Abstract
Laminin-332 (Ln-332) is an extracellular matrix molecule that regulates cell adhesion, spreading, and migration by interaction with cell surface receptors such as α3β1 and α6β4. Previously, we developed a function-blocking monoclonal antibody against rat Ln-332, CM6, which blocks hemidesmosome assembly induced by Ln-332-α6β4 interactions. However, the location of its epitope on Ln-+332 has remained unclear. In this study, we show that the CM6 epitope is located on the LG2 module of the Ln-332 α3 chain. To specify the residues involved in this epitope, we produced a series of GST-fused α3 LG2 mutant proteins in which rat-specific acids were replaced with human acids by a site-directed mutagenesis strategy. CM6 reactivity against these proteins showed that CM6 binds to the 1089NERSVR1094 sequence of rat Ln-332 LG2 module. In a structural model, this sequence maps to an LG2 loop sequence that is exposed to solvent according to predictions, consistent with its accessibility to antibody. CM6 inhibits integrin-dependent cell adhesion on Ln-332 and inhibits cell spreading on both Ln-332 and recombinant LG2 (rLG2; but not rLG3), suggesting the presence of an α3β1 binding site on LG2. However, we were unable to show that rLG2 supports adhesion in standard assays, suggesting that LG2 may contain a “weak” integrin binding site, only detectable in spreading assays that do not require washes. These results, together with our previous findings, indicate that binding sites for α3β1 and α6β4 are closely spaced in the Ln-332 LG domains where they regulate alternative cell functions, namely adhesion/migration or hemidesmosome anchoring.
Keywords: laminin, integrin, function-blocking, antibody, cell adhesion
Introduction
Laminin-332 (Ln-332; formerly named laminin-5, epiliglin, kalinin, ladsin, and nicein) is a heterotrimer consisting of α3, β3, and γ2 chains (Miyazaki et al., 1993; Kunneken et al., 2004), which regulates various significant physiological events such as cell adhesion, migration, spreading, proliferation, differentiation, and survival (Kunneken et al., 2004; Manohar et al., 2004; Klees et al., 2005). It is well known that there are two major integrin receptors for Ln-332, α6β4 and α3β1, which are expressed by various cells (Carter et al., 1991; Niessen et al., 1994). In the basement membrane (BM) of skin, Ln-332 anchors the epidermis to underlying collagen matrix via the formation of hemidesmosomes, which are multi-protein cell adhesion devices revolving around the interaction of integrin α6β4 and intermediate filaments (Carter et al.,1991; Borradori et al., 1998; Jones et al., 1998; Hirako, 1998). In wound-healing situations, keratinocytes surrounding a wound site upregulate the expression and secretion of Ln-332; this activity has also been correlated with the induction of cell migration via α3β1 (Zhang and Kramer, 1996; Nguyen et al., 2001). The interactions between Ln-332 and integrin receptors, and their involvement in many significant physiological processes, have been intensely studied. Clearly, there is great interest in understanding the mechanism(s) by which Ln-332 interacts with these receptors.
Similar to other laminin α chains, the Ln-332 α3 chain harbors a large C-terminal globular (G) domain composed of five laminin G-like (LG) modules, designated LG1 through LG5, which play a central, if poorly detailed, role in interacting with integrins (Timpl et al., 2000; Tisi et al., 2000; Hintermann and Quaranta, 2004). This role has been clearly established by studies utilizing recombinant single or combined LG module proteins or recombinant laminins with deleted LG modules (Mizushima et al., 1997; Shang et al., 2001; Utani et al., 2001; Okamoto et al., 2003; Kim et al., 2005). An alternative strategy for elucidating cell binding sites within the G domain of laminin is the use of function-blocking monoclonal antibodies (mAb) prepared against laminin isoforms to detect cell-binding sites. This is especially useful for Ln-332, which harbors binding sites for two functionally distinct integrins (Carter et al., 1991; Niessen et al., 1994). α3β1 interacts with actin cytoskeleton, supports adhesion and motility, engages Ln-332 in wounded or remodeling epithelia, and is confined to lateral plasma membranes in resting epithelia. In contrast, α6β4 interacts with keratins, supports static adhesion via hemidesmosomes, engages Ln-332 in resting epithelia, and is excluded from Ln-332 interactions in remodeling epithelia.
To study spatial relationships between Ln-332 binding sites for integrins α3β1 and α6β4, respectively, we previously developed a function-blocking mAb against Ln-332 purified from the rat bladder carcinoma 804G cell line (Plopper et al., 1996). This antibody, named CM6, inhibits hemidesmosome assembly and, according to rotary shadowing electron microscopy, is likely to bind around the G domain of the Ln-332 α3 chain (Baker et al., 1996), suggesting that the CM6 epitope on Ln-332 is involved with integrin binding sites, especially α6β4. Therefore, characterization of the CM6 epitope may shed light on structural determinants of integrin-Ln-332 interactions.
In the current study, we first mapped the CM6 epitope to a short amino-acid sequence in the LG2 domain of Ln-332, 1089NERSVR1094, by testing with recombinant GST- fused LG module proteins followed by a site-directed mutagenesis strategy. We then found that, while recombinant LG2 (rLG2) or 1089NERSVR1094 do not support cell adhesion, in a mild, no-washing assay rLG2 and Ln-332 promoted cell spreading to a similar extent, which was specifically inhibited by CM6. We therefore conclude that LG2 harbors a “weak” integrin binding site and that CM6 may be a useful tool for future studies aimed at dissecting the relative contribution of this site to integrin α3β1 versus α6β4 engagement.
Materials and Methods
Cell line and culture conditions
MCF10A cells were purchased from American Type Culture Collection (ATCC, Manassas, VA), and maintained in DMEM/F12 media (Invitrogen, Carlsbad, CA) containing 5% horse serum (Invitrogen, Carlsbad, CA), 0.1 μg/ml cholera toxin (Calbiochem, EMD Chemicals, Gibbstown, NJ), 10 μg/ml insulin (Invitrogen), 0.5 μg/ml hydrocortisone (Sigma, St. Louis, MO), and 20 ng/ml epidermal growth factor (Invitrogen) at 37°C in a humidified, 5% CO2, 95% air atmosphere.
Antibodies and reagents
An anti-His mAb was purchased from Qiagen (Valencia, CA), anti-glutathione-S-transferase mAb (G1160) from Sigma (St. Louis, MO), polyclonal antibody (pAb) against the C-terminus of Ln-332 β3 chain (sc-20775; H-300) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and secondary anti-rabbit and mouse-IgG HRP mAbs were purchased from GE Healthcare (Chalfont St. Giles, UK). All antibodies were used according to manufacturer’s instructions. CM6 mAb and TR1 were prepared in house as previously described (Plopper et al., 1996). For western blots, PVDF membranes (Perkin Elmer, Waltham, MA) were used to transfer protein from gels, and the ECL +plus system (Perkin Elmer Life and Analytical Science, Shelton, CT) was used for visualization of protein bands. Celltracker™ green CMFDA was purchased from Invitrogen.
Fluorescent labeling of cells
One day before fluorescent labeling, MCF10A cells were seeded in T25 flasks. After approximately 24 h, media was aspirated and CelltrackerTM green CMFDA in D-phosphate buffered saline (PBS; labeling solution) was added and incubated for 30 min at 37°C. After initial incubation, labeling solution was removed, and 10 ml of complete media was added and allowed to incubate for 30 min at 37°C. After final incubation, complete medium was again added and allowed to incubate overnight at 37°C until use.
Measurement and quantification of cell spreading in single-cell microscopy assays
Sterile 96-well plates (BD Biosciences Falcon™, San Jose, CA) were coated with Ln-332 (1 μg/ml; purified from 804G cells in house) or collagen-I (10 μg/ml; Sigma) at 4°C overnight. Labeled MCF10A cells were detached with TrypLE™ Express (Invitrogen) and suspended in DMEM growth media and plated (5000 cells in 100 μl per well). Normal mouse IgG (100 μg/ml; Invitrogen), custom-made CM6 (100 μg/ml), or custom-made TR1 (50 μg/ml) were added to cells when plated where indicated. Cells were incubated for ~7 h, and images were collected every 6 min using a BD Pathways 855 Bioimager (BD, Franklin Lakes, NJ).
To quantify cell spreading (via surface area), grayscale TIFF images were imported into MATLAB (The MathWorks, Natick, MA) and converted to binary images using a local thresholding algorithm to eliminate any artifacts due to uneven illumination. Binary images were segmented using watershed segmentation to separate touching cells. Areas and centroids for each cell at each time point were computed from the segmented image. An external particle tracking toolbox (physics.georgetown.edu/matlab) was used to assign cell centroids to individual cell IDs across multiple time points. Cell IDs along with area and centroid position information were saved to comma-separated text files for statistical analysis. Our custom MATLAB code also generated composite images consisting of overlays of the detected areas and automatically generated cell IDs combined with the original grayscale images. These composite images were used to visually validate the results of the automated area detection. Prior to quantification, any objects that were <300 or >3000 pixels were thresholded from dataset to remove extraneous noise and clumps of cells from analysis (range determined based on manual inspection).
Cell spreading inhibition assays
Sterile 96-well microplates were coated with Ln-332 (1 μg/ml), GST (50 μg/ml), rLG2 (50 μg/ml), or rLG3 (50 μg/ml) at 4°C overnight. MCF10A cells were detached with trypsin and neutralized with complete medium. 100 ml of media containing 5000 cells was added to wells and incubated for 5 h at 37°C in a humidified, 5% CO2, 95% air atmosphere. After incubation, attached cells were fixed with 3.7% paraformaldehyde for 10 min at 37°C and permeabilized with 0.1% Triton X-100 in PBS for 10 min at RT and then immediately blocked with PBS containing 10% goat serum and 1% BSA for 40 min at RT. Cells were stained with Hoechst and phalloidin in PBS containing 3% BSA for 2 h at RT. Cell spreading was quantified by counting spread cells in five microscope fields.
Construction, expression, and purification of Ln-332 α3 LG modules
Individual LG1-5 modules of rat Ln-332 were expressed as recombinant, GST-fused proteins (rLG1-5) using the pGEX-2T expression vector (Pharmacia, Piscataway, NJ). Rat Ln-332 α3 cDNA was cloned from an 804G lambda zap library (Baker et al., 1996), and two overlapping rat laminin α3 cDNA clones, pCR5C5 (bps 874–3180) and pCRα3α4 (bps 3034–5178) were used as templates for PCR amplification of the LG modules using the Taq polymerase (Boehringer Mannheim, Indianapolis, IN).
Induction of protein expression in Escherichia coli (E. coli) JM109 strain grown to mid-log phase in Luria Bertani medium was carried out with 1.0 mM isopropyl-β-D-thiogalactopyranoside (Boehringer Mannheim). After protein induction for 5 h at 34°C, cells were harvested by centrifugation at 6000 rpm for 10 min (Sorvall, GSA rotor). Cell pellets were stored at −80°C until use. For protein purification, pellets were thawed and resuspended in ice-cold lysis buffer (Tris-HCl, 20 mM, ph 7.8; glycerol, 10%; NaCl, 0.15 M; imidazole, 5 mM; Triton-X-100, 1%) containing 1 mM phenylmethylsulphonyl fluoride (PMSF; Sigma). Cell lysis was carried out by passage of cell suspension through French Press (SLM Instruments, Urbana, IL) at 1,500 psi twice. Clarified cell lysates were applied to Ni-NTA columns (Qiagen, Valencia, CA). The column was washed with wash buffer (Tris-HCl, 20 mM, pH 7.8; glycerol, 10%; NaCl, 0.5 M; imidazole, 60 mM). Protein elution was carried out with 300 mM imidazole and 1 mM dithiothreitol (Sigma) in wash buffer.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the overlap extension method (Offord, 1987). Four primers were used for cloning of each mutant. Two general primers, each corresponding to the 5′-start and 3′-end sequences of the laminin α3LG2 module, were used for all mutants. In addition, two mutant-specific primers, each corresponding to the sense and anti-sense sequences encompassing the mutation site were also used. The primers used are included in Supplemental figure 1; mutated codons are underlined. Plasmid DNA of mutant clones were sequenced from both the 5′ and 3′ ends to confirm the presence of the intended mutation without any additional, unwanted mutations.
Solid phase binding assays
Sterile 96-well microplates were coated with Ln-332 (1 μg/ml), GST (50 μg/ml), rLG2 (50 μg/ml), or synthetic peptides: NERSVR, DVQNTI, or GLLFFA (100 μg/ml, Biopeptide Co. Inc., San Diego, CA) at 4°C overnight. After coating, wells were blocked with 5% skim milk in PBS at 37°C for 1 h. CM6 was added as a primary antibody to wells (50 μg/ml) and incubated for 2 h at RT and washed five times with TBST (TBS buffer containing 0.01% Tween 20). Anti-mouse IgG conjugated with HRP was added as a secondary antibody to wells and incubated for 1 h at RT and washed five times with TBST. After washing, 100 ml of chromogen was added to wells and incubated for 10 min at RT, at which time 100 ml of stop solution was added to wells. Bound antibody was detected by measuring O.D. (450 nm).
Protein tertiary structure predictions
To create the tertiary structure predictions of the rat Ln-332 LG2 module, we used the homology detection and structure prediction by hidden Markov model (HMM)-HMM comparison (HHpred; Soding, 2005; Soding et al., 2005), a tool available on the Max Plank Institute for Developmental Biology website (http://toolkit.tuebingen.mpg.de/hhpred). Upon completion of the job, Modeller—under a free academic license key—was used with published laminin α2 templates (Tisi et al., 2000; Carafoli et al., 2009), template scores >99%) to compute the tertiary structure (Eswar et al., 2006). Finally, Modeller-generated PDB files were rendered using PyMOL (DeLano Scientific LLC, Palo Alto, CA), to highlight the region of interest and export the visualizations.
Statistical analysis
All data are presented in box-and-whisker plots representing the mean for each treatment (bold horizontal line), with 25th and 75th quartiles (box), 95% confidence intervals (whiskers), and outliers (scatter). Statistical analysis was performed using SPSS, version 17 (SPSS Inc., Chicago, IL). Distributions of data were examined using Shapiro Wilks W tests for normality. Differences between treatments were examined using Mann-Whitney U tests, and were considered significant when P < 0.05.
Results
CM6 mAb recognizes the α3 chain of Ln-332
Ln-332 is a heterotrimer that consists of α3, β3, and γ2 chains (Fig. 1A), which can be resolved into individual bands by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions (Fig. 1B). Previously published N-terminal sequencing results have shown that these bands, with apparent molecular weights of approximately 150, 140, and 135 kDa, correspond to the α3, γ2, and β3 chains of Ln-332, respectively (Falk-Marzillier et al., 1998). Western blotting analysis of the Ln-332 chains with CM6 indicates that it specifically recognizes the ~150 kDa band, which corresponds to the α3 chain (Fig. 1C). In contrast, a commercially available anti-β3 antibody recognizes the ~135 kDa band, or the β3 chain as expected. And TR1, a non-functional mAb to Ln-332, recognized the ~140 kDa and ~80 kDa bands (i.e., both forms of γ2 chain, as expected) (Plopper et al., 1996). Taken together, these results indicate that the CM6 epitope exists within the α3 chain, but not the β3 or γ2 chains of Ln-332.
Fig. 1. CM6 recognizes α3 chain, but not β3 and γ2 of Ln-332.

(A) Ln-332 is composed of α3, β3, and γ2 chains assembled into α3β3γ2 heterotrimer via a three-s tranded α-helical coiled-coil domain. A globular domain (G domain) uniquely exists at the C-terminal region of the α3 chain, which consists of five tandem laminin G-like (LG) repeats module (LG1-LG5). The linker region between LG3 and LG4 (indicated by arrowhead) is the target for processing by serine proteases such as plasmin. (B) SDS-PAGE of Ln-332. Purified Ln-332 was run on 4–12 % gradient gel under reducing conditions. Separated Ln-332 chains were stained with Coomassie Brilliant Blue. The gel includes bands identified as the α3 (~150 kDa), β3 (~135 kDa), and γ2 (~140 kDa and ~80 kDa) chains. (C) Immunoblot analysis was carried out against Ln-332 chains using CM6, anti-β3, anti-γ2 antibody (TR1). CM6, anti-β3 pAb, and TR1 were used as a first antibody at the diluted ratio of 1: 100, 1: 1000, 1: 200, respectively. Anti-rabbit or mouse HRP conjugated IgG was used as a secondary antibody at the diluted ratio of 1: 1000. These results suggest that CM6 recognizes the α3 chain, but not β3 and γ2 chains, of Ln-332.
CM6 reacts specifically with the LG2 module of Ln-332
As shown in Fig. 2A, the Ln-332 α3 chain consists of a number of distinct protein modules, including laminin type EGF-like repeats (domain IIIa), coiled-coil domains (domains I/II), and tandem LG modules (domains LG1-LG5) (Baker et al., 1996). In particular, the five LG modules at the C-terminus of the α3 chain are known to play a significant role in regulating cell behavior by interacting with various cell surface receptors, such as integrins (Hintermann and Quaranta, 2004). Having identified that CM6 is specific to the Ln-332 α3 chain, we further investigated the exact location of the CM6 epitope. Individual LG modules of the α3 chain were expressed in E. coli as double-tagged fusion proteins, comprised of a GST moiety at the N-terminus and a His tag at the C terminus, to provide convenient handles for protein purification and to enhance solubility (Fig. 2A). Purified, individual, recombinant LG proteins (rLG1-5) were separated by SDS-PAGE under reducing conditions and visualized with Coomassie Brilliant Blue (Fig. 2B). To confirm location of the purified products, western blot analysis using an anti-His antibody was also carried out against the identical gel (Fig. 2C, top). As expected, this antibody detected all individual rLG proteins, suggesting that all were purified to near homogeneity because of their appearance as single bands on the stained gels. To further identify the LG module for which CM6 is specific, a western blot using CM6 was also performed against the identical gel (Fig. 2C, bottom). This result revealed that LG2 is the only module reactive with CM6 among all LG modules within the C-terminal domain of Ln-332.
Fig. 2. The epitope of CM6 exists within the LG2 module of Ln-332.
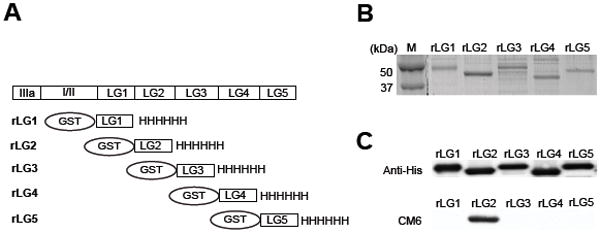
(A) Schematic diagram of recombinant LG modules (rLG1-5) used in this study. Individual modules were expressed in bacteria as a fusion protein with GST tag at the N-terminus and with his tag at C-terminus. (B) SDS gel electrophoresis of Ln-332 rLG modules. Modules were purified by nickel agarose beads and run on a 12 % gel under reducing conditions. Protein bands were visualized by Coomassie Brilliant Blue staining. (C) Immunoblot analysis was carried out using the mAb against His6 tag (top) and CM6 (bottom). Identical gel was transferred to PVDF membrane. mAbs against His6 and CM6 was used as a first antibody at the diluted ratio of 1: 2000 and 1: 100, respectively. Anti-mouse HRP conjugated IgG was used as a secondary antibody, at the diluted ratio of 1: 1000. These results suggest that the epitope of CM6 exists within the LG2 module of Ln-332.
Localization of CM6 epitope to a hexapeptide sequence within Ln-332 α3LG2 module
We further refined localization of the CM6 epitope with the following strategy. Since CM6 is a rat-specific antibody and does not react with human Ln-332, mutation of rat-specific amino acid residues to corresponding human residues should negatively affect CM6 reactivity with rat rLG2. When the primary sequence of rat LG2 is aligned to the human sequence, a total of 29 single residue substitutions are revealed (Fig. 3A). To narrow down screening by mutagenesis, multiple mutations were introduced concurrently into a single mutant if the sites were contiguous, based on the rLG2 molecule used in Fig. 3A (i.e., reducing 29 residues to 17 sites; shown in grey). Out of a total of 17 mutants expressed, only 2 mutants were found to be CM6 negative by western blotting (Fig. 3B). These two mutants both contain mutation sites encompassing a continuous stretch where the rat sequence, 1089NERSVR1094, was changed to the human sequence, DVQNTI (i.e., sites 14–15). These results suggest that the CM6 epitope contains the hexapeptide, 1089NERSVR1094, within rat laminin α3 LG2 module. Therefore, we investigated whether CM6 binds to NERSVR sequence using the following synthetic peptides: NERSVR (rat 1089–1094), DVQNTI (human 1089–1094), and GLLFFA (rat 1025–1030). As shown in Fig. 3C, CM6 bound the synthetic peptide sequence NERSVR similarly to intact Ln-332 (N=2, in triplicate; p=0.394) and rLG2 module (N=2, in triplicate; p=0.240). In contrast, compared to Ln-332, CM6 had almost no reactivity to the human sequence DVQNTI (N=2, in triplicate; p=0.002) or the rat LG2 module sequence GLLFFA (N=2, in triplicate; p=0.002), both of which resulted in measurements similar to antibody on GST or PBS controls. Taken together, these results suggest that CM6 specifically recognizes 1089NERSVR1094 sequence on rat LG2 module.
Fig. 3. Mapping of CM6 epitope by site-directed mutagenesis.
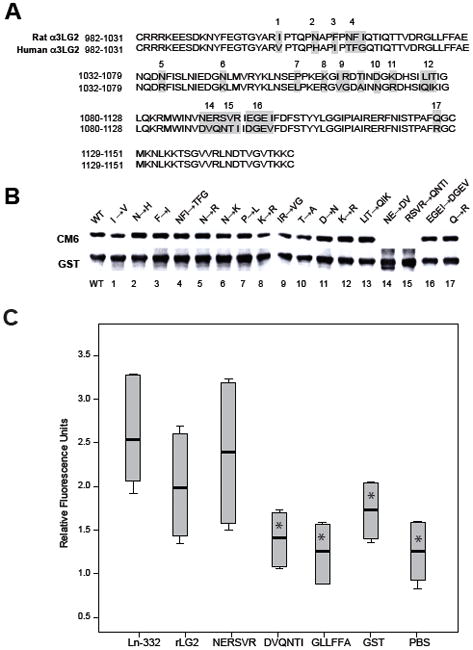
(A) Sequence alignment of human and rat laminin α3 LG2 module. The rat laminin α3 LG2 sequence was aligned to that of human laminin α3 LG2. There is a difference of total 29 amino acid residues between them. To delimitate the epitope of CM6, a total of 17 mutants were generated by replacing rat amino acid residues for human amino acid residues within the rat laminin α3 LG2 module using a site directed mutagenesis method. (B) In site-directed mutagenesis, pGEX-2T LG2 vector (which encodes rat laminin α3 LG2 module) was used as a template to generate 17 mutant LG2 proteins. Wild type and 17 mutant LG2 proteins were purified by the Nickel agarose beads. Purified recombinant wild type (lane 1) and 17 mutant LG2 proteins (lanes 2–17) were run on 12 % gel and separated gel was immunoblotted by CM6 mAb at the diluted ratio of 1: 200 to identify the epitope sequences. Antibody against GST was also used as a loading control. Out of a total of 17 mutants expressed, only 2 mutants (NE→DV, lane 14 and RSVR→QNTI, lane15) were negative against CM6 antibody. (C) To determine the specificity of CM6 binding, 96-well microplates were coated with Ln-332 (1 μg/ml), GST (50 μg/ml), rLG2 (50 μg/ml), or a synthetic peptide: NERSVR, DVQNTI, or GLLFFA (100 μg/ml) at 4°C overnight. CM6 mAb was added as a primary antibody to wells (50 μg/ml) and incubated for 2 h at RT. Anti-mouse IgG conjugated with HRP was added as a secondary antibody to well and incubated for 1 h at RT. Bound antibody was detected by measuring O.D. (450 nm). CM6 bound the synthetic peptide NERSVR similarly to intact Ln-332 (N=2, in triplicate; p=0.394) and rLG2 module (N=2, in triplicate; p=0.240). In contrast, compared to Ln-332, CM6 had almost no reactivity to the human sequence DVQNTI (N=2, in triplicate; p=0.002*) or the rat LG2 module sequence GLLFFA (N=2, in triplicate; p=0.002*), both of which resulted in measurements similar to antibody on GST or PBS.
In order to determine whether the localization of the 1089NERSVR1094 sequence made sense with respect to antibody binding, we modeled the 3-D structure of rat α3LG2 by comparison with the published crystal structure of mouse laminin α2LG5 (Hohenester et al., 1999), a homologous LG domain (Fig. 4A). This comparison placed the 1089NERSVR1094 sequence on a loop sequence connecting β-strands I and J, as well as a major part of the β-strand J (Fig. 4B). These findings suggest that the location of the CM6 epitope is exposed to solvent and therefore likely accessible by antibodies.
Fig. 4. Sequence/structure relation of CM6 epitope sequence on LG2 module of rat Ln-332.
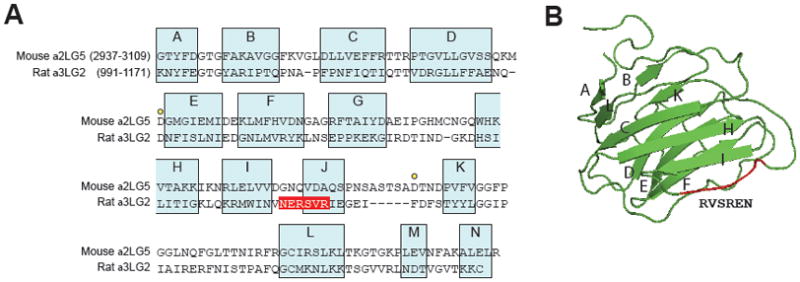
(A) Laminin α3 LG2 module sequence alignment. The rat laminin α3 LG2 sequence (residues 991–1171) is aligned to that of the mouse laminin α2 LG5 sequence (residues 2937- 3109). The assignment to β-strands A to N (blue boxes) is based on molecular modeling. A hexapeptide sequences of epitope are printed in white on red. Yellow dots mark the two aspartic acid residues of the α2 LG5 implicated in calcium binding. (B) A 3-D model of laminin α3 LG2 module was generated with HHpred. β-strands are visualized by arrows. The localization of hexapeptide sequences involved in the epitope is shown in red.
CM6 mAb reduces MCF10A cell spreading on Ln-332
In a previous study, we showed that CM6 is a Ln-332 function-blocking antibody that disrupts hemidesmosome formation (Plopper et al., 1996), i.e., cell anchoring that is mediated by the keratin-binding integrin α6β4. Another integrin receptor for Ln-332, α3β1, is instead involved in cell adhesion, spreading, and migration on Ln-332 (Hintermann et al., 2004). Therefore, we investigated the effect of CM6 mAb on single-cell spreading on Ln-332 (and Coll-1 for comparison) using the epithelial breast cell line MCF10A. Representative bright-field images were captured using a BD Bioimager equipped with a 20x objective (Fig. 5A). As shown in Fig. 5B, CM6 significantly reduced MCF10A cell spreading (i.e., surface area) on Ln-332 compared to cells in the absence of antibody (N=2, in triplicate; p<0.001), while TR1 (mAb against γ2 chain) had almost no effect on the same substrate (N=2, in triplicate; p=0.984). Further, we found that CM6 had almost no effect on cell surface area when plated on Coll-I (Fig. 5B; N=2; in triplicate; p=0.118). Taken together, these results indicate that, in addition to disrupting hemidesmosome formation via integrin α6β4, CM6 also blocks cell spreading on Ln-332 via integrin α3β1, suggesting that the binding sites for these integrins are nearby.
Fig. 5. CM6 inhibits cell spreading on Ln-332.
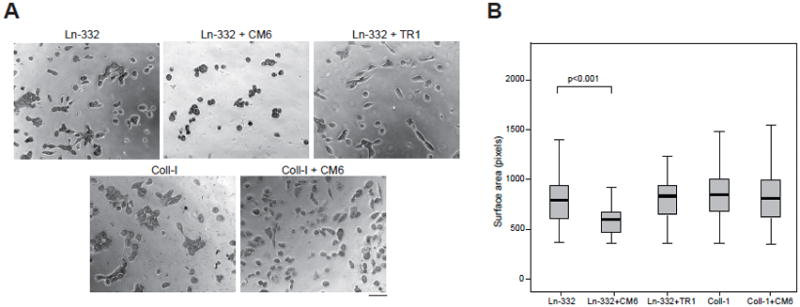
(A) 96-well microplates were coated with Ln-332 or collagen-I at 4°C overnight. MCF10A cells were labeled with Celltracker™ green CMFDA in PBS (labeling solution) and incubated for 30 min at 37°C. After a washing step, cells were seeded in microplates (5000 cells in 100 μl per well), and allowed to incubate (and spread) for ~7 h, imaging every 6 min using a BD Pathways 855 Bioimager. Normal mouse IgG, CM6, or TR1 were also added where indicated. Representative images from each treatment are shown. Scale bar is equal to 50 μm. (B) Box-and-whisker plots representing mean cell speed (bold horizontal line), 25th and 75th quartiles (box), and 95% confidence intervals (whiskers) are shown (N=70–750). MCF10A cells treated with CM6 exhibited significantly reduced spreading when plated on Ln-332 (p<0.001), but not on collagen-I (p=0.118). In contrast, non-function-blocking antibody TR1 had almost no affect on spreading (p=0.984).
CM6 mAb inhibits MCF10A cell spreading on Ln-332 LG2 module
Next, we tested whether rLG2 by itself could support cell adhesion and spreading. Even at high concentrations, rLG2 was negative in cell adhesion assays (data not shown). However, in spreading assays that do not require washing and therefore are more sensitive to weak cell adhesion, MCF10A cells were clearly spread on rLG2 (Fig. 6A). The CM6 mAb, as shown in Fig. 6B, significantly reduced cell spreading on rLG2 (N=5; p=0.008) and Ln-332 (N=5; p=0.008). In contrast, CM6 had negligible effects on cell spreading on rLG3 module (N=5; p=0.222). Taken together, these results suggest that CM6 interferes with “weak” integrin binding and can specifically block cell spreading on LG2 module.
Fig. 6. CM6 inhibits MCF10A cell spreading on the LG2 module of rat Ln-332.
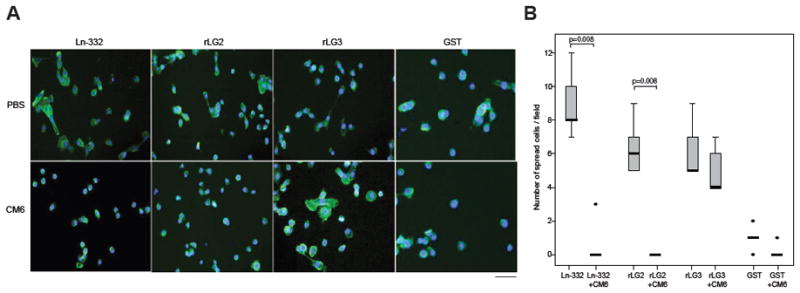
(A) 96-well microplates were coated with Ln-332 (1 μg/ml), GST (50 μg/ml), rLG2 (50 μg/ml), rLG3 (50 μg/ml) at 4°C overnight. MCF10A cells were added to wells (5000/well) and incubated for 5 h at 37°C in a humidified, 5% CO2, 95% air atmosphere. After incubation, attached cells were fixed and stained with Hoechst (blue) and phalloidin (green) in PBS containing 3% BSA for 2 h at RT. Cell spreading was quantified by manually counting spread cells in five representative microscope fields. (B) CM6 significantly reduced cell spreading on Ln-332 (N=5; p=0.008) and rLG2 (N=5; p=0.008). In contrast, CM6 had little effect on cell spreading on rLG3 module (N=5; p=0.222) or GST (N=5; p=0.095).
Discussion
Ln-332 is a glycoprotein expressed mainly in epithelial cells, which mediates cell adhesion, migration, and spreading by interacting with integrins α3β1 and α6β4 (Hintermann et al., 2004). Interactions of Ln-332 with its receptors alter cellular phenotypes with respect to motility and anchoring to the BM (Hintermann et al., 2004), and therefore have been investigated with a variety of approaches (Kunneken et al., 2004; Utani et al., 2001; Okamoto et al., 2003; Shang et al., 2001; Mizushima, 1997; Kim et al., 2005; Hirosaki et al., 2000; Bachy et al., 2008), including characterization of rLG modules and function-blocking antibodies. However, the binding sites of α3β1 and α6β4 to Ln-332 remain poorly understood. In this study we determine that the epitope of the Ln-332 function-blocking mAb, CM6, critically requires the 1089NERSVR1094 sequence within the rat Ln-332 LG2 module, and use this information to shed light on possible arrangements of these binding sites.
We previously showed that CM6 disrupts hemidesmosome assembly induced by the interaction of Ln-332 with integrin α6β4 (Baker et al., 1996). In this paper, we further investigated the effect of CM6 mAb on cell spreading on Ln-332, which is induced via integrin α3β1 (Shang et al., 2001; Hintermann et al., 2004). Our results show that CM6 mAb significantly reduced cell spreading on Ln-332, but not on Coll-I, suggesting that CM6 interferes with the binding sites of both integrin α3β1 and α6β4 on Ln-332. Immunoblot analysis showed that CM6 mAb recognized the Ln-332 α3, but not β3 or γ2 subunits.
This result is consistent with our previous study, in which rotary shadowing electron microscopy showed that CM6 mAb is likely to bind around the G domain of the α3 chain of Ln-332 (Baker et al., 1996). Using rLG modules, we also determined that CM6 mAb is specifically reactive to LG2 module of rat Ln-332 α3 chain. Furthermore, site-directed mutagenesis and solid phase binding assays using synthetic peptides revealed that the rat LG2 hexapeptide sequence, 1089NERSVR1094, is critical for CM6 recognition. Accordingly, a structural modeling approach revealed that this sequence is located on a loop sequence connecting β-strands I and J, as well as a major part of the β-strand J on the LG2 module, which is predicted to be accessible to solvent and antibody binding (Timpl et al., 2000). Previous studies have shown that many of these loops contain epitopes for heparin, proteoglycans, and other molecules (Timpl et al., 2000), which have also been shown to have high homology with fibronectin RGD-containing integrin binding regions (Dickenson, 1994).
Based on these clear-cut CM6 binding results, it would be tempting to conclude that the hexapeptide sequence, 1089NERSVR1094, is central for integrin binding to Ln-332. However, we believe this is not the case based on the following considerations: i) cell adhesion assays failed to show that the hexapeptide can, by itself, support cell adhesion, spreading, or migration; ii) the sequence NERSVRis rat-specific (in fact, it is likely the basis for its immunogenicity in mice, the host species for CM6 production), which is in conflict with the well-known fact that integrin binding to ligand is independent of species-specificity; and iii) studies with recombinant laminins, in which domains were swapped between isoforms, do not support a linear peptide, but rather a domain paradigm for integrin binding to laminins. With respect to the latter point, current hypothetical models propose that the LG1-3 region of Ln-332 interacts with α3β1 cooperatively by forming a complex together with amino acid residues at the C-terminus of laminin β3 and γ2 chains. This region (LG1-3) is also necessary for the binding with integrin α6β4 (Ido et al., 2007; Taniguchi et al., 2009; Carafoli et al., 2009). Thus, it is likely that the interaction between integrins and Ln-332 involves several contact points distributed on all three subunits. It is however likely that a primary role is played by sites on the LG domains. We previously showed that the LG3 module harbors an integrin α3β1 binding site such that rLG3 supports, on its own, cell adhesion and spreading, albeit primarily in fibrosarcoma, not epithelial cells (Shang et al., 2000). In that study, we were unable to demonstrate any cell adhesion to LG1 or LG2 modules. Since the CM6 epitope is on LG2, it would be appealing to conclude that its cell adhesion blocking activity is entirely due to steric hindrance of integrin binding sites located elsewhere on the Ln-332 molecule. However, we investigated this matter with cell spreading assays, which differ from cell adhesion assays by the absence of any washing step, and are in a sense more suitable to detect weak integrin-laminin interactions. In these assays, both LG2 and LG3 supported cell spreading, but CM6 only inhibited LG2-based spreading. Interestingly, these gentle assays were easily performed with epithelial cells, which even in our previous study (Shang et al., 2000) performed poorly in the harsher cell adhesion assays on LG3.
In summary, it is tempting to speculate that our data point to the existence of several integrin contact sites on Ln-332, and that, while CM6 interferes by steric hindrance, it is nonetheless a useful probe to characterize these sites. Other researchers have reported studies examining the epitope of various function-blocking antibodies that disrupt interactions of integrins with laminins. For example, Ido et al. previously showed that 4C7, a function-blocking antibody against human Ln-511, recognizes LG1 module of laminin α5 chain (Ido et al., 2006). McMillan et al. have demonstrated that BM165, a function-blocking antibody against human Ln-332, recognizes LG1 module of human laminin α3 chain (McMillan et al., 2003). The ability of LG1 to support cell spreading was not tested in these studies, leaving open the possibility that the LG1 domain may also contain an integrin binding site.
Our favorite interpretation of the data is that Ln-332 LG2 harbors a “weak” integrin binding site, where “weak” signifies that it can be detected only by gentle assay, such as a no-wash spreading assay. Nonetheless, by no means should “weak” be interpreted as inconsequential, since the role of such integrin contact site(s) in the context of the native Ln-332 molecule remains to be determined. In this respect, a special feature of the Ln-332-integrin interaction is the “switch” that occurs between α3β1 and α6β4 engagement, as epithelia transition from a remodeling state to a resting state, respectively, and vice versa. The consequences of this switch to cellular organization are not entirely understood, but they are major since α3β1 connects to the microfilament cytoskeleton, whereas α6β4 connects to the intermediate filament cytoskeleton (Borradori and Sonnenberg, 1999). This transition between distinct cellular adhesive systems is regulated, at least in part, at the level of ligand (Goldfinger et al., 1998). Since CM6 both blocks cell spreading (α3β1-dependent) and disassembles hemidesmosomes (α6β4-dependent), we submit it will be a useful reagent to define the Ln-332 structural determinants involved in integrin switching. For example, it already suggests that the α3β1 and α6β4 binding sites on Ln-332 are closely spaced or structurally linked, since one antibody inhibits both in independent assays. However, further studies are needed to determine whether “weak” integrin contact sites on Ln-332 may be involved in the Ln-332 integrin switch.
Supplementary Material
Site-directed mutagenesis was performed using the overlap extension method. Four primers were used for cloning of each mutant. Two general primers, each corresponding to the 5′-start and 3′-end sequences of the laminin α3LG2 module, were used for all mutants. In addition, two mutant specific primers, each corresponding to the sense and anti-sense sequences encompassing the mutation site were also used. Mutated codons are underlined. Plasmid DNA of mutant clones were sequenced from both the 5′ and 3′ ends to confirm the presence of the intended mutation without any additional, unwanted mutations.
Acknowledgments
This work was supported by National Institutes of Health-NCI grant 5R01GM067221-03 awarded to VQ.
References
- Andrew SM, Titus JA. Fragmentation of immunoglobulin G. Curr Protoc Immunol. 2001;Chapter 2(Unit 28) doi: 10.1002/0471142735.im0208s21. [DOI] [PubMed] [Google Scholar]
- Bachy S, Letourneur F, Rousselle P. Syndecan-1 interaction with the LG4/5 domain in laminin-332 is essential for keratinocyte migration. J Cell Physiol. 2008;214(1):238–249. doi: 10.1002/jcp.21184. [DOI] [PubMed] [Google Scholar]
- Baker SE, Hopkinson SB, Fitchmun M, Andreason GL, Frasier F, Plopper G, Quaranta V, Jones JC. Laminin-5 and hemidesmosomes: role of the alpha 3 chain subunit in hemidesmosome stability and assembly. J Cell Sci. 1996;109 (Pt 10):2509–20. doi: 10.1242/jcs.109.10.2509. [DOI] [PubMed] [Google Scholar]
- Baudoin C, Fantin L, Meneguzzi G. Proteolytic processing of the laminin alpha3 G domain mediates assembly of hemidesmosomes but has no role on keratinocyte migration. J Invest Dermatol. 2005;125(5):883–8. doi: 10.1111/j.0022-202X.2005.23881.x. [DOI] [PubMed] [Google Scholar]
- Beck K, Dixon TW, Engel J, Parry DA. Ionic interactions in the coiled-coil domain of laminin determine the specificity of chain assembly. J Mol Biol. 1993;231(2):311–23. doi: 10.1006/jmbi.1993.1284. [DOI] [PubMed] [Google Scholar]
- Borradori L, Chavanas S, Schaapveld RQ, Gagnoux-Palacios L, Calafat J, Meneguzzi G, Sonnenberg A. Role of the bullous pemphigoid antigen 180 (BP180) in the assembly of hemidesmosomes and cell adhesion--reexpression of BP180 in generalized atrophic benign epidermolysis bullosa keratinocytes. Exp Cell Res. 1998;239(2):463–76. doi: 10.1006/excr.1997.3923. [DOI] [PubMed] [Google Scholar]
- Borradori L, Sonnenberg A. Structure and function of hemidesmosomes: more than simple adhesion complexes. J Invest Dermatol. 1999;112(4):411–8. doi: 10.1046/j.1523-1747.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- Carafoli F, Clout NJ, Hohenester E. Crystal structure of the LG1-3 region of the laminin alpha2 chain. J Biol Chem. 2009 doi: 10.1074/jbc.M109.026658. 2009 Jun 24. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell. 1991;65(4):599–610. doi: 10.1016/0092-8674(91)90092-d. [DOI] [PubMed] [Google Scholar]
- Champliaud MF, Lunstrum GP, Rousselle P, Nishiyama T, Keene DR, Burgeson RE. Human amnion contains a novel laminin variant, laminin 7, which like laminin 6, covalently associates with laminin 5 to promote stable epithelial-stromal attachment. J Cell Biol. 1996;132(6):1189–98. doi: 10.1083/jcb.132.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognato H, Yurchenco PD. Form and function: the laminin family of heterotrimers. Dev Dyn. 2000;218(2):213–34. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Dickenson CD, eerapandian B, Dai XP, Hamlin RC, Zuong NH, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J Mol Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Ekblom M, Falk M, Salmivirta K, Durbeej M, Ekblom P. Laminin isoforms and epithelial development. Ann N Y Acad Sci. 1998;857:194–211. doi: 10.1111/j.1749-6632.1998.tb10117.x. [DOI] [PubMed] [Google Scholar]
- Engel J. Laminins and other strange proteins. Biochemistry. 1992;31(44):10643–51. doi: 10.1021/bi00159a001. [DOI] [PubMed] [Google Scholar]
- Engvall E, Davis GE, Dickerson K, Ruoslahti E, Varon S, Manthorpe M. Mapping of domains in human laminin using monoclonal antibodies: localization of the neurite-promoting site. J Cell Biol. 1986;103(6 Pt 1):2457–65. doi: 10.1083/jcb.103.6.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk-Marzillier J, Domanico SZ, Pelletier A, Mullen L, Quaranta V. Characterization of a tight molecular complex between integrin alpha 6 beta 4 and laminin-5 extracellular matrix. Biochem Biophys Res Commun. 1998;251(1):49–55. doi: 10.1006/bbrc.1998.9400. [DOI] [PubMed] [Google Scholar]
- Goldfinger LE, Stack MS, Jones JC. Processing of laminin-5 and its functional consequences: role of plasmin and tissue-type plasminogen activator. J Cell Biol. 1998;141(1):255–65. doi: 10.1083/jcb.141.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermann E, Quaranta V. Epithelial cell motility on laminin-5: regulation by matrix assembly, proteolysis, integrins and erbB receptors. Matrix Biol. 2004;23(2):75–85. doi: 10.1016/j.matbio.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Hirako Y, Owaribe K. Hemidesmosomes and their unique transmembrane protein BP180. Microscopy Research and Technique. 1998;43:207–17. doi: 10.1002/(SICI)1097-0029(19981101)43:3<207::AID-JEMT2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Hirosaki T, Mizushima H, Tsubota Y, Moriyama K, Miyazaki K. Structural requirement of carboxyl-terminal globular domains of laminin alpha 3 chain for promotion of rapid cell adhesion and migration by laminin-5. J Biol Chem. 2000;275(29):22495–502. doi: 10.1074/jbc.M001326200. [DOI] [PubMed] [Google Scholar]
- Hohenester E, Tisi D, Talts JF, Timpl R. The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin. Mol Cell. 1999;4(5):783–92. doi: 10.1016/s1097-2765(00)80388-3. [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36(Database issue):D245–9. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido H, Harada K, Yagi Y, Sekiguchi K. Probing the integrin-binding site within the globular domain of laminin-511 with the function-blocking monoclonal antibody 4C7. Matrix Biol. 2006;25(2):112–7. doi: 10.1016/j.matbio.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Ido H, Nakamura A, Kobayashi R, Ito S, Li S, Futaki S, Sekiguchi K. The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin gamma chains in integrin binding by laminins. J Biol Chem. 2007;282(15):11144–54. doi: 10.1074/jbc.M609402200. [DOI] [PubMed] [Google Scholar]
- Jones JC, Hopkinson SB, Goldfinger LE. Structure and assembly of hemidesmosomes. Bioessays. 1998;20(6):488–94. doi: 10.1002/(SICI)1521-1878(199806)20:6<488::AID-BIES7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Kim JM, Park WH, Min BM. The PPFLMLLKGSTR motif in globular domain 3 of the human laminin-5 alpha3 chain is crucial for integrin alpha3beta1 binding and cell adhesion. Exp Cell Res. 2005;304(1):317–27. doi: 10.1016/j.yexcr.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Klees RF, Salasznyk RM, Kingsley K, Williams WA, Boskey A, Plopper GE. Laminin-5 induces osteogenic gene expression in human mesenchymal stem cells through an ERK-dependent pathway. Mol Biol Cell. 2005;16(2):881–90. doi: 10.1091/mbc.E04-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunneken K, Pohlentz G, Schmidt-Hederich A, Odenthal U, Smyth N, Peter-Katalinic J, Bruckner P, Eble JA. Recombinant human laminin-5 domains. Effects of heterotrimerization, proteolytic processing, and N-glycosylation on alpha3beta1 integrin binding. J Biol Chem. 2004;279(7):5184–93. doi: 10.1074/jbc.M310424200. [DOI] [PubMed] [Google Scholar]
- Manohar A, Shome SG, Lamar J, Stirling L, Iyer V, Pumiglia K, DiPersio CM. Alpha 3 beta 1 integrin promotes keratinocyte cell survival through activation of a MEK/ERK signaling pathway. J Cell Sci. 2004;117(Pt 18):4043–54. doi: 10.1242/jcs.01277. [DOI] [PubMed] [Google Scholar]
- Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007;7(5):370–80. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- McMillan JR, Akiyama M, Shimizu H. Ultrastructural orientation of laminin 5 in the epidermal basement membrane: an updated model for basement membrane organization. J Histochem Cytochem. 2003;51(10):1299–306. doi: 10.1177/002215540305101007. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Kikkawa Y, Nakamura A, Yasumitsu H, Umeda M. A large cell-adhesive scatter factor secreted by human gastric carcinoma cells. Proc Natl Acad Sci U S A. 1993;90(24):11767–71. doi: 10.1073/pnas.90.24.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima H, Takamura H, Miyagi Y, Kikkawa Y, Yamanaka N, Yasumitsu H, Misugi K, Miyazaki K. Identification of integrin-dependent and -independent cell adhesion domains in COOH-terminal globular region of laminin-5 alpha 3 chain. Cell Growth Differ. 1997;8(9):979–87. [PubMed] [Google Scholar]
- Navdaev A, Heitmann V, Desantana Evangelista K, Mörgelin M, Wegener J, Eble JA. The Cterminus of the gamma 2 chain but not of the beta 3 chain of laminin-332 is indirectly but indispensably necessary for integrin-mediated cell reactions. Exp Cell Res. 2008;314(3):489–97. doi: 10.1016/j.yexcr.2007.10.027. [DOI] [PubMed] [Google Scholar]
- Nguyen BP, Ren XD, Schwartz MA, Carter WG. Ligation of integrin alpha 3beta 1 by laminin 5 at the wound edge activates Rho-dependent adhesion of leading keratinocytes on collagen. J Biol Chem. 2001;276(47):43860–70. doi: 10.1074/jbc.M103404200. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Hogervorst F, aspars LH, de Melker AA, Delwel GO, Hulsman EH, Kuikman I, Sonnenberg A. The alpha 6 beta 4 integrin is a receptor for both laminin and kalinin. Exp Cell Res. 1994;211(2):360–7. doi: 10.1006/excr.1994.1099. [DOI] [PubMed] [Google Scholar]
- Offord RE. Protein engineering by chemical means? Protein Eng. 1987;1(3):151–7. doi: 10.1093/protein/1.3.151. [DOI] [PubMed] [Google Scholar]
- Okamoto O, Bachy S, Odenthal U, Bernaud J, Rigal D, Lortat-Jacob H, Smyth N, Rousselle P. Normal human keratinocytes bind to the alpha3LG4/5 domain of unprocessed laminin-5 through the receptor syndecan-1. J Biol Chem. 2003;278(45):44168–77. doi: 10.1074/jbc.M300726200. [DOI] [PubMed] [Google Scholar]
- Parnaud G, Hammar E, Rouiller DG, Armanet M, Halban PA, Bosco D. Blockade of beta1 integrin-laminin-5 interaction affects spreading and insulin secretion of rat beta-cells attached on extracellular matrix. Diabetes. 2006;55(5):1413–20. doi: 10.2337/db05-1388. [DOI] [PubMed] [Google Scholar]
- Plopper G, Falk-Marzillier J, Glaser S, Fitchmun M, Giannelli G, Romano T, Jones JC, Quaranta V. Changes in expression of monoclonal antibody epitopes on laminin-5r induced by cell contact. J Cell Sci. 1996;109 (Pt 7):1965–73. doi: 10.1242/jcs.109.7.1965. [DOI] [PubMed] [Google Scholar]
- Porter RR. Structural studies of immunoglobulins. Science. 1973;180(87):713–6. doi: 10.1126/science.180.4087.713. [DOI] [PubMed] [Google Scholar]
- Shang M, Koshikawa N, Schenk S, Quaranta V. The LG3 module of laminin-5 harbors a binding site for integrin alpha3beta1 that promotes cell adhesion, spreading, and migration. J Biol Chem. 2001;276(35):33045–53. doi: 10.1074/jbc.M100798200. [DOI] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Ido H, Sanzen N, Hayashi M, Sato-Nishiuchi R, Futaki S, Sekiguchi K. The C-terminal region of laminin beta chains modulates the integrin binding affinities of laminins. J Biol Chem. 2009;284(12):7820–31. doi: 10.1074/jbc.M809332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiger CF, Champliaud MF, Pedrosa-Domellof F, Thornell LE, Ekblom P, Gullberg D. Presence of laminin alpha5 chain and lack of laminin alpha1 chain during human muscle development and in muscular dystrophies. J Biol Chem. 1997;272(45):28590–5. doi: 10.1074/jbc.272.45.28590. [DOI] [PubMed] [Google Scholar]
- Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19(4):309–17. doi: 10.1016/s0945-053x(00)00072-x. [DOI] [PubMed] [Google Scholar]
- Tisi D, Talts JF, Timpl R, Hohenester E. Structure of the C-terminal laminin G-like domain pair of the laminin alpha2 chain harbouring binding sites for alpha-dystroglycan and heparin. Embo J. 2000;19(7):1432–40. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utani A, Nomizu M, Matsuura H, Kato K, Kobayashi T, Takeda U, Aota S, Nielsen PK, Shinkai H. A unique sequence of the laminin alpha 3 G domain binds to heparin and promotes cell adhesion through syndecan-2 and -4. J Biol Chem. 2001;276(31):28779–88. doi: 10.1074/jbc.M101420200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kramer RH. Laminin 5 deposition promotes keratinocyte motility. Exp Cell Res. 1996;227(2):309–22. doi: 10.1006/excr.1996.0280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Site-directed mutagenesis was performed using the overlap extension method. Four primers were used for cloning of each mutant. Two general primers, each corresponding to the 5′-start and 3′-end sequences of the laminin α3LG2 module, were used for all mutants. In addition, two mutant specific primers, each corresponding to the sense and anti-sense sequences encompassing the mutation site were also used. Mutated codons are underlined. Plasmid DNA of mutant clones were sequenced from both the 5′ and 3′ ends to confirm the presence of the intended mutation without any additional, unwanted mutations.


