI] [PubMed] [Google Scholar]
- 69.Hazell LJ, Davies MJ, Stocker R. Biochem J. 1999;339(Pt 3):489–495. [PMC free article] [PubMed] [Google Scholar]
- 70.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 71.Ischiropoulos H. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 72.DeFelippis MR, Murthy CP, Faraggi M, Klapper MH. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- 73.Sanaullah, Wilson S, Glass RS. J Inorg Biochem. 1994;55:87–99. doi: 10.1016/0162-0134(94)85031-3. [DOI] [PubMed] [Google Scholar]
- 74.Zhang H, Xu Y, Joseph J, Kalyanaraman B. Methods Enzymol. 2008;440:65–94. doi: 10.1016/S0076-6879(07)00804-X. [DOI] [PubMed] [Google Scholar]
- 75.Andrekopoulos C, Zhang H, Joseph J, Kalivendi S, Kalyanaraman B. Biochem J. 2004;378:435–447. doi: 10.1042/BJ20031466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.