Abstract
The immediate-early effector gene Arc/Arg3.1 is robustly upregulated by synaptic activity associated with learning and memory. Here we show in primary cortical neuron culture that diverse stimuli induce Arc expression through new transcription. Searching for regulatory regions important for Arc transcription, we found nine DNaseI-sensitive nucleosome-depleted sites at this genomic locus. A reporter gene encompassing these sites responded to synaptic activity in an NMDA receptor–dependent manner, consistent with endogenous Arc mRNA. Responsiveness mapped to two enhancer regions ∼6.5 kb and ∼1.4 kb upstream of Arc. We dissected these regions further and found that the proximal enhancer contains a functional and conserved “Zeste-like” response element that binds a putative novel nuclear protein in neurons. Therefore, activity regulates Arc transcription partly by a novel signaling pathway. We also found that the distal enhancer has a functional and highly conserved serum response element. This element binds serum response factor, which is recruited by synaptic activity to regulate Arc. Thus, Arc is the first target of serum response factor that functions at synapses to mediate plasticity.
Keywords: plasticity, brain-derived neurotrophic factor, Zeste, serum response factor, Arc, gene transcription
Introduction
Activity-dependent, long-term changes in synaptic strength that mediate learning and memory require new gene expression (Kandel, 2001). Among genes induced during learning, the activity-regulated cytoskeletal immediate-early gene (IEG) Arc/Arg3.1 (Link et al., 1995; Lyford et al., 1995) is essential for memory consolidation (Plath et al., 2006). Cellular models of plasticity depend on Arc too. If Arc is suppressed, long-term potentiation (LTP) is induced but not maintained (Plath et al., 2006; Messaoudi et al., 2007). Arc regulates AMPA receptor (AMPAR) trafficking (Chowdhury et al., 2006) at synapses and actin polymerization in spines (Messaoudi et al., 2007). Thus, Arc offers a unique opportunity to investigate relationships between synaptic activity, gene expression, and memory.
Arc induction may be tuned to specific signals from synaptic activity associated with learning and memory. Compared with two other activity-dependent IEGs, zif268 and c-fos, Arc expression in the hippocampus correlates better with spatial task acquisition (Guzowski et al., 2006). Learning associated with novelty may produce activity-dependent signals that evoke Arc. In the hippocampus, Arc levels are significantly higher in newly trained rats than in pseudotrained or overtrained animals (Kelly and Deadwyler, 2002). Exposing rats to the same environment triggers neural activity, but Arc induction becomes uncoupled from that activity with repeated exposures (Guzowski et al., 2006). Similarly, patterns of visual experience–induced Arc expression in the occipital cortex depend on stimulus novelty (Wang et al., 2006). In situ detection of Arc mRNA is thus used to monitor behaviorally relevant synaptic activity and to map neuronal ensembles in vivo (Guzowski et al., 1999).
How extracellular stimuli regulate Arc production is incompletely understood. Behaviorally induced synaptic activity initiates new Arc transcription in vivo (Guzowski et al., 2006), but the transcription factors that mediate rapid Arc transcription remain unknown. Arc apparently lacks consensus binding sites for the well known Ca2+- and cAMP-response element binding protein (CREB) (Montminy and Bilezikjian, 1987; Sheng et al., 1990), suggesting that stimuli regulate Arc transcription through other signaling pathways (Waltereit et al., 2001). To discover how activity regulates Arc production, the effects of specific stimuli, including brain-derived neurotrophic factor (BDNF) (Rao et al., 2006), cAMP (Waltereit et al., 2001), and Ca2+ influx (Waltereit et al., 2001; Ichikawa et al., 2003), on Arc were examined. Yet it remains unclear which signaling pathways are specifically recruited by synaptic activity to regulate Arc expression and whether transcription initiation is a shared mechanism of Arc regulation for these pathways. Arc induction can be distinguished from that of at least some other IEGs and is closely correlated with learning, raising the intriguing possibility that Arc transcription is controlled by yet-to-be discovered activity-dependent signaling pathways and transcription factors.
We used DNaseI hypersensitivity assays to screen the endogenous chromatin structure flanking the Arc gene locus for discrete “open” nucleosome-depleted regions important for transcription. Our results provide important insights into the molecular mechanisms underlying Arc gene expression and have broad implications for how specific patterns of synaptic activity regulate neuronal gene expression to mediate long-term plasticity and memory.
Materials and Methods
Cell cultures.
Neonatal pup cortices were dissected and treated with papain (10 units/ml, 30 min; Worthington Biochemical) and then with a trypsin inhibitor (10 mg/ml, 15 min; Sigma). After trituration, dissociated neurons were plated on plastic tissue-culture plates (3.4 × 105 cells/cm2) coated with laminin (BD Biosciences) and poly-d-lysine (Chemicon). After 2 h, cells were transferred into a neuronal culture medium (NCM) based on Basal Medium Eagle (Invitrogen) (Rao et al., 2006). Undifferentiated rat pheochromocytoma (PC12) cells were maintained in standard growth medium consisting of Dulbecco's modified Eagle medium with 4.5 g/L glucose and 25 mm HEPES (Mediatech), supplemented with 5% calf serum, 5% horse serum (Hyclone), 1% penicillin-streptomycin (10 mg/ml), and 1% 200 mm l-glutamine (Invitrogen).
Pharmacology.
BDNF was from Amgen. d-2-amino-5-phosphonovaleric acid (APV) and UO126 were from Tocris. Tetrodotoxin (TTX) was from Calbiochem. H89, forskolin, and 3-isobutyl-1-methylxanthine (IBMX) were from Sigma. NGF was from Austral Biologicals.
Quantitative fluorogenic reverse-transcription PCR.
Cortical neurons (11 d in vitro [DIV]) were transferred to serum-free medium (SFM, otherwise identical to NCM described above). Cortical neurons challenged with the TTX-withdrawal paradigm were transferred into SFM supplemented with TTX (2 μm). At 45–48 h after transfer (13 DIV), neurons were stimulated either with drugs or by TTX washout for 6 h, lysates were collected, and RNA was isolated (QIAshredder Kit, RNeasy Plus Kit; Qiagen). Reverse transcription (TaqMan Reverse Transcription Reagents; Applied Biosystems) and qfPCR were performed with SYBR Green PCR Master Mix (Applied Biosystems) and an ABI Prism 7700 sequence detector (Applied Biosystems). All qfPCR primers have been described, including those for GAPDH (internal control gene for normalization) (Rao et al., 2006). Genomic DNA background was excluded by testing control samples that were not reverse transcribed. “No template” controls were also tested to exclude background signal from primer dimers.
In silico analyses.
To identify the orthologous regions of the human and mouse genomes flanking Arc, we used the University of California, Santa Cruz, Genome Bioinformatics website to query the Blast-like alignment tool (BLAT) (Kent, 2002) with the highly conserved coding region of rat Arc. These upstream and downstream sequence sets were then entered into the online EVOPRINTER software (Odenwald et al., 2005) to identify nucleotides conserved among these three species for both sequence and relative genomic position. The BLAT and EVOPRINTER analyses were performed with the default program settings (http://evoprinter.ninds.nih.gov/evopoverview.htm). The short conserved nucleotide stretches identified by this approach were then used to query the Transcription Element Search System (TESS) (Overton and Schug, 1997) for transcription factor DNA binding elements. The Arc locus conservation map was generated using default settings for human BLAT analysis (http://genome.ucsc.edu/cgi-bin/hgBlat?command = start).
DNaseI hypersensitivity assays.
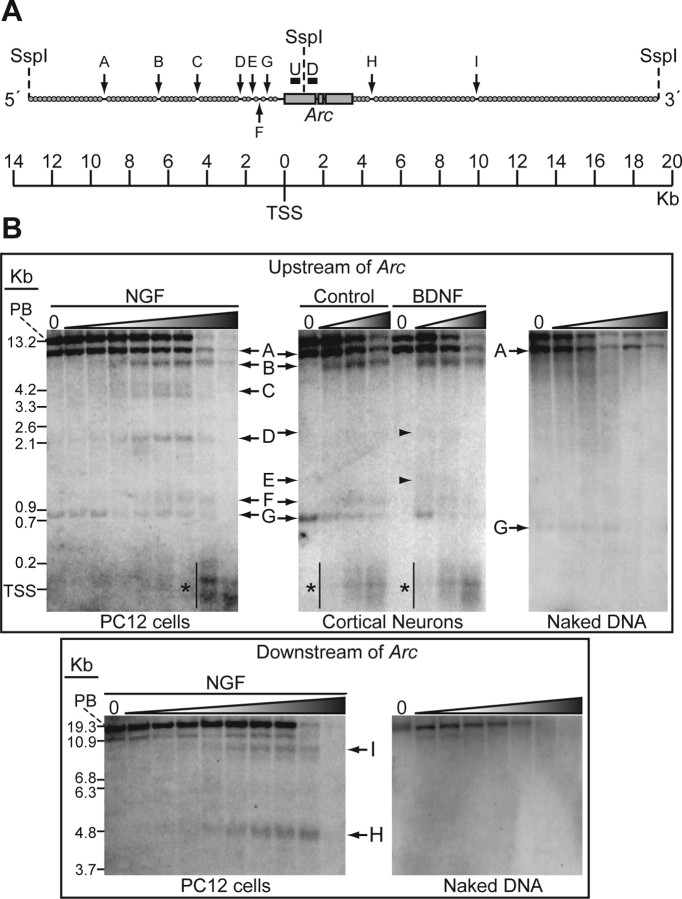
DNaseI hypersensitivity assays (DHAs) were performed as described (Verdin, 1991), with the following exceptions. PC12 cells (∼90% confluent) and cortical neurons (11 DIV) were transferred to SFM, and the next-day stimulations were performed for 3 h. Nuclei were isolated in ice-cold buffer to preserve in vivo chromatin architecture, and aliquots were subjected to a log1.3 concentration gradient of DNaseI (New England Biolabs): 0, 2, 2.6, 3.3, 4.4, 5.7, 7.4, 9.6, 12.5, and 16.25 units/∼3.25 × 106 nuclei. Genomic DNA (∼30 μg) was isolated from each DNaseI-treated aliquot of nuclei and digested overnight with SspI (New England Biolabs), a restriction enzyme with cut sites throughout the Arc locus: ∼13.2 kilobases (kb) upstream of, 925 base pairs (bp) downstream of, and ∼19.3 kb downstream of the Arc transcription start site (TSS). DNA samples were then run on a 1.2% agarose gel at 40 V for 16 h and transferred to Hybond-N+ membrane (Amersham Biosciences). Southern blot probes (see Fig. 3A) were amplified by PCR from bacterial artificial chromosome (BAC) CH230–208M7 (Children's Hospital Oakland Research Institute, Oakland, CA) with the following primers: 5′-gagagctgaaagggttgcacag-3′ and 5′-ttccaggtggctcaggaactc-3′ for the upstream probe (500 bp), 5′-tcccagatccagaaccacatga-3′ and 5′-cctagttgacaaaagcc-acagac-3′ for the downstream probe (537 bp). The upstream DNA ladder was produced by digesting BAC 208M7 with SspI and then performing secondary digests with AflII (5137 bp), BamHI (4296 bp), EcoRI (3544 bp), XhoI (3058 bp), HindIII (1830 bp), DraIII (1667 bp), BssHII (1149 bp), SgrAI (941 bp), or SacII (657 bp). The downstream ladder was created by using many of the same secondary enzymes: EcoRI (9992 bp), HindIII (5834 bp), XhoI (5338 bp), SgrAI (3852 bp), BamHI (2747 bp), DraIII (2162 bp), or AflII (1996 bp).
Figure 3.
DHAs identify nucleosome-depleted regions flanking Arc. A, Probes used to map hypersensitive sites at the Arc locus. Genomic DNA fragments were detected by Southern blot probes that annealed either immediately upstream (“U”; black bar) or downstream (“D”; black bar) of the SspI restriction site in Arc. Arrows indicate the relative positions of the nine DNaseI HSs (A–I) identified at the Arc locus; gray circles represent DNaseI-inaccessible nucleosome-packaged regions. B, Top, DHAs identify seven discrete open regions (bands A–G; arrows) upstream of Arc. Some are present in both rat cortical neurons (RCNs) and PC12 cells (bands A, B, D, F, G); others are visible only in RCNs (band E) or only in PC12 cells (band C). There is a larger open smear (asterisks) at the TSS. The “naked” DNA negative control, performed using both PC12 and RCN DNA, exhibits only bands A and G, suggesting that they are either DHA artifacts or atypical sites and that the remaining bands and smear reflect the native chromatin structure upstream of Arc. Bottom, DHAs identify two discrete open regions (bands H, I) downstream of Arc in PC12 cells, but not in RCNs (data not shown). The naked DNA control demonstrates that both bands represent native chromatin structure. Blot contrasts were chosen to best represent all observed bands. Bands D and E (arrows and arrowheads) are more clearly visible in higher-contrast images (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). All Southern blot images are representative of at least three experiments. Distances are in kb from the Arc TSS. The DNaseI concentration gradient, indicated above each blot, increases from left to right and includes a “no DNaseI” negative control in lane “O” (see Materials and Methods) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). PB: primary band resulting from SspI restriction cuts.
Plasmids.
For the new full-length Arc promoter reporter gene, BAC 208M7 was used to PCR amplify the genomic regions referred to as A (560 bp; primers: 5′-taagtcctgtgtggtagcctctcc-3′ and 5′-ctggtcccaacttccaggatgg-3′), B (653 bp; primers: 5′-tgggtggtggcaggctctgc-3′ and 5′-caggatacagg-gagacgaattgagg-3′), C (521 bp; primers: 5′-tcccagcctccattgttgtgtgg-3′ and 5′-tccttcctgagccacaagaacatg-3′), H (483 bp; primers: 5′-agcagggcta-gccaggaatgg-3′ and 5′-gaaacacagagccatgtctcaaatcg-3′), and I (308 bp; primers: 5′-aacagctctcaggagtggagtgg-3′ and 5′-cccgactctggctcacaagg-3′) with a high-fidelity proof-reading DNA polymerase (PfuUltra; Stratagene). These amplicons were subcloned into TOPO vectors (Invitrogen), sequence verified, and subcloned into a 2.6-kb Arc promoter reporter gene described previously (Ichikawa et al., 2003). Plasmid pSRE-Luc (SRE-Luciferase), used to measure serum response factor (SRF) activity, has five tandem serum response elements (SREs) driving firefly luciferase (Stratagene). Plasmid pRL0-Renilla, used to control for transfection efficiency and sample handling in the reporter assays, was constructed by removing promoter elements from pRL-SV40 (Promega). The wild-type (pCS2+/hSRF-VP16) and mutant (SRFΔM-VP16) SRF-VP16 constructs have been described (Schratt et al., 2002), as have the anti-SRF siRNAs (Yin et al., 2006).
Transient transfections and reporter gene assays.
Cortical neurons (11 DIV) were cotransfected with one of the Arc reporter gene plasmids and pRL0-Renilla (1:1 molar ratio) with Lipofectamine 2000 (Invitrogen) and transferred into SFM. At 20–24 h after transfection, neurons were stimulated with drugs for 8 h, and lysates were collected and assayed with a Dual Luciferase Kit (Promega) and a luminometer (Thermo Electron). Cortical neurons challenged by TTX withdrawal were transferred into SFM supplemented with TTX (2 μm) after transfection. At 40–42 h after transfer (13 DIV), neurons were stimulated by TTX washout, and lysates were harvested after 8 h. Each condition was tested in triplicate, and measured values were averaged. To ensure that reporter gene activity accurately reflects transcriptional activity, the DNA quality of every construct was tested for degradation and supercoiling, which affect transfection efficiency. Furthermore, two separate plasmid maxipreps were tested for each construct to ensure consistent reporter activities.
Chromatin immunoprecipitations.
These experiments were performed with a commercial kit (Active Motif) and the manufacturer's protocol. Primary cultured rat cortical neurons (11 DIV) (∼5 × 106 cells yielding ∼30 μg of genomic DNA) were cross-linked with 1% formaldehyde. Chromatin was enzymatically sheared to a final size of 500–1500 bp and incubated with 10 μg of rabbit anti-SRF antibody (Santa Cruz Biotech, s.c.-335X) or IgG control antibody. The following primer pairs were used for semi-quantitative PCR: Arc SRE1 (forward, 5′-gaagctccttgctgcgtcatgg-3′; reverse, 5′-gttttcctgctgttctctgcaattcc-3′; 130 bp amplicon), c-fos SRE (forward, 5′-tccccccttgcgctgcaccctcaga-3′; reverse, 5′-caacagggaccggccgtggaaacct-3′; 130 bp amplicon), and GAPDH promoter (forward, 5′-aaacaagttcaccaccatgtgaaa-3′; reverse, 5′-ccagggattgaccaaaggtgagtt-3′; 180 bp amplicon). PCR products were separated on 1.5% agarose gels.
Western blots.
These assays were performed as described (Rao et al., 2006) with the following antibodies: phospho-p44/42 MAPK (Cell Signaling, 9101; 1:2000), p44/42 MAPK (Cell Signaling, 9102; 1:2000), β-actin (Sigma, clone AC15; 1:10,000), GAPDH (Abcam, ab9485; 1:5000), and SRF (Santa Cruz Biotech, s.c.-335X; 1:1000); Recombinant Arc protein was expressed and purified as described (Lyford et al., 1995), and rabbit immunization and antibody purification was performed through Invitrogen (1:5000). AMAXA nucleofections for SRF-VP16 overexpression were performed as described (Gärtner et al., 2006). Briefly, 5–6 × 106 cortical neurons and 3 μg of DNA were used for each nucleofection reaction using program O-004. Recombinant lentivirus was generated by cotransfecting the transfer vector (FUGW) with two helper plasmids, delta8.9 (packaging vector) and VSV-G (envelope vector), into 293T cells (American Type Culture Collection). Supernatant was collected and virus was precipitated with PEG-it (System Biosciences). Cortical neurons were infected at 4 DIV with lentivirus expressing control or anti-SRF siRNA. At 6 d postinfection, the neurons were stimulated with BDNF, and lysates were harvested after 4 h.
Electrophoretic mobility shift assays.
Nuclear extracts from cortical neurons were prepared as described (Tao et al., 1998). The binding reaction was performed in a 16-μl-mixture containing 3.75 mm HEPES, pH 7.8, 3.75 mm MgCl2, 37.5 mm KCl, 0.375 mm DTT, 7.5% glycerol, 8 μg of poly(dI-dC), 2 μg of salmon sperm DNA, ∼10 μg of nuclear extract protein, and 3–5 fmol of 32P-labeled probe. Probe Z1 was synthesized as complementary oligodeoxyribonucleotide strands (22 bp; 5′-agagggccgtgagtggggggca-3′ and 5′-tgccccccactcacggccctct-3′) and labeled with 32P-ATP with T4 polynucleotide kinase, according to standard procedures. Probe mZ1 was synthesized as complementary oligodeoxyribonucleotide strands (22 bp; 5′-agagggccgtatatggggggca-3′ and 5′-tgccccccatatacggccctct-3′). After a labeled probe was added, the mixture was incubated for 20 min at 25°C. Fifteen microliters of the binding reaction mixture was then loaded into a 5% TBE Criterion precast gel (Bio-Rad) and electrophoresed in 0.5× TBE buffer at 85 V for 1.75 h. After electrophoresis, the gels were dried and exposed to film (BioMax MS Film; Kodak) at –80°C in a cassette with enhancer screens.
Statistical analyses.
Student's paired t tests and one-way ANOVA with post hoc Tukey's t tests were performed with Prism (GraphPad Software).
Results
Diverse stimuli induce new Arc transcription
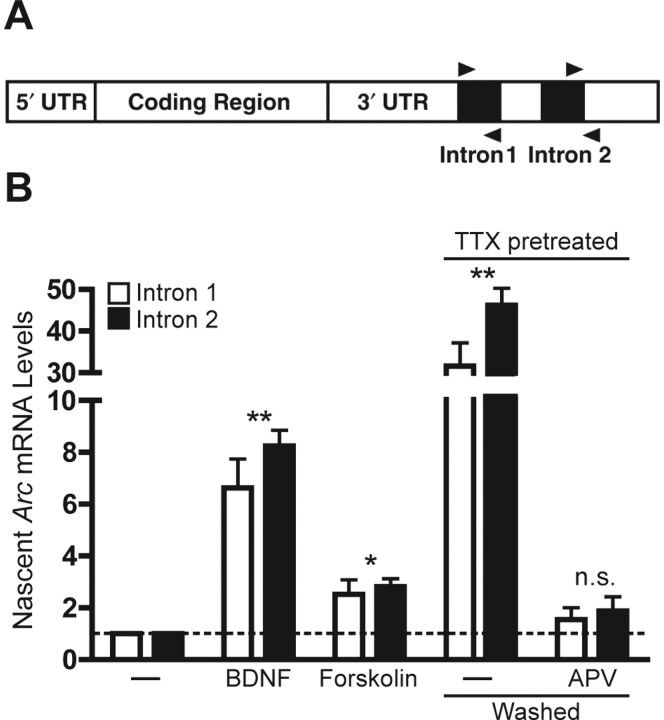
To determine whether distinct physiologically relevant stimuli induce new Arc mRNA transcription in neurons, we used intronic quantitative fluorogenic reverse-transcription PCR (qfRT-PCR) to detect nascent Arc pre-mRNA. The rat Arc gene contains two introns within the 3′ untranslated region (UTR) that are spliced out of Arc pre-mRNA to form the mature transcript (Giorgi et al., 2007). We designed and validated two qfRT-PCR primer pairs that anneal entirely within intron 1 or across the intron 2–exon 3 boundary to detect Arc pre-mRNA (Rao et al., 2006) (Fig. 1A).
Figure 1.
Diverse stimuli induce new Arc transcription. A, Diagram of Arc pre-mRNA and the qfRT-PCR primer pairs (arrowheads) used to detect its levels. B, Induction of nascent Arc pre-mRNA was measured by qfRT-PCR after 6 h stimulations with either BDNF (100 ng/ml), the adenylyl cyclase activator forskolin (50 μm) and the phosphodiesterase inhibitor IBMX (100 μm), or 6 h after washout of chronic TTX (2 μm, 48 h; TTX pretreated). This induction was inhibited by APV (100 μm) added after washout. Values are mean ± SEM. N = 3 distinct cDNA pools, each amplified from a separate neuronal culture prep. Paired t test: **p < 0.01, *p < 0.05 versus control; n.s., not statistically significant.
We first confirmed that synaptic activity directly regulates Arc mRNA synthesis in vitro, as seen in vivo (Guzowski et al., 2006). To evoke synaptic activity in vitro, we used a method that triggers rebound synaptic activity after a period of synaptic inhibition with TTX (Turrigiano et al., 1998; Riegle and Meyer, 2007). This widely used paradigm has been especially useful for studying mechanisms of synaptic homeostasis, which may be particularly relevant to Arc function (Shepherd et al., 2006). Briefly, synaptic activity in mature cultures was suppressed for ∼2 d by bath application of TTX, a sodium-channel antagonist. Treated neurons were washed into TTX-free medium, producing a rebound of synaptic activity and an induction (∼40-fold) of nascent Arc mRNA transcripts (Fig. 1B). In vivo, addition of the NMDA receptor (NMDAR) antagonist APV blocked increases in Arc levels induced by synaptic activity (Steward et al., 1998). Here, we found that addition of APV after TTX washout blocked new Arc transcription, indicating that synaptic activity drives Arc transcription initiation in an NMDAR-dependent manner in vitro. We then tested whether BDNF and cAMP induce Arc expression (Waltereit et al., 2001; Wibrand et al., 2006) by initiating new Arc transcription. Indeed, the synthesis of nascent Arc transcripts was increased approximately sevenfold by BDNF and approximately threefold by forskolin (an adenylyl cyclase activator that produces cAMP) (Fig. 1B).
In silico analysis identifies conserved regions flanking Arc
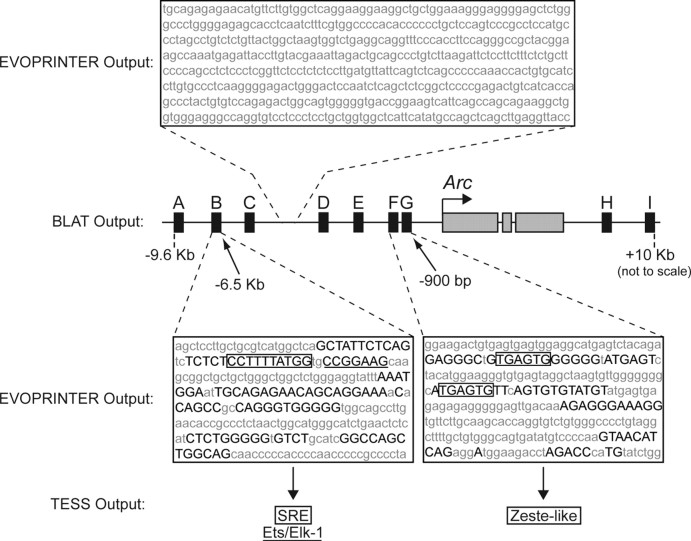
To help identify conserved DNA response elements in the Arc promoter that might mediate this new transcription, we used the University of California, Santa Cruz (UCSC) BLAT server (Kent, 2002) and EVOPRINTER freeware (Odenwald et al., 2005) to align the orthologous regions of the human, mouse, and rat genomes flanking Arc. The output was used to identify nucleotides throughout these alignments whose sequence and relative position in the genome were conserved. We found seven discrete clusters (Fig. 2, regions A–G) of short conserved nucleotide stretches dispersed throughout the ∼13-kb genomic region upstream and two clusters (Fig. 2, regions H, I) within the ∼16-kb genomic region downstream of Arc. In contrast, genomic sequences between these clusters largely lacked sequence and position conservation. The high degree of conservation of regions A–I suggests a physiologically important role in Arc transcription.
Figure 2.
In silico identification of putative regulatory regions containing conserved transcription factor binding sites. The online UCSC BLAT and the online EVOPRINTER analysis tools were used to identify seven upstream (A–G) and two downstream (H, I) regions (black rectangles) flanking Arc (gray rectangles) that are enriched in evolutionarily conserved sequences. Nucleotides within region B and regions F/G that are conserved in both sequence and relative genomic position are shown in black capital letters. In contrast, a portion of the genomic region between regions C and D demonstrates the relative lack of such conservation in the surrounding areas (see supplemental Fig. 3, available at www.jneurosci.org as supplemental material, for additional analysis). TESS was used to screen these conserved sequence elements for consensus binding sites, revealing a putative SRE (boxed) and an adjacent putative Ets/Elk-1 element (underlined) in region B as well as two putative Zeste-like binding sites (boxed) in regions F/G. All distances are from the Arc TSS.
Biochemical identification of putative regulatory regions flanking Arc
We next sought to determine whether the putative regulatory sites identified in silico correspond to open genomic regions with chromatin structure accessible to transcription factors (Elgin, 1988). The DHA identifies gene regulatory regions throughout large genomic expanses by screening for discrete open nucleosome-depleted regions that are hypersensitive to DNaseI cleavage (Pipkin and Lichtenheld, 2006) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Since regulatory regions have been found 5′ and 3′ to sites of transcription initiation (Kikuchi et al., 1992), we used DHAs to examine the upstream and downstream genomic domains flanking Arc (Fig. 3A).
We first performed DHAs on undifferentiated PC12 cell cultures, which upregulate Arc mRNA in response to multiple stimuli, including nerve growth factor (NGF) (Lyford et al., 1995). PC12 cell cultures were stimulated with NGF (100 ng/ml), genomic DNA was isolated, and DNaseI hypersensitive sites (HSs) were identified. The ∼13-kb region upstream of Arc contained six discrete open DNaseI HSs (Fig. 3B, bands A–D, F, G). The more distal sites, referred to as A, B, and C, reside ∼9.3 kb, ∼6.5 kb, and ∼4.5 kb (respectively) 5′ of the Arc transcription start site (TSS). These sites are upstream of regions examined in previous studies of Arc transcription (Waltereit et al., 2001; Ichikawa et al., 2003). Two additional DNaseI HSs (Fig. 3B, bands H, I) were found in the ∼16-kb region downstream of Arc. A ∼200-bp tract of accessible genomic DNA occurred at the Arc TSS.
To rule out sample handling as a source of these DHA sites, we tested whether chromatin that had been stripped of all DNA-binding proteins (“naked” DNA), but was otherwise handled in the same manner, would yield similar DNaseI cleavage sites. Bands A and G persisted in the naked DNA control (Fig. 3B), indicating that they might be artifacts or produced by DNaseI cleavage at atypical HSs (Bode et al., 2003). None of the other previously identified HSs were present, indicating that most of the HSs we identified result from DNaseI cleavage at hypersensitive open domains within the native chromatin structure.
To determine whether the same chromatin architecture is present in a more physiological system, DHAs were performed on rat cortical neuron cultures. Neurons were stimulated or not with BDNF (100 ng/ml). We found upstream HSs in locations similar to those in PC12 cells (Fig. 3B), but we did not identify any downstream HSs (data not shown). Since the intensity of DHA bands may vary with the extent to which a bound protein factor affects DNaseI accessibility, the presence rather than the intensity of a band is the critical indicator of an open genomic region (Gross and Garrard, 1988) (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). The open sites we identified were present under both basal and BDNF-stimulated conditions (and after chronic TTX washout, data not shown), indicating that they are static and suggesting that stimulus-induced Arc transcription does not result from changes in chromatin accessibility. We conclude that neurons and PC12 cells contain a common set of open genomic regions at the Arc locus. The positions of these regions correlated well with those of the conserved nucleotide clusters predicted by in silico analysis (Fig. 4A). These observations strongly suggest that these regions are physiologically important and likely contain response elements that regulate Arc transcription.
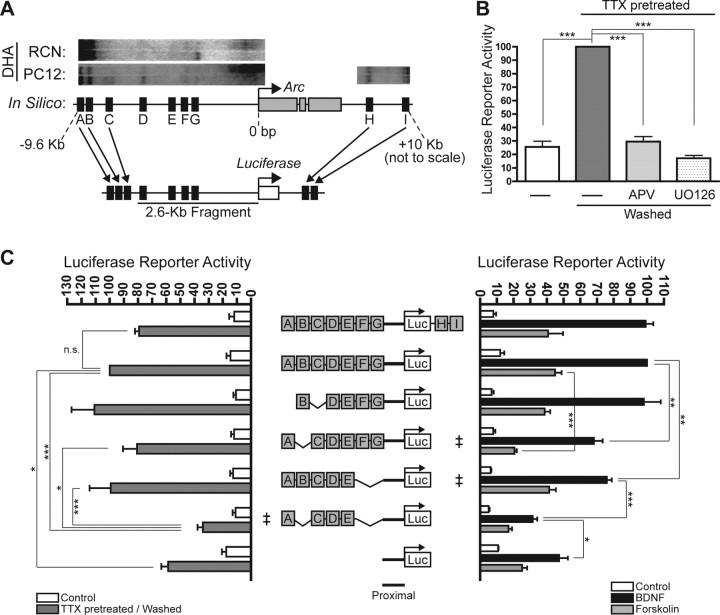
Figure 4.
Stimulus-inducible distal and proximal enhancer regions. A, A strong correlation was observed between the genomic positions of the DHA HSs and the evolutionarily conserved regions, suggesting potential involvement in Arc transcription. A reporter with a 2.6-kb genomic fragment containing the proximal Arc promoter, which already contained four of the nine putative regulatory sites (D–G), was supplemented with the remaining upstream (A–C) and downstream (H, I) HSs. B, The new full-length Arc reporter is induced by synaptic activity. In agreement with activity-regulated expression of endogenous Arc mRNA, this reporter response is completely blocked by the NMDAR inhibitor APV or the ERK activation inhibitor UO126. C, A systematic deletional analysis of the new full-length Arc reporter. Left, “‡” indicates the reporter construct with the minimal deletions (ΔB and ΔF/G together) that result in a reduced response to synaptic activity. “Proximal” indicates the ∼900-bp basal promoter region immediately upstream of Arc. All values are percentages (mean ± SEM) of the maximum measurement for reporter ABCDEFG. N = 4, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001, *p < 0.05; n.s., not statistically significant. Right, “‡” indicates the reporter constructs with the minimal deletions (ΔB or ΔF/G individually) that result in a reduced stimulus-mediated response. This side-by-side construct comparison highlights how these two regions differentially contribute to BDNF- and forskolin-induced reporter activity. All values are percentages (mean ± SEM) of the maximum BDNF measurement for reporter ABCDEFG. N = 5, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001, **p < 0.01, *p < 0.05.
In silico identification of putative regulatory elements within DHA regions
Open genomic regions are often enriched in relevant transcription factor binding sites (Gilbert et al., 2004). We analyzed the short conserved nucleotide stretches in regions A–I above for consensus transcription factor binding sites with the online TESS query database (Overton and Schug, 1997). Region B contained many conserved binding sites, including adjacent sites for SRF and Elk-1, which together form a ternary complex factor (TCF) involved in gene regulation (Norman et al., 1988; Buchwalter et al., 2004) (Fig. 2; supplemental Fig. 3, available at www.jneurosci.org as supplemental material). The other regions (A, C–I) contained putative binding sites for factors implicated in neuronal plasticity, including nuclear factor of activated T cells (Groth and Mermelstein, 2003), nuclear factor κB (Meffert et al., 2003), and myocyte-specific enhancement factor 2 (Flavell et al., 2006) (data not shown). Putative binding sites were found for other factors, including Nkx-2.5 (Moskowitz et al., 2007), N-Oct-3 (Blaud et al., 2004), and Zeste (Biggin et al., 1988) (data not shown). Although putative CREB binding sites were found, they did not meet the stringent cross-species conservation criteria of this in silico method.
Identification of novel distal and proximal enhancer regions
Previous reporter gene studies of Arc have been inconclusive (Waltereit et al., 2001; Ichikawa et al., 2003), perhaps because they focused on proximal regions of the 5′ Arc promoter that lacked many of the HSs we found. To assess the functional role of these putative regulatory regions, we made a new Arc reporter gene containing all nine DNaseI HSs (A–I). We began construction with a 2.6-kb genomic fragment of the proximal Arc promoter, which contains four of the HSs (D–G) (Ichikawa et al., 2003). We appended the remaining upstream HSs (A–C) in a tandem array immediately upstream of the 2.6-kb fragment, and the remaining downstream HSs (H, I) immediately downstream of the firefly luciferase reporter gene to maintain their relative positions in vivo (Fig. 4A). This reporter design likely mimics how genomic DNA normally flexes to bring distal and proximal regulatory regions together, enabling protein binding factors to interact and regulate transcription (West and Fraser, 2005).
In cortical neurons, this new reporter gene responded to synaptic activity in an NMDAR- and ERK-dependent manner (Fig. 4B), like endogenous Arc in vitro (Fig. 1B) and in vivo (Lyford et al., 1995; Ying et al., 2002). The reporter also responded to neuronal stimulation with BDNF (∼10-fold relative to control) and forskolin (∼fourfold relative to control) (Fig. 4C, right) in a manner that closely paralleled the stimulus-mediated inductions of endogenous Arc mRNA (Fig. 1B). Thus, this new Arc reporter gene, whose construction was based on the structure of chromatin at the Arc locus in vivo, faithfully recapitulates important features of the induction of endogenous Arc mRNA. These findings also indicate that one or more of these promoter regions contain DNA response element(s) that mediate stimulus-dependent Arc transcription.
To assess the role of each regulatory region, we created a set of reporter constructs with systematic deletions of DHA regions A–I and tested them in neurons. Region B and regions F/G mediated responses to synaptic activity (Fig. 4C, left). Region B was also a major mediator of both BDNF- and forskolin-induced reporter gene responses (Fig. 4C, right). Furthermore, regions F/G were required to mediate the reporter's full response to BDNF but not to forskolin (Fig. 4C, right). Although regions F and G appear as distinct DHA bands (Fig. 3B, top), they are physically too close to be clearly separated and were considered together here and below.
Identification of a distal functional SRE
Region B of the Arc promoter is required to mediate responses induced by synaptic activity, BDNF, and forskolin, but is it sufficient? To answer this question, we determined whether region B can enhance the transcription of a distinct promoter. Indeed, when inserted ectopically into a luciferase vector driven by the SV40 promoter (pGL3-Promoter; Promega), region B increased the promoter's response to synaptic activity; the enhancement was completely blocked by the NMDAR inhibitor APV or the PKA inhibitor H89 (Fig. 5A), suggesting that Ca2+ influx and PKA activation are required for synaptic activity to induce region B. Region B also enhanced the reporter's response to forskolin (Fig. 5B), confirming that PKA induces region B. Furthermore, region B enhanced reporter responses to BDNF. Thus, it may be sensitive to ERK signaling. In contrast, region D, which is similar in length to region B, failed to enhance stimulus-induced activity of the SV40 reporter gene (Fig. 5B). Thus, region B is a bona fide enhancer that contains one or more functional DNA elements.
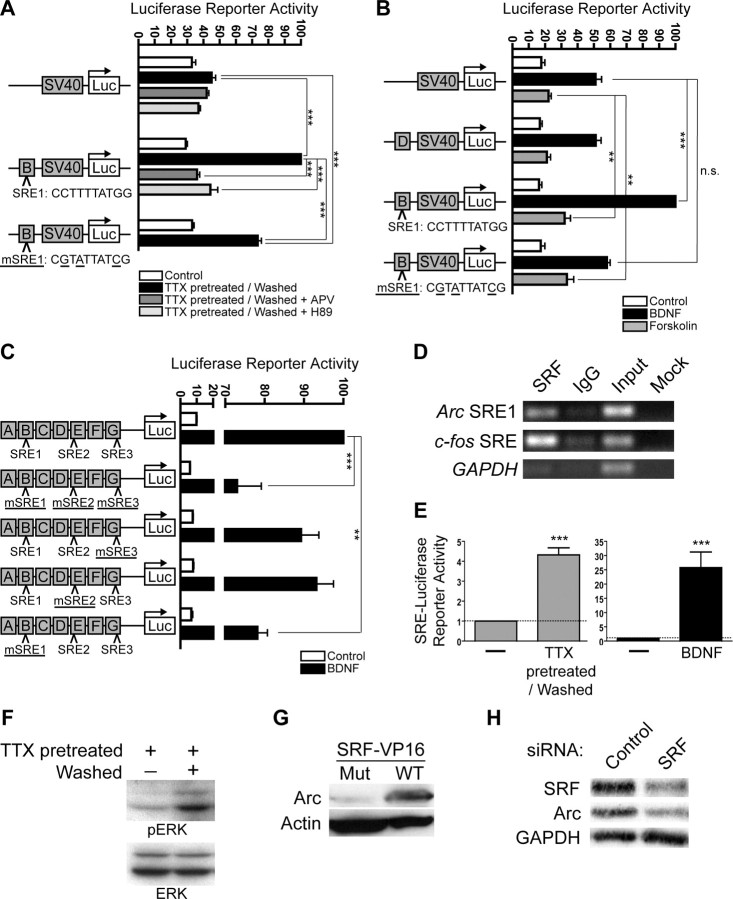
Figure 5.
SRF mediates enhancer activity of distal region B. A, Region B is sufficient to enhance SV40 promoter activity in response to synaptic activity, and inactivating point mutations of SRE1 (mSRE1, with mutated base pairs underlined) reduce this response by ∼50%. This enhancer activity is completely blocked by the NMDAR inhibitor APV or the PKA inhibitor H89. All values are percentages (mean ± SEM) of the maximum induction for B. N = 4, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001. B, Region B is sufficient to enhance SV40 promoter activity in response to both BDNF and forskolin (consistent with Fig. 4C, right). SRE1 is necessary to mediate the response to BDNF but is not important for the response to forskolin. In contrast, region D has no effect on BDNF- or forskolin-induced SV40 promoter activity. All values are percentages (mean ± SEM) of the maximum for B. N = 5, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001, **p < 0.01; n.s., not statistically significant. C, Three putative SREs identified in silico were systematically inactivated by subtle point mutations (mSRE) in the context of the full-length Arc reporter. SRE1, located within region B, mediates ∼20% of the response to BDNF and is the only functional element identified. All values are percentages (mean ± SEM) of the maximum for ABCDEFG (top construct). N = 3, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001, **p < 0.01. D, ChIPs demonstrate that SRF physically associates in vivo with SRE1. Semi-quantitative PCR shows that an SRF-specific antibody enriches for genomic region B compared with a nonspecific control IgG. “Input” refers to the genomic input DNA used for the immunoprecipitation. “Mock” refers to a no-template control PCR. Gel images are representative of three experiments. E, Stimulation paradigms that induce Arc also activate SRF. BDNF treatment or chronic TTX pretreatment and washout induce SRE-Luciferase reporter gene activity, indicating that SRF mediates transcriptional responses to both stimuli. Values are mean ± SEM. N = 2, unpaired t test: ***p < 0.001. F, Synaptic activity induced by chronic TTX washout (“Washed”) leads to the phosphorylation of ERK1/2 (“pERK”), an upstream regulator of SRF-containing SRE-bound complexes. Total ERK1/2 expression levels are unaffected (“ERK”). All Western blot images are representative of three experiments. G, Constitutively active SRF (SRF-VP16) is sufficient to induce endogenous Arc protein expression. Wild-type SRF-VP16 (“WT”) robustly induces Arc expression, while a SRF-VP16 mutant (“Mut”) lacking the SRE DNA-binding domain of SRF does not. Actin served as a loading control. All Western blot images are representative of two experiments. H, SRF is important for the expression of endogenous Arc. Specific knockdown of SRF expression causes a corresponding decrease in BDNF-induced Arc levels, compared with a negative control siRNA that has no effect on SRF levels or BDNF-induced Arc expression. GAPDH served as a loading control. All Western blot images are representative of two biological experiments performed in triplicate.
We reviewed the list of conserved transcription factor binding sites in region B (Fig. 2) and focused on the consensus SRE (referred to as SRE1). ERK activation is required for Arc induction (Rao et al., 2006), and SRE can mediate responses to ERK through bound TCF and SRF. However, ERK activates many transcription factors in addition to TCF and SRF (Johnson and Lapadat, 2002), so a direct role for SRE/SRF in Arc regulation remains to be established. Arc levels in the brains of SRF-null mice were normal in one study (Etkin et al., 2006) but significantly reduced in another (Ramanan et al., 2005). Since SRF was absent during development, it is hard to tell from these studies whether SRF regulates Arc directly or indirectly or whether compensatory changes have obscured its role. Previous efforts to directly identify a functional SRE within the Arc promoter have not been successful (Waltereit et al., 2001).
Therefore, to test the functional significance of SRE1 in Arc expression, we introduced inactivating point mutations. Mutating SRE1 reduced the response to synaptic activity by ∼50% (Fig. 5A) and completely blocked BDNF-mediated, but not forskolin-mediated, enhancement of SV40 reporter activity by region B (Fig. 5B). TESS analysis also identified a conserved SRE in region E (SRE2) and one in region G (SRE3) (Fig. 5C). To further test the functional significance of these SREs in the context of the Arc promoter, we created a panel of full-length Arc reporters with inactivating point mutations of SRE1–3, either individually or all together. SRE2 and SRE3 were dispensable for BDNF-induced reporter activity (Fig. 5C), as expected from our deletion analysis (Fig. 4C, right) and previous findings (Waltereit et al., 2001). In contrast, inactivation of SRE1 reduced BDNF-induced reporter activity to nearly the same extent as deletion of region B from the full-length reporter (Fig. 4C, right). Thus, SRE1 is a functional transcription enhancer element that responds to synaptic activity in an NMDAR-, PKA-, and ERK-dependent manner (Figs. 4B, 5A). However, these data also suggest a separate component of region B enhancer activity that is dependent on synaptic activity and PKA.
If SRE1 functions as a response element for Arc in vivo, then its cognate factor, SRF, should be bound at this genomic region. To determine whether SRF is bound to SRE1 at the endogenous Arc promoter, we performed chromatin immunoprecipitations (ChIPs). An SRF-specific antibody immunoprecipitated genomic DNA fragments from neurons that contain Arc SRE1, as shown by semi-quantitative PCR (Fig. 5D). The ChIP was specific, as the SRF-specific antibody also immunoprecipitated the genomic region with a known SRE from the c-fos promoter (Norman et al., 1988), but did not enrich for promoter regions of GAPDH, which lacks an SRE. These results show SRF binds SRE1 in vivo, further establishing SRE1 as a functional response element that is physiologically relevant for Arc expression.
To demonstrate that SRF functionally links stimulation to SRE1-mediated Arc expression, we tested whether SRF itself is regulated by synaptic activity or BDNF. Both significantly induced an SRE-driven firefly luciferase reporter gene (SRE-Luciferase) that directly measures SRF activity (Fig. 5E). Synaptic activity also led to the phosphorylation of ERK (Fig. 5F), a kinase that activates SRF-containing promoter complexes and SRE-dependent gene transcription and is required for Arc induction (Rao et al., 2006). These data show that SRF mediates transcription in response to patterns of synaptic activity or BDNF stimulation that induce endogenous Arc expression (Fig. 1B).
We therefore tested whether SRF is necessary and whether activated SRF is sufficient for Arc transcription. In neuronal culture, transient transfection of a constitutively active version of SRF fused to the VP16 transactivation domain (SRF-VP16) drove robust expression of endogenous Arc protein even in the absence of stimulation. In contrast, a mutant version of SRF-VP16 lacking the SRE DNA-binding domain of SRF did not induce Arc (Fig. 5G). Since active SRF was sufficient to induce Arc expression, we next tested whether SRF is required for stimulus-dependent Arc expression. Transduction of cultured neurons with anti-SRF siRNA led to a specific and reproducible reduction in SRF levels, which correlated with a reproducible and significant reduction in BDNF-dependent Arc induction. A negative control siRNA had no effect on SRF levels or BDNF-induced Arc expression (Fig. 5H). The incomplete knockdown of SRF might explain why Arc induction was not blocked completely; however, an independent contribution from the Zeste-like response element (described below) could also explain, at least in part, the remaining Arc expression. Together, the above findings clearly link synaptic activity, SRF activity, SRE1, and Arc expression.
A putative novel neuronal transcription factor regulates Arc
Next, we investigated the contribution of regions F/G to synaptic activity- and BDNF-induced Arc transcription. When subcloned into the SV40 promoter reporter, regions F/G enhanced reporter gene responses to synaptic activity (Fig. 6A). APV or H89 blocked this enhanced response (Fig. 6A), indicating that NMDAR and PKA activation are required for synaptic activity to induce regions F/G. Regions F/G also enhanced the reporter's response to BDNF but not to forskolin (Fig. 6B), suggesting regions F/G respond to ERK-mediated signaling. Together with our deletion analysis (Fig. 4C, right), these findings establish that regions F/G contain one or more DNA elements that selectively mediate responses to synaptic activity and BDNF but not to forskolin.
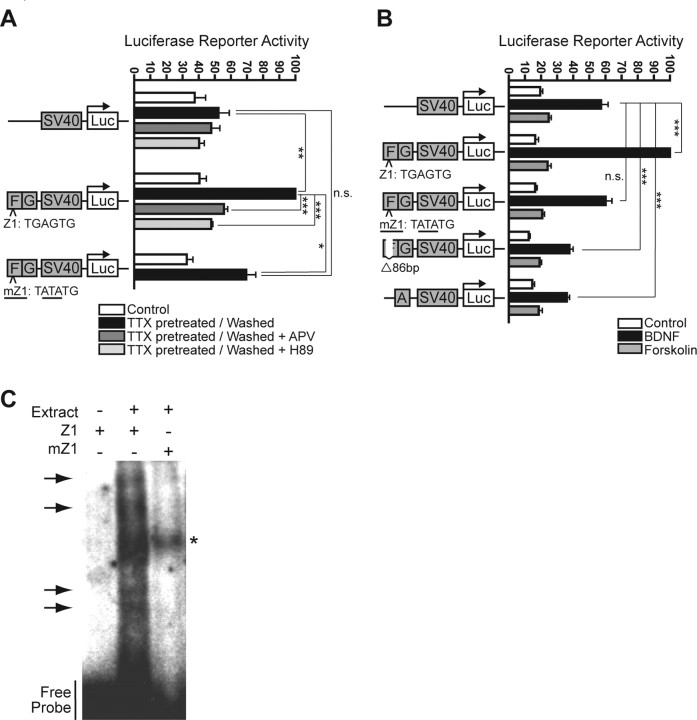
Figure 6.
A putative novel factor regulates proximal enhancer regions F/G. A, Regions F/G are sufficient to enhance SV40 promoter activity in response to synaptic activity. This enhancer activity is blocked by the NMDAR inhibitor APV or the PKA inhibitor H89. Inactivating point mutations of Zeste1 (mZ1, with mutated base pairs underlined) demonstrate that this site is necessary to mediate this enhanced response. All values are percentages (mean ± SEM) of the maximum induction for regions F/G. N = 3, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001, **p < 0.01, *p < 0.05; n.s., not statistically significant. B, Regions F/G are sufficient to enhance SV40 promoter activity in response to BDNF but not to forskolin (consistent with Fig. 4C, right). Inactivating point mutations of Zeste1 demonstrate that this site is necessary to mediate the enhanced response to BDNF. Deletion of an 86-bp stretch containing the Zeste1 and Zeste2 elements (Δ86bp) reduces BDNF-induced SV40 promoter activity below baseline, suggesting the presence of possible repressor elements in the remaining region. Region A also suppresses BDNF-induced SV40 promoter activity. All values are percentages (mean ± SEM) of the maximum induction for regions F/G. N = 5, one-way ANOVA and post hoc Tukey's t tests: ***p < 0.001; n.s., not statistically significant. C, EMSAs demonstrate that a nuclear protein factor or factors physically associate with the Zeste1 element. Several distinct mobility shifts (arrows) are observed when the wild-type Zeste1 probe (Z1) is pre-incubated with nuclear protein extracts (“Extract”), while the mutant Zeste1 probe (mZ1, containing the same point mutations that eliminate enhancer activity) did not show these corresponding shifts. The single shift observed for mZ1 (indicated by an asterisk) might represent a nonspecific binding event. Blot images are representative of three experiments.
Bioinformatic analysis of regions F/G identified two conserved and closely spaced consensus binding sites for Zeste (Fig. 2). Efficient Zeste binding and function often require multiple binding sites in close proximity (Mansukhani et al., 1988), suggesting that the sites we found in Arc might be important. However, Zeste is an invertebrate transcription factor without a known mammalian ortholog (Levine et al., 2002), and it has not been implicated in plasticity-related neuronal gene expression (Biggin et al., 1988). We hypothesized that a novel “Zeste-like” mammalian factor binds to these sites and is important for Arc expression.
To investigate the functional significance of the Zeste-like sites, we first took a mutagenesis-based approach. We made point mutations in the upstream site (Zeste1) in the Arc promoter that would be expected from studies in Drosophila (Moses et al., 2006) to disrupt Zeste binding. If a mammalian Zeste-like factor binds that site with similar specificity, these inactivating point mutations should block the enhancer activity of regions F/G. Indeed, a subtle 3-bp point mutation of Zeste1 eliminated the enhancer activity of this ∼500-bp region in response to synaptic activity (Fig. 6A) and BDNF (Fig. 6B). With the data above, these findings indicate that the novel Zeste1 site mediates transcriptional responses to synaptic activity in an NMDAR-, PKA-, and ERK-dependent manner (Figs. 4B, 6A).
To further investigate how these sites contribute to the transcriptional responses conferred by regions F/G, we deleted an 86-bp stretch that contains both of the putative Zeste-like binding sites. This deletion eliminated the BDNF-induced enhancement of SV40 promoter activity by regions F/G (Fig. 6B), further supporting the notion that this 86-bp stretch contains at least one binding site for a transcriptional enhancer. The deletion also reduced reporter gene activity to a level below that produced by the SV40 promoter alone, indicating that the portions of regions F/G surrounding the Zeste-like binding sites contain a transcriptional repressor element or elements. Region A also repressed BDNF-induced SV40 promoter activity, consistent with the observation that deleting both region B and regions F/G from the full-length reporter (but retaining region A) led to lower BDNF-induced levels of activity than were obtained with the ∼900-bp proximal Arc promoter alone (Fig. 4C, right).
If Zeste-like elements mediate synaptic activity- and BDNF-induced Arc transcription through a protein-bound factor, the corresponding DNA sequence should bind one or more nuclear proteins in neurons. Since this putative transcription factor is unknown, we cannot perform ChIPs, so we performed electrophoretic mobility shift assays (EMSAs). Short radiolabeled oligonucleotides encoding the wild-type Zeste1 site (probe Z1) or a version with point mutations (probe mZ1) that eliminate the enhancer activity of Zeste1 (Figs. 6A,B) were incubated with nuclear lysates from rat cortical neurons and analyzed by electrophoresis. The mobility of probe Z1 was significantly retarded by the formation of several distinct higher-molecular-weight species between probe Z1 and neuronal nuclear factor(s) (Fig. 6C). If the factor(s) from rat cortical neurons that binds Z1 behaves similarly to Zeste from Drosophila, these results might be expected. In Drosophila, Zeste forms large oligomeric protein complexes in vitro (Chen and Pirrotta, 1993), and the availability of excess binding sites in the EMSA reaction might favor the formation of different oligomeric species. Importantly, mutations in the Zeste1 element (probe mZ1) that disrupt its ability to mediate transcription induced by synaptic activity and BDNF also eliminate nearly all binding of the neuronal nuclear factor(s) to the probe. The one apparent higher-molecular-weight band seen for probe mZ1 might represent a nonspecific binding event.
Discussion
Whether signaling pathways triggered by synaptic activity, such as Ca2+, cAMP, or receptor tyrosine kinase activation, induce Arc chiefly through new transcription has been debated (Waltereit et al., 2001; Ichikawa et al., 2003; Giorgi et al., 2007). Our results show that these diverse stimuli induce the synthesis of nascent Arc mRNA. In silico analysis and DHAs of genomic DNA from PC12 cells and neurons revealed nine conserved nucleosome-depleted regions at the Arc locus. A reporter gene controlled by these nine promoter regions recapitulated patterns of endogenous Arc induction; two of the regions contained enhancer activity, and two contained repressor activity. The region located ∼6.5 kb upstream of Arc enhanced transcription induced by synaptic activity, BDNF, and forskolin. This region contains an SRE required to mediate responses to synaptic activity and BDNF, but not to forskolin, and it binds SRF in vivo. In turn, SRF is recruited by synaptic activity to directly regulate Arc transcription. The region located ∼1.4 kb upstream of Arc enhances synaptic activity- and BDNF- but not forskolin-induced reporter gene activity. The region contains two conserved Zeste-like elements that must be present to mediate such responses. Although there is no known mammalian ortholog of Zeste, subtle mutations that would be predicted to disrupt Zeste binding blocked the ability of the element to mediate stimulus-induced transcription in neurons and the binding of a putative novel neuronal nuclear protein in vitro.
The discovery that SRF/SRE is a critical regulator of Arc transcription suggests a signaling pathway that links behavior, synaptic activity, and gene expression. Mice lacking SRF show significant but selective defects in learning and memory (Etkin et al., 2006) and alterations in the expression of many genes (Ramanan et al., 2005). It is unknown which genes with altered expression in SRF−/− mice are direct SRF targets, and which of those are responsible for the memory defects. SREs have been found in several other genes that are induced by synaptic activity, including c-fos and zif268 (Rivera et al., 1990; DeFranco et al., 1993). However, Arc appears to be the first gene regulated directly by SRE/SRF that functions at synapses and is required for learning and memory. SRE-dependent transcription can be activated by stimuli such as BDNF and synaptic activity (Fig. 5E) through the Ras/ERK pathway. ERK1/2 activates Elk-1, a transcription cofactor that forms a ternary complex with a dimer of SRF (Buchwalter et al., 2004), which recruits and activates the basal transcriptional machinery. An Elk-1 binding site is located 3 bp downstream of SRE1, positioned to mediate SRE1-dependent Arc transcription. Such a mechanism is consistent with the observations that synaptic activity induces phosphorylation of ERK1/2 (Fig. 5F) and that inhibitors of ERK1/2 activation block stimulus-induced Arc expression (Rao et al., 2006). SRF activity can also be regulated independently of Ras by MAL, a member of the myocardin-related transcription factor family (Tabuchi et al., 2005). It is unknown whether MAL is important for Arc regulation. How region B of the Arc promoter mediates PKA-dependent transcription is also unclear. Mutations in SRE1 that disrupt SRF binding did not eliminate the response of region B to forskolin or to synaptic activity, suggesting that other response elements must exist within region B.
We also discovered novel Zeste-like elements within the Arc promoter that contribute almost as much as SRE1 to synaptic activity- and BDNF-induced Arc transcription. They are identical to the cognate binding sequence of the Drosophila transcription factor Zeste (Biggin et al., 1988), but no mammalian ortholog is known (Levine et al., 2002). Although several bioinformatic screens failed to find a mammalian candidate (data not shown), our data show that a nuclear factor from rat cortical neurons binds to this element. Thus, Arc transcription may be controlled partly by a novel Zeste-like mammalian neuronal transcription factor bound to this element. Such a factor may play a wide role in neuronal gene expression and function. Three Zeste-like elements reside within a known enhancer region of human BACE1 (Sambamurti et al., 2004), a molecule implicated in Alzheimer's disease (Rossner et al., 2006). BACE1 also regulates amyloid precursor protein cleavage under conditions associated with learning, memory, and synaptic plasticity (Ma et al., 2007). We also found Zeste-like elements in the promoter of Homer1a (data not shown), which plays a key regulatory role in synaptic plasticity (Van Keuren-Jensen and Cline, 2006). Arc and Homer1a are expressed in the same neuronal subpopulations (Vazdarjanova et al., 2002); they may be part of a program of activity-dependent gene expression that specifically elicits long-term adaptive responses through activity-dependent structural and functional synaptic modifications (Park et al., 2008; Ronesi and Huber, 2008; Waung et al., 2008). This scenario would imply that these genes (and perhaps others) are regulated at least in part by a specific shared set of transcription factors that may include a Zeste-like factor.
Identifying the first functional response elements required for rapid Arc transcription made it possible to elucidate the specific signaling pathways that impinge upon them to induce Arc expression in response to synaptic activity, BDNF, and forskolin (Fig. 7). This study revealed that synaptic activity must induce both the PKA and ERK pathways to regulate region B. PKA directly regulates some yet unidentified transcription factor (or factors) in region B. It remains unclear, however, whether ERK signaling regulates SRF-dependent transcription in parallel with or downstream of PKA signaling. Although the ERK activation inhibitor UO126 completely blocked the full reporter's response to synaptic activity (Fig. 4B), it was not possible to further assess how ERK directly regulates region B, as UO126 also blocked the basal activity of the SV40 promoter (data not shown). We also show that synaptic activity regulates Zeste-like factor-dependent transcription in regions F/G by recruiting both the PKA and ERK pathways. Curiously, the PKA inhibitor H89 blocked the enhancer activity of F/G (Fig. 6A), but forskolin alone did not induce a response from F/G (Fig. 6B) [in contrast to region B, where forskolin drove transcription (Fig. 5B)]. This could reflect a requirement for PKA activity in basal promoter activity or for cross- talk between ERK and PKA signaling for proper regulation of regions F/G (Impey et al., 1998). The insights gathered here will help answer important questions regarding how synaptic activity regulates specific sets of neuronal genes.
Figure 7.
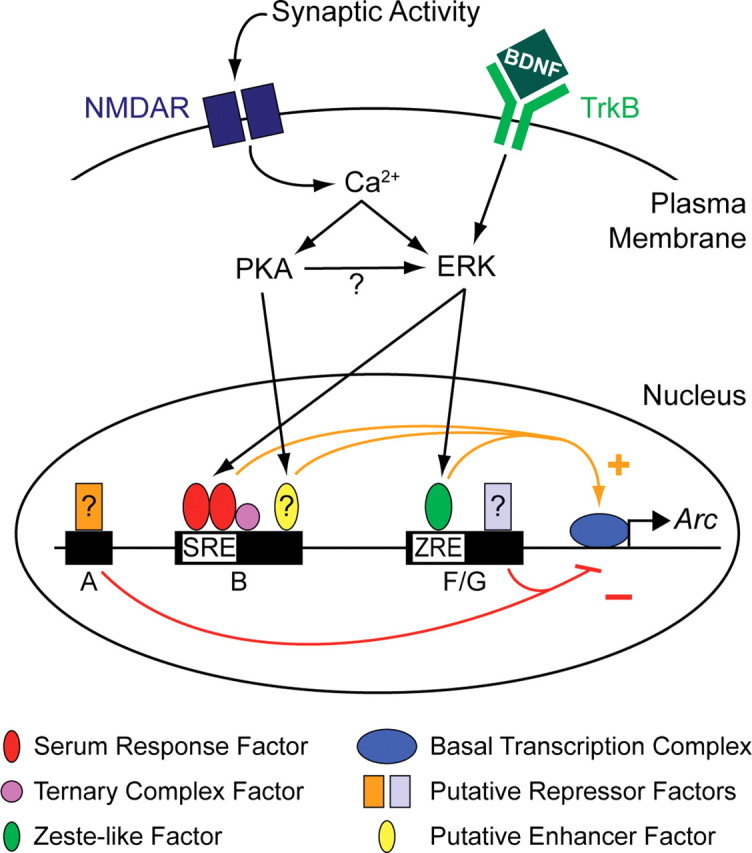
A model of Arc transcriptional regulation, showing how the activity-dependent PKA and ERK signaling pathways contribute to Arc transcription. Synaptic activity drives Arc transcription by inducing SRF/TCF activity in an NMDAR-, PKA-, and ERK-dependent manner (Figs. 4B, 5A, 5B, 5E–H). SRF binds to a conserved SRE in distal open region B (Figs. 3, 5D), and a conserved TCF binding site is found immediately downstream (Fig. 2). An unidentified enhancer factor likely also binds to region B to mediate the remaining response to synaptic activity and PKA (Figs. 5A, 5B). Synaptic activity also induces Arc transcription through a Zeste-like response element (ZRE) in proximal open regions F/G (Figs. 3, 6C) in an NMDAR-, PKA-, and ERK-dependent manner (Figs. 4B, 6A, 6B). Tyrosine receptor kinase B (TrkB) activation by BDNF induces Arc transcription via SRE and ZRE in an ERK-dependent manner (Figs. 4B, 5B, 6B). Some unidentified repressor factors likely bind to region A (Figs. 4C, 6B) and to regions F/G (Fig. 6B) to regulate stimulus-induced inhibition of reporter activity. The role of these putative repressors in Arc expression remains unclear.
The combined use of DHAs, in silico analyses, and reporter genes in primary neurons was critical to identify regulatory elements in Arc. Previous studies of Arc focused on proximal 5′ promoter regions because it is difficult to incorporate large genomic stretches into reporter genes. However, cis-acting regulatory regions can be located 5′ and 3′ to, as well as long distances from, a basal promoter (West and Fraser, 2005). Although this study appears to be the first to apply DHAs to neuronal IEG expression, they have been used for over 25 years to find candidate regulatory regions of genes within large genomic expanses (Pipkin and Lichtenheld, 2006). In addition, DHAs can be applied to native chromatin structure found in vivo in neurons, increasing the chances of finding physiologically relevant regulatory regions. Our DHAs and in silico analyses defined region B in the Arc promoter and led to the discovery of SRE1. SRE1 is located farther upstream than sequences examined in previous studies of Arc, and farther upstream than any element known to regulate the transcription of a plasticity-related IEG. Our DHAs and in silico analyses also identified regions F/G in the Arc promoter, which led to the discovery of a novel synaptic activity- and BDNF-responsive Zeste-like element. More widespread use of these techniques in neuroscience may help provide critical new insights into the broader mechanisms underlying neuronal gene expression.
Footnotes
S.A.P. is supported by a National Research Service Award from the National Institute of Neurological Disorders and Stroke. S.F. is supported by National Institutes of Health (NIH) Grants 2NS R01 039074 from the National Institute of Neurological Disorders and Stroke and 2P01 AG022074 from the National Institute on Aging, and The J. David Gladstone Institutes. The animal care facility was partly supported by a NIH Extramural Research Facilities Improvement Program Project. We are grateful to Amgen, Inc. (Thousand Oaks, CA) for providing recombinant human BDNF, Dr. N. Miki (Osaka University Medical School, Osaka, Japan) for providing the 2.6-kb Arc promoter reporter gene, Dr. A. Nordheim (University of Tübingen, Tübingen, Germany) for providing the SRF-VP16 constructs, and Dr. B. P. Herring (Indiana University School of Medicine, Indianapolis, IN) for providing the anti-SRF siRNA. We thank Dr. J. Kent (University of California, Santa Cruz, CA) and Dr. T. Brody (National Institute of Neurological Disorders and Stroke, NIH) for insightful discussions regarding in silico analyses. We are also grateful to Dr. T. Aragón, Dr. J. Santiago-García, Dr. A. O'Mahony, and M. Montano (University of California, San Francisco, CA) for their technical assistance. We thank G. Howard and S. Ordway for editorial assistance, K. Nelson for administrative assistance, and members of the Finkbeiner laboratory for helpful comments.
References
- Biggin MD, Bickel S, Benson M, Pirrotta V, Tjian R. Zeste encodes a sequence-specific transcription factor that activates the Ultrabithorax promoter in vitro. Cell. 1988;53:713–722. doi: 10.1016/0092-8674(88)90089-x. [DOI] [PubMed] [Google Scholar]
- Blaud M, Vossen C, Joseph G, Alazard R, Erard M, Nieto L. Characteristic patterns of N Oct-3 binding to a set of neuronal promoters. J Mol Biol. 2004;339:1049–1058. doi: 10.1016/j.jmb.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Bode J, Goetze S, Heng H, Krawetz SA, Benham C. From DNA structure to gene expression: mediators of nuclear compartmentalization and dynamics. Chromosome Res. 2003;11:435–445. doi: 10.1023/a:1024918525818. [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Chen JD, Pirrotta V. Multimerization of the Drosophila zeste protein is required for efficient DNA binding. EMBO J. 1993;12:2075–2083. doi: 10.1002/j.1460-2075.1993.tb05856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco C, Damon DH, Endoh M, Wagner JA. Nerve growth factor induces transcription of NGFIA through complex regulatory elements that are also sensitive to serum and phorbol 12-myristate 13-acetate. Mol Endocrinol. 1993;7:365–379. doi: 10.1210/mend.7.3.8483478. [DOI] [PubMed] [Google Scholar]
- Elgin SC. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988;263:19259–19262. [PubMed] [Google Scholar]
- Etkin A, Alarcón JM, Weisberg SP, Touzani K, Huang YY, Nordheim A, Kandel ER. A role in learning for SRF: deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron. 2006;50:127–143. doi: 10.1016/j.neuron.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Gärtner A, Collin L, Lalli G. Nucleofection of primary neurons. Methods Enzymol. 2006;406:374–388. doi: 10.1016/S0076-6879(06)06027-7. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Groth RD, Mermelstein PG. Brain-derived neurotrophic factor activation of NFAT (nuclear factor of activated T-cells)-dependent transcription: a role for the transcription factor NFATc4 in neurotrophin-mediated gene expression. J Neurosci. 2003;23:8125–8134. doi: 10.1523/JNEUROSCI.23-22-08125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci U S A. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H, Fujimoto T, Taira E, Miki N. The accumulation of Arc (an immediate early gene) mRNA by the inhibition of protein synthesis. J Pharmacol Sci. 2003;91:247–254. doi: 10.1254/jphs.91.247. [DOI] [PubMed] [Google Scholar]
- Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, Deloulme JC, Chan G, Storm DR. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kelly MP, Deadwyler SA. Acquisition of a novel behavior induces higher levels of Arc mRNA than does overtrained performance. Neuroscience. 2002;110:617–626. doi: 10.1016/s0306-4522(01)00605-4. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Bichell DP, Rotwein P. Chromatin changes accompany the developmental activation of insulin-like growth factor I gene transcription. J Biol Chem. 1992;267:21505–21511. [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Ma H, Lesné S, Kotilinek L, Steidl-Nichols JV, Sherman M, Younkin L, Younkin S, Forster C, Sergeant N, Delacourte A, Vassar R, Citron M, Kofuji P, Boland LM, Ashe KH. Involvement of beta-site APP cleaving enzyme 1 (BACE1) in amyloid precursor protein-mediated enhancement of memory and activity-dependent synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:8167–8172. doi: 10.1073/pnas.0609521104. Epub 2007 Apr 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansukhani A, Crickmore A, Sherwood PW, Goldberg ML. DNA-binding properties of the Drosophila melanogaster zeste gene product. Mol Cell Biol. 1988;8:615–623. doi: 10.1128/mcb.8.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Messaoudi E, Kanhema T, Soulé J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including Id2, Tbx5, and Nkx2–5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci U S A. 2005;102:14700–14705. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton GC, Schug J. TESS: Transcription element search software on the WWW. 1997. Technical report CBIL-TR (1997) 1001-v0.1 ( http://www.cbil.upenn.edu/tess) of the Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania, Philadelphia (1997)
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Lichtenheld MG. A reliable method to display authentic DNase I hypersensitive sites at long-ranges in single-copy genes from large genomes. Nucleic Acids Res. 2006;34:e34. doi: 10.1093/nar/gkl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bösl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schütz G, Linden DJ, Ginty DD. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- Rao VR, Pintchovski SA, Chin J, Peebles CL, Mitra S, Finkbeiner S. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat Neurosci. 2006;9:887–895. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- Riegle KC, Meyer RL. Rapid homeostatic plasticity in the intact adult visual system. J Neurosci. 2007;27:10556–10567. doi: 10.1523/JNEUROSCI.1631-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Sheng M, Greenberg ME. The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum stimulation. Genes Dev. 1990;4:255–268. doi: 10.1101/gad.4.2.255. [DOI] [PubMed] [Google Scholar]
- Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–547. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression—implications for Alzheimer's disease. Prog Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Sambamurti K, Kinsey R, Maloney B, Ge YW, Lahiri DK. Gene structure and organization of the human beta-secretase (BACE) promoter. FASEB J. 2004;18:1034–1036. doi: 10.1096/fj.03-1378fje. [DOI] [PubMed] [Google Scholar]
- Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 Mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Estevez M, Henderson JA, Marx R, Shiota J, Nakano H, Baraban JM. Nuclear translocation of the SRF co-activator MAL in cortical neurons: role of RhoA signalling. J Neurochem. 2005;94:169–180. doi: 10.1111/j.1471-4159.2005.03179.x. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Van Keuren-Jensen K, Cline HT. Visual experience regulates metabotropic glutamate receptor-mediated plasticity of AMPA receptor synaptic transmission by homer1a induction. J Neurosci. 2006;26:7575–7580. doi: 10.1523/JNEUROSCI.5083-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci. 2002;22:10067–10071. doi: 10.1523/JNEUROSCI.22-23-10067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J Virol. 1991;65:6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J Neurosci. 2001;21:5484–5493. doi: 10.1523/JNEUROSCI.21-15-05484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- Wibrand K, Messaoudi E, Håvik B, Steenslid V, Løvlie R, Steen VM, Bramham CR. Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur J Neurosci. 2006;23:1501–1511. doi: 10.1111/j.1460-9568.2006.04687.x. [DOI] [PubMed] [Google Scholar]
- Yin F, Hoggatt AM, Zhou J, Herring BP. 130-kDa smooth muscle myosin light chain kinase is transcribed from a CArG-dependent, internal promoter within the mouse mylk gene. Am J Physiol Cell Physiol. 2006;290:C1599–C1609. doi: 10.1152/ajpcell.00289.2005. [DOI] [PubMed] [Google Scholar]
- Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neurosci. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]