Abstract
Genetic association studies have demonstrated the importance of variants in the CHRNA5-CHRNA3-CHRNB4 cholinergic nicotinic receptor subunit gene cluster on chromosome 15q24-25.1 in risk for nicotine dependence, smoking, and lung cancer in populations of European descent. We have now carried out a detailed study of this region using dense genotyping in both European- and African-Americans.
We genotyped 75 known single-nucleotide-polymorphisms (SNPs) and one sequencing-discovered SNP in an African-American (AA) sample (N = 710) and European-American (EA) sample (N = 2062). Cases were nicotine-dependent and controls were non-dependent smokers.
The non-synonymous CHRNA5 SNP rs16969968 is the most significant SNP associated with nicotine dependence in the full sample of 2772 subjects (p = 4.49×10−8, OR 1.42 (1.25–1.61)) as well as in AAs only (p = 0.015, OR = 2.04 (1.15–3.62)) and EAs only (p = 4.14×10−7, OR = 1.40 (1.23–1.59)). Other SNPs that have been shown to affect mRNA levels of CHRNA5 in EAs are associated with nicotine dependence in AAs but not in EAs. The CHRNA3 SNP rs578776, which has low correlation with rs16969968, is associated with nicotine dependence in EAs but not in AAs. Less common SNPs (frequency ≤ 5%) also are associated with nicotine dependence.
In summary, multiple variants in this gene cluster contribute to nicotine dependence risk, and some are also associated with functional effects on CHRNA5. The non-synonymous SNP rs16969968, a known risk variant in European-descent populations, is also significantly associated with risk in African-Americans. Additional SNPs contribute in distinct ways to risk in these two populations.
Keywords: genetic association, smoking, cholinergic nicotinic receptors, nicotinic acetylcholine receptors
INTRODUCTION
Cigarette smoking is a major public health problem and the single largest cause of preventable death in the world, contributing to over 5 million deaths a year [1]. In the United States, smoking causes approximately 443,000 deaths each year including an estimated 82% of lung cancer deaths [2].
Cigarette smoking is common in European-Americans and African-Americans, with similar prevalence of current smoking among adults: 21.4% (95% confidence interval 20.4–22.4) in European-Americans and 19.8% (19.2–21.4) in African-Americans [3]. In past-month smokers, rates of current nicotine dependence, defined by the Fagerstrom Test for Nicotine Dependence (FTND) [4], are also similar though slightly lower in African-Americans: 57.3% (55.1–59.5) versus 61.7% (61.0–62.4) in European-Americans [5]. Despite these similarities, African-Americans have a higher incidence of lung cancer than European-Americans (82.7 per 100,000 versus 64.3 per 100,000) and higher lung cancer mortality (64.1 per 100,000 versus 54.1 per 100,000) [6]. Many issues may contribute to these differences in lung cancer, including discrepancies in quit rates and disparities in medical care [7, 8]. Genetic factors such as differences in allele frequencies and linkage disequilibrium (LD) also may play a role.
We previously carried out a high-density whole-genome and candidate gene association study of nicotine dependence in a European-descent sample [9, 10]. Among the top association signals were chromosome 15q24-25.1 variants, including a biologically compelling finding at rs16969968, a nonsynonymous SNP in the alpha5 cholinergic nicotinic receptor subunit gene (CHRNA5) [10]. The risk allele at rs16969968 causes an amino acid change (D398N, aspartate to asparagine) in the alpha5 subunit and functional changes in the receptor in vitro [11]. Independent replication of this association with nicotine dependence or smoking has now been confirmed in multiple populations of European descent, using rs16969968 or SNPs highly correlated (r2 ≥ 0.8) with rs16969968 [11–19]. Fine-mapping in our original sample continued to highlight rs16969968 and other highly correlated SNPs, which extend across CHRNA5-CHRNA3-CHRNB4 and into neighboring genes IREB2, PSMA4 and LOC123688 [20].
Also in this region, a locus tagged by rs578776 and rs3743078 is significantly associated with nicotine dependence and smoking in European-descent samples [10, 12, 14, 16]. This locus has been described as protective because the minor allele is more frequent in controls than in cases. There is evidence that this locus is statistically distinct from rs16969968: in two independent European-ancestry samples the correlation between rs578776 and rs16969968 is low (r2 = 0.2), with D′ = −1.0 indicating repulsion phase (the minor allele at one locus co-occurs with the major allele at the other), and both SNPs are significant in joint analyses [14, 20].
Genome-wide studies of lung cancer report strong association with rs16969968 and its r2 correlates [16, 21–23]. The locus tagged by rs578776 is also associated with lung cancer. Thus there has been a convergence of genetic findings for nicotine dependence and for lung cancer.
The association studies cited above were carried out in European-descent samples. Studying this region in individuals of African descent provides an opportunity to confirm its role in an important and understudied population. Also, the contrasting genetic architecture in Africans and Europeans can be leveraged to narrow down the potential functional source of the disease associations [24]. In the European ancestry population, many highly correlated SNPs often can detect the same association signal, and determining which variant(s) are “causal” is not straightforward. LD and allele frequencies can differ between European and African populations. For instance, in HapMap, rs16969968 is non-polymorphic in the YRI (Yoruba in Ibadan, Nigeria) sample; for rs3743078, the minor allele in YRI (“C”, 33%) is the major allele (77%) in CEU (CEPH, Utah residents with ancestry from northern and western Europe) [25].
Here we present an association study of nicotine dependence across the CHRNA5-CHRNA3-CHRNB4 region in a new African-American (AA) sample and an expanded European-American (EA) sample. We also analyze novel variants discovered from DNA resequencing of EAs in a 26 kb region between CHRNA5 and CHRNA3. Our goal is to determine whether any variants consistently affect nicotine dependence risk in both EAs and AAs. Additionally, we are interested in evidence of variants that affect risk in the AA sample alone.
MATERIALS AND METHODS
Study design and sample
All individuals (N = 2772) were recruited by the Collaborative Genetic Study of Nicotine Dependence (COGEND), a United States multi-site project. All cases and controls reported smoking ≥100 cigarettes lifetime, the threshold classically used to define a smoker [26]. In recruitment of new subjects, cases were nicotine dependent, defined by current FTND≥4, and controls were required to have lifetime FTND of 0 or 1. The AA genetic sample in COGEND consists of 461 cases and 249controls. The EA genetic sample of 1063cases and 999 controls consists of 454 new subjects (266 cases, 188 controls) together with 1608 original COGEND EA individuals previously analyzed in [9, 10] (797cases (FTND≥4), 811 controls (FTND=0)).
The study complies with the Code of Ethics of the World Medical Association and obtained informed consent from participants and approval from the appropriate institutional review boards.
Genotyping and quality control
Thirty-three SNPs covering the CHRNA5-CHRNA3-CHRNB4 region were genotyped in the additional subjects by the Genome Center at Washington University1. These SNPs were genotyped using Illumina GoldenGate assay as part of a set of 1536 SNPs that included SNPs for checking population stratification and SNPs to follow up several genomic regions. Cleaning was performed using all 1536 SNPs. Four DNA samples had poor call rates (≤63% of the 1536 SNPs) and were dropped. All remaining DNA samples had call rates above 90% and were retained; 99.5% of DNA samples had call rates ≥95%. Twenty samples were included in duplicate. SNPs with more than 1 non-concordant duplicate sample genotype were dropped; the 33 SNPs reported here were 100% concordant. SNPs were also required to pass a call rate threshold of 98%. Self-reported race was verified using EIGENSTRAT [27]. An additional 42 SNPs were genotyped using Sequenom MassArray iPLEX technology2. These SNPs passed a call rate threshold of 95%. Map positions were obtained from the National Center for Biotechnology Information (NCBI) Human Reference Build 36.2 and dbSNP build 129.
These 75 SNPs were selected to include the same SNPs used to cover this region in our previous publications [9, 10, 20]. Forty-nine of the genotyped SNPs lie between 76642961 bp and 76722642 bp, encompassing CHRNA5-CHRNA3-CHRNB4 and the 2kb flanking either side (5′ of CHRNA5 and 5′ of CHRNB4), matching the dbSNP definition of in or near a gene. These SNPs provide good coverage of the 72 SNPs in this region that are common (minor allele frequency (MAF) ≥ 0.05) in either the HapMap CEU or YRI samples. Of the 61 SNPs that have MAF ≥ 5% in CEU, 54 (93%) are tagged by at least one genotyped SNP at an r2 ≥ 0.8 according to CEU LD data; all 61 are tagged at an r2 ≥ 0.5. Of the 65 SNPs in this region that have MAF ≥ 5% in YRI, 46 (71%) are tagged at an r2 ≥ 0.8 in the YRI LD data; 60 (92%) are tagged at an r2 ≥ 0.5 (HapMap release 23a). Twenty-six additional SNPs flanking the cluster or in IREB2, LOC123688, and PSMA4 were included because of the high r2 extending from CHRNA5-CHRNA3-CHRNB4.
Genotype data were then combined for this new sample and the previously genotyped sample of 1608 EAs. Allele correspondence was checked. All association analyses are reported using the major allele in the EA sample as the reference allele.
DNA Resequencing in the CHRNA5 and CHRNA3 intergenic region
Human CHRNA3 and CHRNA5 are physically linked and partially overlap tail-to-tail [28]. We targeted a 26-kb region spanning exon 5 of CHRNA5 to exon 4 of CHRNA3 and sequenced 10 cases and 12 controls of European descent. The goal was to identify novel variants, genotype them in the larger sample, and test them for association with nicotine dependence.
Sequencing was performed at the Genome Center at Washington University using standard protocols3. We used Sequencher (Gene Codes, Ann Arbor, Michigan) to assemble and scan sequence traces for variation from the reference sequence. Novel variants were genotyped using Sequenom MassArray iPLEX technology.
Linkage disequilibrium
LD was calculated in EAs and AAs using PLINK [29] and Haploview [30]. LD (r2) plots were generated using WGAviewer4 [31].
Genetic association analyses
Association analysis of case-control status was performed on the entire sample using logistic regression with gender and race as covariates. This analysis highlights SNPs that have similar effect in both EAs and AAs. The full genetic model is:
| (Equation 1) |
where P is the probability of being a case, g is gender (0=male, 1=female), s is race (0=EA, 1=AA), and G is genotype, coded as the number of copies of the minor allele in the EA sample, corresponding to a log-additive (multiplicative) model. The term β3G is tested for significance by the standard likelihood chi-square with one degree of freedom. We then analyzed AAs and EAs separately with gender as a covariate. Association tests were carried out using PLINK [29] and SAS (Cary, NC).
Cross-population heterogeneity analysis
Heterogeneity of odds ratio between EAs and AAs was tested by adding the interaction term β4Gs to Equation 1 [24]. This test is of interest for SNPs that are associated with disease in at least one population. If the interaction term is significant (e.g. p ≤ 0.05, unadjusted for multiple tests), we conclude the genetic effect is different in the two population samples.
A significant difference in odds ratio may occur because the genetic effect is present in only one of the two populations. In this case, one interpretationis that this SNP is not biologically causal, though in the associated population it may tag the functional SNP. This interpretation is based on the hypothesis that the underlying biological processes leading to disease risk are shared in common across human populations [32].
RESULTS
Genetic association analyses
Supplementary Table 1 shows the allele frequencies of the 75 known SNPs and one novel SNP from our SNP discovery project, in the EA and AA samples; selected SNPs, including those most associated with nicotine dependence in each sample, are in Table 1. Sixty-three SNPs show significant allele frequency differences between EAs and AAs. Nevertheless, for most SNPs the minor allele is the same in both populations; notable exceptions are rs578776 and rs3743078, which are known to be associated with nicotine dependence in European-Americans.
Table 1.
Genotyped SNPs and allele frequencies, sorted by position.
| Snp | Chr | Position (bp) | Gene | Reference allele1 | Minor allele | Reference allele frequency | ||
|---|---|---|---|---|---|---|---|---|
| EA sample | AA sample | EA sample | AA sample* | |||||
| rs17483548 | 15 | 76517368 | IREB2 | G | A | A | 0.662 | 0.941* |
| rs17405217 | 15 | 76518204 | IREB2 | C | T | T | 0.662 | 0.944* |
| rs2656052 | 15 | 76527987 | IREB2 | T | G | G | 0.653 | 0.565* |
| rs17484235 | 15 | 76548469 | IREB2 | C | G | G | 0.662 | 0.944* |
| rs7164594 | 15 | 76590112 | LOC123688 | C | T | T | 0.785 | 0.584* |
| rs8034191 | 15 | 76593078 | LOC123688 | T | C | C | 0.648 | 0.840* |
| rs10519203 | 15 | 76601101 | LOC123688 | A | G | G | 0.643 | 0.697* |
| rs2036534 | 15 | 76614003 | LOC123688 | T | C | C | 0.789 | 0.778 |
| rs12916483 | 15 | 76619452 | PSMA4 | G | A | A | 0.575 | 0.835* |
| rs3813570 | 15 | 76619887 | PSMA4 | A | G | G | 0.789 | 0.739* |
| rs11858230 | 15 | 76622607 | PSMA4 | G | A | A | 0.574 | 0.771* |
| rs2036527 | 15 | 76638670 | C | T | T | 0.645 | 0.778* | |
| rs667282 | 15 | 76650527 | CHRNA5 | T | C | C | 0.779 | 0.704* |
| rs588765 | 15 | 76652480 | CHRNA5 | C | T | T | 0.576 | 0.705* |
| rs6495306 | 15 | 76652948 | CHRNA5 | A | G | G | 0.576 | 0.702* |
| rs17486278 | 15 | 76654537 | CHRNA5 | A | C | C | 0.648 | 0.710* |
| rs601079 | 15 | 76656634 | CHRNA5 | T | A | A | 0.574 | 0.578 |
| rs680244 | 15 | 76658343 | CHRNA5 | G | A | A | 0.573 | 0.576 |
| rs637137 | 15 | 76661031 | CHRNA5 | T | A | A | 0.779 | 0.701* |
| rs951266 | 15 | 76665596 | CHRNA5 | C | T | T | 0.649 | 0.888* |
| rs555018 | 15 | 76666297 | CHRNA5 | T | C | C | 0.576 | 0.699* |
| rs647041 | 15 | 76667536 | CHRNA5 | C | T | T | 0.578 | 0.704* |
| rs16969968 | 15 | 76669980 | CHRNA5 | G | A | A | 0.650 | 0.948* |
| rs660652 | 15 | 76674887 | CHRNA3 | G | A | A | 0.632 | 0.717* |
| rs578776 | 15 | 76675455 | CHRNA3 | C | T | C2 | 0.726 | 0.451* |
| rs6495307 | 15 | 76677376 | CHRNA3 | C | T | T | 0.580 | 0.595 |
| rs1051730 | 15 | 76681394 | CHRNA3 | C | T | T | 0.651 | 0.882* |
| rs3743078 | 15 | 76681814 | CHRNA3 | C | G | C2 | 0.774 | 0.416* |
| rs3743077 | 15 | 76681951 | CHRNA3 | G | A | A | 0.579 | 0.847* |
| rs1317286 | 15 | 76683184 | CHRNA3 | A | G | G | 0.645 | 0.753* |
| rs938682 | 15 | 76683602 | CHRNA3 | T | C | C | 0.778 | 0.711* |
| rs11637630 | 15 | 76686774 | CHRNA3 | A | G | G | 0.778 | 0.697* |
| GSC3_26 | 15 | 76693692 | CHRNA3 | A | G | G | 0.938 | 0.986* |
| rs7177514 | 15 | 76694461 | CHRNA3 | C | G | G | 0.777 | 0.698* |
| rs6495308 | 15 | 76694711 | CHRNA3 | T | C | C | 0.777 | 0.702* |
| rs8042059 | 15 | 76694914 | CHRNA3 | A | C | C | 0.777 | 0.702* |
| rs8042374 | 15 | 76695087 | CHRNA3 | A | G | G | 0.780 | 0.750 |
| rs4887069 | 15 | 76696125 | CHRNA3 | A | G | G | 0.772 | 0.698* |
| rs8192479 | 15 | 76696453 | CHRNA3 | C | T | T | 0.980 | 0.999* |
| rs6495309 | 15 | 76702300 | CHRNB4 | C | T | T | 0.794 | 0.748* |
| rs12914008 | 15 | 76710560 | CHRNB4 | G | A | A | 0.955 | 0.994* |
| rs17487223 | 15 | 76711042 | CHRNB4 | C | T | T | 0.629 | 0.897* |
| rs1996371 | 15 | 76743861 | A | G | G | 0.587 | 0.929* | |
Reference allele is major allele in EA sample.
Minor allele differs in EAs and AAs.
Allele frequencies differ significantly between EAs and AAs by Fisher’s exact test (p < 10−3).
Table 2 shows association results for the Table 1 SNPs; Supplementary Table 2 shows all 76 SNPs. The most significant SNP in the combined sample is the non-synonymous CHRNA5 SNP rs16969968 (p = 4.49×10−8, OR = 1.42 (95% CI 1.25–1.61)). Furthermore, rs16969968 is the most significant of the 76 SNPs in the AAs alone (p = 0.0147, OR = 2.04 (1.15–3.62)) and in the EAs alone(p = 4.14×10−7, OR = 1.40 (1.23–1.59)). The same allele (A) is elevated in EA and AA cases. Additional SNPs correlated with rs16969968 in EAs and AAs are also significant.
Table 2.
Chromosome 15 association results for selected SNPs, sorted by chromosomal position. Odds ratios are for each copy of the non-reference allele.
| Snp | AA sample | EA sample | Full sample | Cross-population heterogeneity p-value | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-value | OR (95% CI) | p-value | OR (95% CI) | p-value | ||
| rs17483548 | 1.58(0.95–2.61) | 7.98E-2 | 1.36(1.19–1.54) | 5.95E-6 | 1.36(1.20–1.54) | 1.88E-6 | 6.17E-01 |
| rs17405217 | 1.78(1.04–3.04) | 3.45E-2 | 1.35(1.18–1.54) | 7.50E-6 | 1.37(1.20–1.55) | 1.55E-6 | 3.56E-01 |
| rs2656052 | 0.97(0.78–1.20) | 7.90E-1 | 1.27(1.11–1.44) | 3.84E-4 | 1.18(1.06–1.32) | 3.05E-3 | 5.38E-02 |
| rs17484235 | 1.79(1.05–3.05) | 3.36E-2 | 1.36(1.19–1.54) | 5.25E-6 | 1.37(1.21–1.55) | 1.06E-6 | 3.62E-01 |
| rs7164594 | 1.00(0.80–1.25) | 9.79E-1 | 0.74(0.64–0.86) | 1.10E-4 | 0.81(0.71–0.91) | 1.04E-3 | 3.35E-02 |
| rs8034191 | 1.32(0.96–1.80) | 8.84E-2 | 1.37(1.20–1.56) | 2.30E-6 | 1.36(1.21–1.53) | 5.59E-7 | 8.47E-01 |
| rs10519203 | 1.13(0.88–1.44) | 3.45E-1 | 1.33(1.17–1.51) | 1.62E-5 | 1.29(1.15–1.44) | 1.65E-5 | 2.98E-01 |
| rs2036534 | 0.89(0.69–1.15) | 3.88E-1 | 0.75(0.64–0.87) | 2.77E-4 | 0.79(0.69–0.89) | 3.40E-4 | 2.56E-01 |
| rs12916483 | 0.86(0.64–1.15) | 3.16E-1 | 0.94(0.83–1.06) | 3.57E-1 | 0.93(0.83–1.04) | 2.11E-1 | 5.15E-01 |
| rs3813570 | 0.82(0.64–1.04) | 1.09E-1 | 0.75(0.64–0.87) | 2.35E-4 | 0.77(0.68–0.87) | 7.39E-5 | 5.36E-01 |
| rs11858230 | 0.86(0.67–1.10) | 2.37E-1 | 0.93(0.82–1.05) | 2.65E-1 | 0.92(0.82–1.02) | 1.30E-1 | 5.37E-01 |
| rs2036527 | 1.36(1.04–1.78) | 2.37E-2 | 1.37(1.21–1.56) | 1.81E-6 | 1.37(1.22–1.53) | 1.41E-7 | 9.57E-01 |
| rs667282 | 0.90(0.71–1.14) | 3.99E-1 | 0.75(0.64–0.87) | 1.68E-4 | 0.79(0.69–0.89) | 2.03E-4 | 2.28E-01 |
| rs588765 | 0.80(0.63–1.00) | 5.48E-2 | 0.91(0.80–1.02) | 1.18E-1 | 0.88(0.79–0.98) | 2.56E-2 | 3.56E-01 |
| rs6495306 | 0.80(0.63–1.00) | 5.29E-2 | 0.90(0.80–1.02) | 1.16E-1 | 0.88(0.79–0.98) | 2.44E-2 | 3.45E-01 |
| rs17486278 | 1.28(1.01–1.63) | 4.43E-2 | 1.40(1.23–1.59) | 5.74E-7 | 1.37(1.22–1.54) | 6.77E-8 | 6.12E-01 |
| rs601079 | 0.86(0.69–1.06) | 1.61E-1 | 0.90(0.79–1.02) | 1.03E-1 | 0.89(0.80–0.99) | 3.56E-2 | 6.65E-01 |
| rs680244 | 0.87(0.70–1.08) | 2.08E-1 | 0.90(0.79–1.01) | 8.68E-2 | 0.89(0.80–0.99) | 3.66E-2 | 8.03E-01 |
| rs637137 | 0.91(0.71–1.15) | 4.31E-1 | 0.74(0.64–0.86) | 1.03E-4 | 0.78(0.69–0.88) | 1.64E-4 | 1.76E-01 |
| rs951266 | 1.45(1.01–2.06) | 4.15E-2 | 1.39(1.22–1.57) | 9.83E-7 | 1.39(1.23–1.57) | 1.25E-7 | 8.29E-01 |
| rs555018 | 0.79(0.63–0.99) | 4.50E-2 | 0.91(0.80–1.02) | 1.22E-1 | 0.88(0.79–0.98) | 2.43E-2 | 3.30E-01 |
| rs647041 | 0.80(0.64–1.01) | 6.74E-2 | 0.91(0.80–1.02) | 1.27E-1 | 0.89(0.79–0.98) | 3.15E-2 | 4.04E-01 |
| rs16969968 | 2.04(1.15–3.62) | 1.47E-2 | 1.40(1.23–1.59) | 4.14E-7 | 1.42(1.25–1.61) | 4.49E-8 | 2.37E-01 |
| rs660652 | 0.80(0.63–1.00) | 5.85E-2 | 0.95(0.83–1.08) | 4.49E-1 | 0.92(0.82–1.02) | 1.37E-1 | 2.27E-01 |
| rs578776 | 1.01(0.80–1.26) | 9.58E-1 | 0.72(0.62–0.82) | 3.21E-6 | 0.79(0.70–0.88) | 7.09E-5 | 1.26E-02 |
| rs6495307 | 0.86(0.69–1.08) | 2.06E-1 | 0.91(0.81–1.03) | 1.62E-1 | 0.90(0.81–1.00) | 7.22E-2 | 6.86E-01 |
| rs1051730 | 1.38(0.98–1.95) | 6.76E-2 | 1.40(1.23–1.59) | 5.88E-7 | 1.39(1.23–1.57) | 1.16E-7 | 9.39E-01 |
| rs3743078 | 1.05(0.84–1.30) | 6.82E-1 | 0.76(0.65–0.87) | 2.68E-4 | 0.84(0.74–0.94) | 4.48E-3 | 1.82E-02 |
| rs3743077 | 0.80(0.59–1.06) | 1.28E-1 | 0.91(0.80–1.02) | 1.20E-1 | 0.89(0.79–0.99) | 3.97E-2 | 3.42E-01 |
| rs1317286 | 1.27(0.98–1.63) | 6.56E-2 | 1.36(1.19–1.54) | 4.44E-6 | 1.33(1.19–1.49) | 1.10E-6 | 5.91E-01 |
| rs938682 | 0.90(0.71–1.15) | 4.07E-1 | 0.76(0.65–0.87) | 2.87E-4 | 0.79(0.70–0.89) | 3.20E-4 | 2.59E-01 |
| rs11637630 | 0.91(0.71–1.15) | 4.22E-1 | 0.75(0.65–0.87) | 2.10E-4 | 0.79(0.70–0.89) | 2.90E-4 | 2.12E-01 |
| GSC3_26 | 0.73(0.29–1.84) | 5.07E-1 | 0.77(0.59–0.99) | 4.69E-2 | 0.76(0.59–0.97) | 3.17E-2 | 7.94E-01 |
| rs7177514 | 0.97(0.76–1.23) | 7.87E-1 | 0.76(0.65–0.88) | 3.10E-4 | 0.81(0.71–0.92) | 1.21E-3 | 9.29E-02 |
| rs6495308 | 0.98(0.77–1.25) | 8.82E-1 | 0.76(0.65–0.87) | 2.64E-4 | 0.81(0.71–0.92) | 1.34E-3 | 7.01E-02 |
| rs8042059 | 1.00(0.78–1.28) | 9.94E-1 | 0.76(0.65–0.88) | 3.23E-4 | 0.82(0.72–0.92) | 2.01E-3 | 5.70E-02 |
| rs8042374 | 1.01(0.79–1.29) | 9.15E-1 | 0.77(0.66–0.89) | 5.21E-4 | 0.83(0.73–0.93) | 3.33E-3 | 5.55E-02 |
| rs4887069 | 1.01(0.79–1.29) | 9.31E-1 | 0.76(0.65–0.87) | 2.40E-4 | 0.82(0.72–0.92) | 1.83E-3 | 4.36E-02 |
| rs8192479 | 0.54(0.03–8.65) | 6.62E-1 | 1.31(0.84–2.03) | 2.40E-1 | 1.27(0.82–1.97) | 2.79E-1 | 5.16E-01 |
| rs6495309 | 0.94(0.72–1.21) | 6.13E-1 | 0.75(0.64–0.87) | 2.67E-4 | 0.79(0.70–0.90) | 5.41E-4 | 1.67E-01 |
| rs12914008 | 0.53(0.13–2.12) | 3.67E-1 | 0.73(0.54–0.97) | 3.53E-2 | 0.71(0.53–0.95) | 2.21E-2 | 5.39E-01 |
| rs17487223 | 1.59(1.07–2.36) | 2.07E-2 | 1.35(1.19–1.54) | 3.92E-6 | 1.37(1.21–1.54) | 3.58E-7 | 4.66E-01 |
| rs1996371 | 1.10(0.72–1.69) | 6.55E-1 | 1.15(1.01–1.30) | 3.46E-2 | 1.14(1.01–1.28) | 3.95E-2 | 6.81E-01 |
Boldface denotes SNPs discussed in the text
In European-descent samples, a locus distinct from rs16969968, tagged by rs578776, has been consistently associated with nicotine dependence [10, 14]. In the full sample, rs578776 is significantly associated (p = 7.09×10−5, OR = 0.79 (0.70–0.88)) and is the most strongly associated of the correlated SNPs representing this “protective” locus. However, this association is driven by the EA sample (p =3.21×10−6, OR = 0.72 (0.62–0.82)); in the AA sample, rs578776 is not associated (p = 0.96, OR = 1.01 (0.80–1.26)). This contrasts with the agreement between EA and AA association results for rs16969968.
In the AA sample, the most significant SNP that confers a protective effect is rs555018 in CHRNA5 (p = 0.045, OR = 0.79 (0.63–0.99)). This SNP is not correlated with rs16969968 in AAs (r2 = 0.02), though |D′| = 1. It is highly correlated with rs588765, which has been shown to affect expression of CHRNA5 in EAs [33, 34] (r2 = 0.99 in our AA sample and 0.999 in EAs). Therefore the evidence for association between rs588765 and nicotine dependence in AAs is similar (p = 0.055, OR = 0.80 (0.63–1.00)) to that found for rs555018. In contrast, neither rs555018 nor rs588765 is associated with nicotine dependence in the EAs.
Finally, a non-synonymous variant rs12914008 in CHRNB4, though rare in EAs (4.5%) and virtually non-polymorphic in AAs (0.6%), is nominally associated with nicotine dependence in EAs (p = 0.035, OR = 0.73 (95% CI 0.54–0.97)). Although this SNP is too rare for a meaningful test in AAs alone, the odds ratio is consistent in the two groups, and combining AAs and EAs improves the association evidence slightly (p = 0.022, OR = 0.71 (0.53–0.95)).
DNA Resequencing in the CHRNA5 and CHRNA3 intergenic region
We detected 29 novel variants. Seven of these have also been reported by other groups while we were validating our sequencing results. While many of these were rare (MAF≤5%), ten had MAF ≥ 10% based on the 44 chromosomes. We genotyped these novel variants in the original 1608 EAs. Eleven were polymorphic in this larger sample and passed quality control (per-SNP call rate ≥ 0.96 and Hardy-Weinberg p ≥ 0.05).
Table 3 shows association results for these 11 SNPs. Three SNPs were highly correlated with already genotyped SNPs (r2 > 0.9) and did not provide new information. The most significant novel SNP, GSC3-26, had a relatively strong odds ratio of 0.67 (0.50–0.89) and an r2 ≤ 0.25 with previously genotyped SNPs, though |D′| was often high (e.g. r2 = 0.23, |D′| = 1.0 with rs3743078).
Table 3.
Results for novel, sequencing-discovered genetic variants in the CHRNA5-CHRNA3 intergenic region
| Novel variant | rs number (if available) | Position (bp) | Minor allele | Major allele | COGEND EA original (N = 1591) | |||
|---|---|---|---|---|---|---|---|---|
| MAF | p-value | Odds ratio (95% confidence interval) | Maximum r2 with previously genotyped SNPs | |||||
| GSC3_2 | 76671157 | TAAG | DEL | 0.0100 | 0.6773 | 1.16 (0.57–2.36) | <0.2 | |
| GSC3_4 | 76671923 | A | T | 0.0028 | 0.351 | 0.51 (0.13–2.09) | 0.46 | |
| GSC3_15 | rs62010327 | 76679839 | A | G | 0.3666 | 0.8474 | 1.02 (0.87–1.18) | 0.94 |
| GSC3_16 | 76679938 | DEL | TACTC | 0.0493 | 0.03726 | 0.71 (0.51 –0.98) | 0.95 | |
| GSC3_17 | rs55958820 | 76681412 | T | G | 0.0098 | 0.1722 | 1.70 (0.79–3.61) | <0.2 |
| GSC3_18 | rs57708953 | 76683263 | T | C | 0.0039 | 0.6057 | 0.74 (0.23–2.36) | 0.35 |
| GSC3_20 | 76684921 | T | DEL | 0.3719 | 0.9045 | 1.01 (0.87–1.17) | 0.97 | |
| GSC3_26 | 76693692 | G | A | 0.0648 | 0.006386 | 0.67 (0.50–0.89) | 0.25 | |
| GSC3_27 | 76694121 | T | G | 0.0060 | 0.7098 | 1.19 (0.48–2.97) | <0.2 | |
| GSC3_28 | 76695823 | T | C | 0.0063 | 0.6532 | 0.81 (0.33– 2.00) | <0.2 | |
| GSC3_29 | 76696413 | G | C | 0.0028 | 0.3521 | 0.51 (0.13–2.09) | 0.46 | |
Logistic regression, additive model
We therefore genotyped GSC3_26 in the remaining EAs and AAs (Tables 1 and 2). GSC3_26 is relatively uncommon (MAF of 0.062 in EAs and 0.014 in AAs). In the combined sample with race and gender as covariates, p = 0.032, OR = 0.76 (0.59–0.97).
LD and cross-population heterogeneity
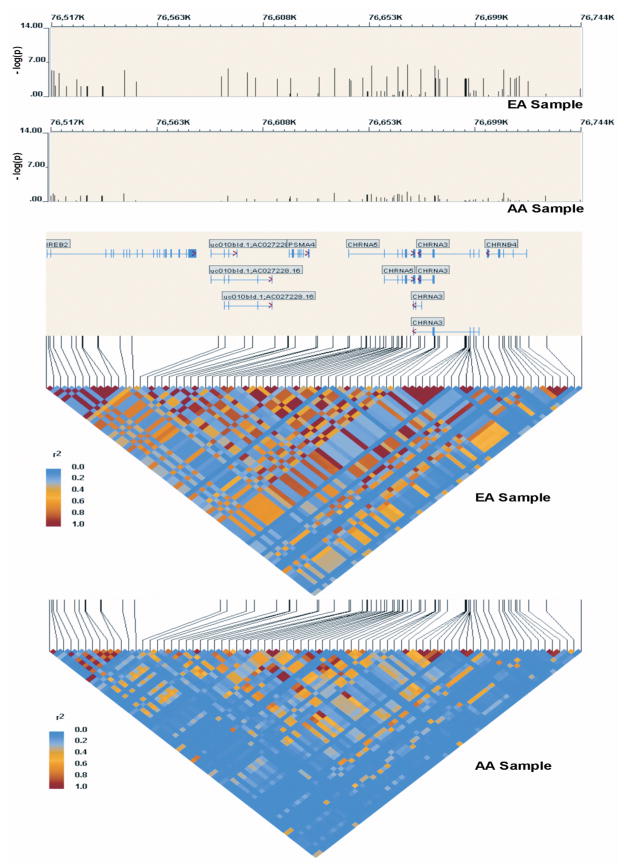
Figure 1 displays EA and AA results and r2 for all 76 SNPs. The LD results confirm that the most significantly associated SNPs in the full sample are highly correlated with rs16969968 in EAs (r2 ranging from 0.68 to 1.00). These SNPs all have OR > 1. In AAs, the LD is reduced (r2 from 0.11 to 0.73). The next group of significantly associated SNPs, which have OR < 1, are highly correlated with rs578776 in EAs.
Figure 1.
Association results and r2 on chromosome 15q24-25.1 for the EA and AA samples.
Three SNPs are of particular interest from the single-SNP association results: rs16969968 (associated in both EAs and AAs); rs555018 (associated in AAs only), and rs578776 (associated in EAs only). Among them, r2 is low to moderate; |D′| is higher. For rs16969968 and rs555018, r2 is 0.39 and |D′| = 0.99 in EAs, while r2 = 0.024 and |D′| = 1 in AAs. For rs16969968 and rs578776, r2 = 0.20 and |D′| = 1 in EAs; r2 = 0.07 and |D′| = 1 in AAs. For rs555018 and rs578776, r2 = 0.075 and |D′| = 0.52 in EAs; r2 = 0.43 and |D′| = 0.90 in AAs.
Fourteen SNPs demonstrate significantly different odds ratios in EAs versus AAs (Table 2 and Supplementary Table 2, last column), including rs578776, which represents the “protective” finding in EAs. All other SNPs displaying significant heterogeneity are at least moderately correlated with rs578776 in EAs (r2 ranges from 0.39 to 0.76) and are nominally associated (p < 0.05) in EAs with OR < 1. The correlation is sharply reduced in AAs (r2 from 0.001 to 0.40). This difference in LD may explain the lack of association for these SNPs in AAs and suggests that the functional variant driving the signal is in LD with these SNPs in EAs.
Notably, at rs16969968 the AA and EA odds ratios are notsignificantly different (heterogeneity p = 0.24).
Multilocus analyses
rs555018 and rs16969968
The association between rs555018 and nicotine dependence in AAs is intriguing because this SNP is one of a group of highly correlated SNPs that are associated with CHRNA5 mRNA levels in EAs [33, 34]. However, in EAs, rs555018 and correlates such as rs588765 are not associated with nicotine dependence. To further investigate these differing results at these potentially functional SNPs, we carried out multilocus analyses to account for the established effects of rs16969968.
First, we tested rs555018 in EAs after controlling for rs16969968 (coded additively) as a covariate in the logistic regression. The goal was to determine if rs555018, which shows no main-effect association in EAs, might be associated after adjusting for rs16969968. After accounting for rs16969968, rs555018 has p-value 0.044, OR = 1.18 (1.01–1.39); rs588765 has p-value 0.038, OR = 1.19 (1.01–1.40). The odds ratios in EAs at these the expression-associated SNPs have opposite direction to those in AAs.
This contrast in the direction of effect is clarified by joint analyses of rs16969968 with rs555018, using the genotype that is homozygous for both major alleles as the reference (Table 4). In the presence of “GG” at rs16969968, having 1 or two copies of the minor “C” allele at rs555018 increases risk in EAs, but confers protection in AAs (Table 4, top row in each panel). For rs16969968, we see a consistent direction of effect of the “A” risk allele in EAs and AAs, regardless of genotype at rs555018.
Table 4.
Joint analysis of rs16969968 and rs555018.*
| EA sample | rs555018 | ||
|---|---|---|---|
| rs16969968 | TT | CT | CC |
| GG | 42/63 1.00 (ref) |
188/211 1.33 (0.86–2.07) |
182/185 1.48 (0.95–2.29) |
| AG | 157/161 1.46 (0.93–2.29) |
317/280 1.70 (1.11–2.59) |
3/0 -- |
| AA | 171/89 2.88 (1.81–4.60) |
1/0 -- |
0/0 -- |
| AA sample | rs555018 | ||
| rs16969968 | TT | CT | CC |
| GG | 199/100 1.00 (ref) |
155/101 0.77 (0.55–1.09) |
37/30 0.62 (0.36–1.06) |
| AG | 37/13 1.43 (0.73–2.81) |
18/3 3.02 (0.87–10.48) |
0/0 -- |
| AA | 1/0 -- |
0/0 -- |
0/0 -- |
The first line of each cell indicates the number of cases and controls with the specific genotype combination; the second line indicates the odds ratio and 95% CI when GG/TT for rs16969968/rs555018 is the reference genotype.
We also tested each SNP within the one available homozygous genotype group of the other. The test cannot be performed in the second homozygous group because there is essentially only one genotype present at the other locus (|D′|>0.99; see also Table 4). In AAs, rs555018 remains significant (p= 0.046, OR = 0.78 (0.61–1.00)) when tested within the 622 “GG” homozygotes at rs16969968; within the 350 rs555018 “TT” homozygotes, rs16969968 has p = 0.22, OR = 1.51 (0.78–2.94). In EAs, rs555018 has p = 0.12 (OR= 1.17 (0.96–1.44)) in the 871 rs16969968 “GGs”, and rs16969968 has p = 2.08 × 10−6, OR = 1.73 (1.38–2.17) in the 683 rs555018 “TTs”.
rs578776 and rs16969968
Both rs16969968 and rs578776 show strong association with nicotine dependence in European-descent populations. There is debate about whether these associations are independent or whether they reflect a single finding.
To explore this question in EAs, we tested for association of rs578776 after controlling for rs16969968 (coded additively). Rs578776 remains significant (p-value 0.0055, OR = 0.80 (0.68–0.94)). We also tested for the effect of one locus in homozygotes of the other. Because |D′| is high, this test can be carried out in only the “GG” genotype at rs16969968 or the “CC” genotype at rs578776. In both cases, the second SNP remains significant in EAs. Among “GGs” at rs16969968, rs578776 has p = 0.018, OR = 0.79 (0.65–0.96)); among “CCs” at rs578776, rs16969968 has p = 0.0039, OR = 1.29 (1.09–1.54)).
DISCUSSION AND CONCLUSIONS
This study contributes several new findings about genetic risk for nicotine dependence. The CHRNA5-CHRNA3-CHRNB4 region clearly plays a role in nicotine dependence risk not only in European-Americans but also in African-Americans, and our evidence suggests that multiple variants are involved.
First, rs16969968, which causes an amino acid change in CHRNA5 and is clearly implicated in risk for nicotine dependence and lung cancer in individuals of European descent, is also significantly associated with nicotine dependence in African-Americans. This SNP is the most important one to test first: it is highly replicated, and in vitro studies demonstrate that α4β2α5 nicotinic receptors with the risk variant at rs16969968 exhibit lower maximal response to a nicotinic agonist than receptors with the other allele [11].
This replication, in African-Americans, of the rs16969968 association adds crucial information because of the different LD and population history for African-Americans, and because of the much lower allele frequency of rs16969968 in this population. The allele “A” at rs16969968 is rare in African-Americans, with a frequency of 5% (6% in cases, 3% in controls), compared to 35% in European-Americans (39% in cases, 31% in controls), and it is not present in the HapMap YRI. Nonetheless, it is a risk factor in our sample of African descent. The odds ratio is 2.04 (1.15–3.62) in African-Americans and 1.40 (1.23–1.59) in European-Americans. These confidence intervals overlap, and the ORs are not significantly different in the two populations (heterogeneity p = 0.24). Thus the “A” allele at rs16969968 is a consistent risk factor in both populations. Furthermore, in each group rs16969968 is the most significant and has the highest odds ratio.
This chromosome 15q24-25.1 region also harbors SNPs that are correlated with each other and associated with brain mRNA levels of CHRNA5 in European-descent samples ([33, 34]). Our second major finding is that in African-Americans, these expression-associated SNPs are associated with nicotine dependence. The evidence is nominally significant (p = 0.045, OR = 0.79 (0.63–0.99) for rs555018) in our modestly powered African-American sample. It is intriguing that in European-Americans, single-SNP association at rs555018 is not seen; however, when rs16969968 is included as a covariate, rs555018 does significantly alter risk. Joint genotype analysis clarifies that in the presence of “GG” at rs16969968, the major allele “T” at rs555018 appears to confer protection in European-Americans; however in African-Americans the minor allele “C” at rs555018 is the one more frequent in controls. These contrasting effects are difficult to interpret, but suggest that mRNA levels of CHRNA5, and thus nicotine dependence risk, may be influenced by additional variants in the region.
The two findings – one at rs16969968 and one at expression-associated SNPs such as rs588765 - have the appealing property of being at least LD proxies for functional effects [11, 33, 34], if not the functional SNPs themselves. It is plausible that these SNPs are responsible for two biological mechanisms affecting nicotine dependence risk: a coding change at CHRNA5 to asparagine (regardless of mRNA level) at rs16969968, and altered brain mRNA expression of the common (aspartate) form of CHRNA5, tagged by rs588765.
Our third important result involves rs578776 and its correlates. These SNPs represent an established association in European-descent populations that has only low correlation with rs16969968 (r2 < 0.25) and whose minor allele is elevated in controls. In our sample, rs578776 is associated with nicotine dependence in European-Americans as expected, but is not associated in African-Americans. Furthermore, we detect significant heterogeneity in the odds ratios for rs578776 and some correlates (p ~ 0.01). These results suggest that those SNPs are not the biologically causal variants explaining the statistical association. Recently, rs6495309 and other SNPs correlated with rs578776 in HapMap CEU but not in HapMap JPT and CHB [35] were associated with lung cancer and smoking in a Chinese population [36].
Our next key observation is that less common SNPs appear to be associated with nicotine dependence, suggesting that rarer variants in this gene cluster may have strong effects on susceptibility. In European-Americans, rs12914008 (MAF = 5%) is associated with nicotine dependence (p = 0.035, OR = 0.73 (0.54–0.97)). In the African-American sample, this SNP is very rare (0.6%) and affords little power to detect association, though in the combined analysis the evidence improves (p = 0.022, OR = 0.71 (0.53–0.95)). This SNP causes a missense change in CHRNB4 (threonine to isoleucine, T91I) and is also in LD (r2 = 0.8 in CEU) with rs8192475, a non-synonymous SNP in CHRNA3 (arginine to histidine, R37H). We also identified a novel SNP that is nominally associated with nicotine dependence in the full sample and has a frequency of 6% in European-Americans and 1% in African-Americans. These rarer variants are intriguing. Though we cannot conclude that these SNPs represent additional independent risk factors, they hint that this region may play a more complex biological role in risk for nicotine dependence than will be explained by common variants.
A major question is whether the multiple SNP associations between CHRNA5-CHRNA3-CHRNB4 and nicotine dependence represent the effect of a single causal locus, or whether multiple, statistically distinct loci are involved. Our data lead us to believe that there are at least two distinct, biologically causal loci in this region. In our analyses, the association at rs578776 in European-Americans is not explained by the association at rs16969968. Similarly, the association at rs555018 in African-Americans is not completely explained by rs16969968. The high |D′| between the seloci, however, does not allow us to determine the effect of each locus in the presence of all possible genotypes at other loci.
Rs16969968 is non-synonymous, functional, and a strong candidate to be a causal allele. It remains unclear what biologically causal variant might explain a potential “second” distinct locus, and we are unable to fully exclude the possibility that a single causal variant underlies all observed signals. Interestingly, our evidence for multiple common nicotine dependence risk alleles in the same gene region is similar to other recent examples of multiple independent loci for other common diseases [37, 38].
At the majority of SNPs examined, allele frequencies differ significantly between European-Americans and African-Americans. Therefore it is important to study associated variants that appear to be less common because of their potential to be higher frequency in certain geographic or racial groups, and therefore of higher impact on a population level.
Our results argue for further examination of this gene cluster, including DNA sequencing and variant discovery, especially in subjects of African descent. Such studies may reveal additional variants that affect nicotine dependence risk or CHRNA5 mRNA expression, and thereby further explain some of the observed contrasts between the two populations.
In summary, rs16969968 represents a risk locus for nicotine dependence in African-Americans as well as in European-Americans. We have evidence of a potentially complex role for gene-expression-associated SNPs such as rs555018 and rs588765 in the two populations. The “protective” association at rs578776 and its correlates is seen in European-Americans but not in African-Americans. Less common variants such as the novel SNP in the CHRNA5-CHRNA3 overlapping region also are associated with risk. Overall, multiple variants in this gene cluster are contributing to nicotine dependence risk and therefore to risk for diseases for which smoking is a major contributing factor.
Supplementary Material
Acknowledgments
Grant Support: P01 CA89392 (LJB) from the National Cancer Institute, R01DA013423 (LJB), R01DA019963 (LJB), K02DA021237 (LJB), R03 DA023166 (RCC), and K01DA024722 (SFS) from the National Institute on Drug Abuse, K01 AA015572 (ALH) from the National Institute on Alcohol Abuse and Alcoholism, K25 GM69590 (RCC) from the National Institute of General Medical Sciences, and IRG-58-010-50 from the American Cancer Society (SFS).
COGEND is a collaborative research group and part of the NIDA Genetics Consortium. Subject collection was supported by CA89392 (COGEND, PI - L Bierut) from the National Cancer Institute. Genotyping of the original COGEND sample at Perlegen Sciences was performed under NIDA Contract HHSN271200477471C; additional genotyping was provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a contract from the NIH to The Johns Hopkins University (HHSN268200782096C). Genotyping of the expanded sample was carried out by the Microarray Core at the Washington University Genome Center and Dr. Alison Goate’s laboratory at Washington University. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS)5. The authors thank Heidi Kromrei and Tracey Richmond for data collection assistance and Nanette Rochberg for data management assistance. We are grateful to Gary Chase for his contributions to COGEND. This work is in his memory, and in memory of Theodore Reich, founding Principal Investigator of COGEND.
Footnotes
Conflict of Interest statement: Drs. Bierut, Goate, Hinrichs, Rice, S. Saccone and Wang are listed as inventors on a patent, “Markers of Addiction,” held by Perlegen Sciences, Inc., covering the use of certain SNPs, including rs16969968, in diagnosing, prognosing, and treating addiction. Dr. N. Saccone is the spouse of S. Saccone, who is listed on the above patent. Dr. Bierut has served as a consultant to Pfizer.
References
- 1.World Health Organization. World Health Statistics Report - 2008. Geneva, Switzerland: World Health Organization Press; 2008. [Google Scholar]
- 2.Centers for Disease Control and Prevention. Smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 2000–2004. MMWR Morb Mortal Wkly Rep. 2008;57:1226–8. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Cigarette smoking among adults--United States, 2007. Morbidity and Mortality Weekly Report. 2008;57:1221–6. [PubMed] [Google Scholar]
- 4.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–27. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 5.Substance Abuse and Mental Health Services Administration. Results from the 2006 National Survey on Drug Use and Health: National Findings (Substance Abuse and Mental Health Services Administration) NSDUH Series H-21, DHHS Publication No. SMA 07-4293. Rockville, MD: Office of Applied Studies; 2007. [Google Scholar]
- 6.Ries L, Melbert D, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD: National Cancer Institute; 2008. [Google Scholar]
- 7.Fu SS, Kodl MM, Joseph AM, et al. Racial/Ethnic disparities in the use of nicotine replacement therapy and quit ratios in lifetime smokers ages 25 to 44 years. Cancer Epidemiol Biomarkers Prev. 2008;17:1640–7. doi: 10.1158/1055-9965.EPI-07-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Z, Alvarado GF, Hatsukami DK, et al. Race differences in nicotine dependence in the Collaborative Genetic study of Nicotine Dependence (COGEND) Nicotine Tob Res. 2008;10:1223–30. doi: 10.1080/14622200802163266. [DOI] [PubMed] [Google Scholar]
- 9.Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berrettini W, Yuan X, Tozzi F, et al. alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherva R, Wilhelmsen K, Pomerleau CS, et al. Association of a SNP in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with “pleasurable buzz” during early experimentation with smoking. Addiction. 2008;103:1544–52. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens VL, Bierut LJ, Talbot JT, et al. Nicotinic Receptor Gene Variants Influence Susceptibility to Heavy Smoking. Cancer Epidemiol Biomarkers Prev. 2008 doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss RB, Baker TB, Cannon DS, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grucza RA, Wang JC, Stitzel JA, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–9. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Chen J, Williamson VS, et al. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saccone NL, Saccone SF, Hinrichs AL, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–66. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–30. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saccone NL, Saccone SF, Goate AM, et al. Refining disease association signals using cross-population contrasts. BMC Genetics. 2008;9:58. doi: 10.1186/1471-2156-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. Tobacco Use Among Adults - United States, 2005. Morbidity and Mortality Weekly Report. 2006;55:1145–8. [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Duga S, Solda G, Asselta R, et al. Characterization of the genomic structure of the human neuronal nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster and identification of novel intragenic polymorphisms. J Hum Genet. 2001;46:640–8. doi: 10.1007/s100380170015. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Ge D, Zhang K, Need AC, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–3. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–8. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 33.Wang JC, Grucza R, Cruchaga C, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14:501–10. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JC, Cruchaga C, Saccone NL, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. doi: 10.1093/hmg/ddp231. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, Hu Z, Yu D, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–72. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- 37.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham RR, Cotsapas C, Davies L, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–61. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.