Controlling the spread of this gene will be difficult.
Keywords: Acinetobacter baumannii, oxacillinase, carbapenems, Tn2006, ISAba1, bacteria, zoonoses, infectious diseases, vector-borne infections, research
Abstract
To assess dissemination of OXA-23–producing strains of Acinetobacter baumannii, we obtained 20 carbapenem-resistant, OXA-23–producing isolates from different regions. Their clonal relationship was assessed by pulsed-field gel electrophoresis and multilocus sequence typing. We identified 8 sequence types, including 4 novel types. All except 2 strains belonged to 2 main European clonal lineages. The blaOXA-23 gene was either located on the chromosome or on plasmids and associated with 4 genetic structures.
Acinetobacter baumannii is a gram-negative organism that is increasingly recognized as a major pathogen causing nosocomial infections, including bacteremia and ventilator-associated pneumonia, particularly in patients admitted to intensive care units (1). Several studies have shown the geographically widespread occurrence of multidrug-resistant A. baumannii strains, which suggested a clonal relatedness of these strains. Three international A. baumannii clones associated with multidrug resistance (European clones I, II, and III) have been reported (2).
Increasing resistance to carbapenems has been observed worldwide in the past decade, frequently mediated by production of class D β-lactamases with carbapenemase activity. Three acquired class D β-lactamases with carbapenemase gene clusters have been described in A. baumannii, which correspond to blaOXA-23-like, blaOXA-40-like, and blaOXA-58-like genes (3). The blaOXA-23 gene, first characterized in Scotland (4), has been increasingly reported worldwide. A. radioresistens was recently identified as the progenitor of the blaOXA-23-like genes (5). Clonal outbreaks of carbapenem-resistant and OXA-23–producing A. baumannii have been reported in many countries, such as Bulgaria (6), People’s Republic of China (7), Brazil (8), Iraq (9), Afghanistan (9), and French Polynesia (10).
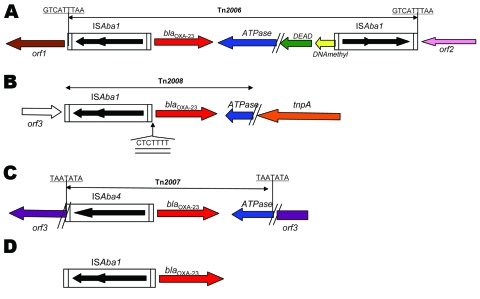
Genetic acquisition of the blaOXA-23 gene was investigated and transposons Tn2006, Tn2007, and Tn2008 were identified as genetic structures harboring this gene (10–12). In Tn2006, the blaOXA-23 gene is flanked by 2 copies of the insertion sequence ISAba1, which are located in opposite orientations (Figure 1). The functionality of Tn2006 has been recently demonstrated (13). Tn2008 is similar to Tn2006 but lacks the second copy of ISAba1 and the blaOXA-23 gene is associated with 1 copy of ISAba4 (which differs from ISAba1) in Tn2007 (Figure 1) (11). As reported for strains from United Arab Emirates and Bahrain, the blaOXA-23 gene can be associated with only 1 copy of ISAba1 (14,15). We studied the clonal relationship and genomic environment of sequences surrounding the blaOXA-23 gene among a collection of OXA-23–producing isolates from 15 countries.
Figure 1.
Genetic structures associated with the blaOXA-23 gene of Acinetobacter baumannii. A) Tn2006 from isolates 240, 512, 810, 859, 883 and AUS (ST22/ST2). B) Tn2008 from isolate 614. C) Tn2007 from isolates Ab14, BEL, and DOS. D) ISAba1 from isolates AS3, 1190, 861, and 877. Boundaries of Tn2006, Tn2007, and Tn2008 are indicated with the target site duplication likely generated by transposition events underlined. The 7-bp difference in the site of insertion of ISAba1 for isolate 614 is double-underlined. The open reading frame 1 (orf1), orf2, and orf3 genes of unknown function is indicated. tnpA, gene encoding a putative transposase; ATPase, gene encoding the putative AAA ATPase; DEAD, gene encoding the putative DEAD (Asp-Glu-Ala-Asp) helicase; DNAmethyl, DNA methylase.
Materials and Methods
Bacterial Strains and Susceptibility Testing
Twenty OXA-23–producing A. baumannii clinical isolates were obtained from 15 countries. These isolates had been obtained from patients hospitalized in intensive care units from December 2003 through March 2008. Isolates were obtained from tracheal aspirates (n = 3), bile (n = 1), urine (n = 4), wounds (n = 1), respiratory tract (n = 1), blood (n = 4), and sputum (n = 1). The isolates were initially chosen after preliminary pulsed-field gel electrophoresis (PFGE)–based typing had identified 13 pulsotypes. Isolates were obtained from France (n = 4), Vietnam (n = 1), New Caledonia (n = 1), Thailand (n = 1), Australia (n = 1), Tahiti (n = 1), Reunion (n = 2), South Africa (n = 1), United Arab Emirates (n = 2), Libya, (n = 1), Bahrain (n = 1), Egypt (n = 1), Belgium (n = 1), Algeria (n = 1), and Brazil (n = 1).
Presence of the blaOXA-23 gene was screened by PCR by using specific primers (OXA-23-A 5′-GGAATTCCATGAATAAATATTTTACTTGC-3′ and OXA-23-B 5′-CGGGATCCCGTTAAATAATATTCAGGTC-3′) and additional sequencing (ABI 3100 sequencer; Applied Biosystems, Foster City, CA, USA). Susceptibility patterns to β-lactam antimicrobial drugs were determined by using a standard disk diffusion method according to published standards (16) and Etest strips (AB Biodisk, Solna, Sweden). Isolates were identified by using 16S rRNA gene sequencing (17).
Clonal Relationships
Isolates were typed by using ApaI macrorestriction analysis and PFGE according to the manufacturer’s recommendations (Bio-Rad, Marnes-la-Coquette, France). Bacteria were grown in a medium appropriate for the strain until an optical density of 0.8 to 1 at 600 nm was reached. One milliliter of cells was centrifuged, washed, and resuspended in 10 mmol/L Tris, pH 7.2, 20 mmol/L NaCl, 50 mmol/L EDTA. Immediately after resuspension, an equal volume of 2% low melting point InCert agarose (Bio-Rad) was added. Solid agarose plugs were lysed at 37°C for 2 h in 1 mL of lysis buffer (10 mmol/L Tris, pH 7.2, 50 mmol/L NaCl, 0.5% sodium laurylsarcosine, 0.2% sodium deoxycholate) supplemented with 20 mg/L of lysozyme. The plugs were then incubated at 55°C for 16 h with proteinase K buffer (100 mmol/L EDTA, pH 8, 0.2% sodium deoxycholate, 1% sodium laurylsarcosine) supplemented with 20 mg/L of proteinase K. Plugs were washed with Tris-EDTA buffer containing 1 mmol/L phenylmethylsulfonyl fluoride (Sigma, St. Louis, MO, USA) and 3× with Tris-EDTA buffer at room temperature.
Whole-cell DNA of A. baumannii isolates was digested with ApaI overnight at room temperature (New England Biolabs, St. Quentin-en-Yvelines, France). Electrophoresis was performed on a 1% agarose gel with 0.5× Tris-borate-EDTA buffer by using a CHEF DRII apparatus (Bio-Rad). Samples were subjected to electrophoresis at 14°C, 6 volts/cm, and a switch angle with 1 linear switch ramp of 3–8 s for 10.5 h, and then for 12–20 s for 10.5 h.
Identification of PCR-based sequence groups was conducted by using 2 multiplex PCR assays designed to selectively amplify group 1 or group 2 alleles of the gene encoding outer-membrane protein A (ompA), the gene encoding part of a pilus assembly system required for biofilm formation (csuE), and the gene encoding the intrinsic carbapenemase gene of A. baumannii) (blaOXA-51) (18). Clonal relationships were established by multilocus sequence typing (MLST) by using 7 standard housekeeping loci (citrate synthase [gltA], gyrase B [gyrB], glucose dehydrogenase B [gdhB], recombination A [recA], chaperone 60 [cpn60], glucose-6-phosphate isomerase [gpi], and RNA polymerase [rpoD]) as described (18). Sequencing of internal fragments was performed by using BigDye fluorescent terminators and primers described (19). Sequences were compared with the A. baumannii database at the MLST Website (http://mlst.zoo.ox.ac.uk). To supplement epidemiologic results, we performed a second MLST typing using the scheme developed by Nemec et al. (20). Sequences of the 7 housekeeping genes were analyzed by using an A. baumannii database (www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html).
Southern Blot Analysis and Location of blaOXA-23 Gene
Southern blot analysis was performed by using total genomic DNA digested with EcoRI, separated by electrophoresis on 0.8% agarose gels, transferred onto Hybond N+ membranes, and hybridized with enhanced chemiluminescence labeled probes overnight at 42°C. The membranes were developed according to the manufacturer’s instructions (GE Healthcare, Saclay, France). Chromosomal or plasmid locations of the β-lactamase gene were assessed by hybridization of I-CeuI–digested genomic DNA with blaOXA-23 and 16S rDNA probes and electrophoresis (20–120 s for 9 h and 60–100 s for 11 h at 14°C and 5 V/cm2) (21). DNA was transferred from an agarose gel to a nylon membrane by capillary transfer. Hybridization, labeling, and detection were conducted as described above. Mating-out assays were performed by using isolates that had plasmid-borne blaOXA-23 as donors and rifampin-resistant A. baumannii BM4547 as recipients as described (22). Transconjugants were selected on trypticase soy agar plates containing ticarcillin (50 mg/L) and rifampin (50 mg/L).
Cloning Experiments
To identify entire transposon structures containing the blaOXA-23 gene in different isolates and determine their location in the target DNA, a cloning procedure was used. Some data had been reported for 6 of 20 isolates (11). Total DNA was digested with either SacI or SalI, ligated into the SacI or SalI sites of plasmid pBK-CMV (kanamycin-resistant cloning vector), and the recombinant plasmids were transformed into Escherichia coli TOP10, as described (14). Recombinant plasmids were selected on trypticase soy agar plates containing amoxicillin (50 mg/L) and kanamycin (30 mg/L). Cloned DNA fragments of several recombinants plasmids were sequenced on both strands by primer walking as described (11).
Results
Clonal Relatedness of the Isolates
Twenty carbapenem-resistant A. baumannii isolates were obtained from 15 countries (Table). All isolates were highly resistant to ticarcillin (MIC >256 mg/L) and showed a high level of resistance to ceftazidime (MIC >256 mg/L), except isolates Ab14 (MIC 4 mg/L) 861 and DOS (MIC 8 mg/L). All isolates were resistant to imipenem and meropenem (MIC >16 mg/L) (Table).
Table. Characteristics of 20 blaOXA-23-positive Acinetobacter baumannii clinical isolates*.
| Isolate | Origin | Date of isolation | Specimen | EC | ST† | Copy no. of blaOXA-23 | Genetic location and size, kb | Genetic structure | MIC, μg/mL |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | IPM | MEM | |||||||||
| 240 | France | 2003 Dec | Tracheal aspirate | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 128 | >32 | >32 |
| 512 | Tahiti | 2004 Mar | Tracheal aspirate | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 64 | >32 | >32 |
| 761 | Vietnam | 2005 May | Bile | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 64 | >32 | >32 |
| 810 | New Caledonia | 2004 Jun | Blood | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 96 | >32 | >32 |
| 863 | Thailand | 2006 Jun | Urine | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 256 | >32 | >32 |
| 883 | Reunion | 2006 Jun | Unknown | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 128 | >32 | >32 |
| Ab13 | France | 2004 Jun | Urine | II | 22/2 | 2 | Chromosome, ≈200,‡ and plasmid, 70 | Tn2006 | 128 | >32 | >32 |
| AUS | Australia | 2004 Oct | Urine | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 96 | >32 | >32 |
| 859 | South Africa | 2006 Jan | Urine | II | 22/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 128 | >32 | >32 |
| 585 | France | 2004 Jul | Tracheal aspirate | II | 53/2 | 1 | Chromosome, ≈200‡ | Tn2006 | 128 | >32 | >32 |
| 614 | Libya | 2004 Oct | Unknown | I | 25/20 | 1 | Plasmid, 130 | Tn2008 | 256 | >32 | 16 |
| AS3 | UAE† | 2006 Oct | Blood | I | 25/20 | 1 | Plasmid, 130 | ISAba1 | 256 | >32 | >32 |
| 1190 | Bahrain | 2008 Mar | Blood | I | 25/20 | 1 | Plasmid, 130 | ISAba1 | 256 | >32 | >32 |
| AS1 | UAE | 2006 Jul | Blood | I | 44/1 | 1 | Chromosome, ≈40‡ | Tn2006 | 256 | >32 | >32 |
| Ab14 | Algeria | 2004 Dec | Unknown | I | 44/1 | 2 | Plasmid, 25, and plasmid, >150 | Tn2007 | 4 | 16 | >32 |
| 910 | Reunion | 2006 Oct | Unknown | I | New1/1 | 1 | Plasmid, 130 | Tn2006 | 256 | 16 | 16 |
| 861 | Egypt | 2005 Nov | Sputum | I | New1/ 1 | 1 | Plasmid, 130 | ISAba1 | 8 | 32 | 32 |
| BEL | Belgium | 2007 Jul | Respiratory tract | I | New2/ 1 | 2 | Plasmid, 25, and plasmid, >150 | Tn2007 | 256 | >32 | >32 |
| DOS | France | 2004 May | Unknown | – | New3/ New | 2 | Plasmid, 25, and plasmid, >150 | Tn2007 | 8 | >32 | >32 |
| 877 | Brazil | 2006 Jul | Wound | – | New4/15 | 1 | Plasmid, 130 | ISAba1 | 96 | >32 | >32 |
*EC, European clone; ST, sequence type; UAE, United Arab Emirates; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem. The MIC for ticarcillin was >256 μg/mL for all 20 isolates. †ST determined by Bartual et al. (19) compared with ST determined by Nemec et al. (20). ‡Size of chromosome band carrying the blaOXA-23 gene, as determined by using the I-CeuI technique.
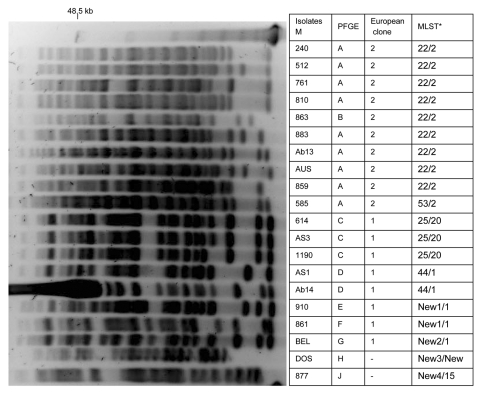
Multiplex PCR for identification of sequence groups showed 10 isolates that belonged to group 1 according to Turton et al. (18), eight that belonged to group 2, and 2 isolates that did not belong to groups 1 or 2. The 10 isolates that belonged to group 1 and corresponded to European clone II (18) were classified into 2 sequence types (STs), ST22 and ST53, according to MLST analysis (18). ST22 (1–3-3–2-2–7-3) was the most frequent type identified. Nine isolates were identified: 2 from France and 1 each from Vietnam, New Caledonia, Thailand, Australia, Tahiti, Reunion, and South Africa. A single European clone II isolate was classified as ST53 (1–3-3–2-2,3-3), a single-locus variant of ST22. Among 10 other isolates, 8 belonged to group 2 (corresponding to European clone I). Four STs were identified: ST25 (10–12–4–11–1–9–5) (Libya, United Arab Emirates, and Bahrain), ST44 (10–12–4–11–4–9–5) (United Arab Emirates and Algeria), and 2 new STs, 1 for isolates from Reunion and Egypt (10–12–4–11–4–16–5) and another related ST identified in the single isolate from Belgium (10–12–4–11–4,4–5). These 4 STs differ by 1 locus. The 2 most recent isolates from France and Brazil did not belong to European clones I or II and corresponded to 2 STs (1–22–3-11–1-9–7 and 12–18–12–1-15–9-19, respectively) (Table). Although 8 STs were identified in this collection, 9 pulsotypes were characterized by PFGE according to the criteria of Tenover et al. (23) (Figure 2).
Figure 2.
Pulsed-field electrophoresis (PFGE) profiles of ApaI-digested genomic DNA from strains of Acinetobacter baumannii. PFGE types, European clone types, and multilocus sequence typing (MLST) results are shown. *ST, sequence type determined by Bartual et al. (19) compared with ST determined by Nemec et al. (20). Lane M, molecular size markers (48.5 kb).
According to MLST analysis developed by Nemec et al. (20), all isolates that belonged to European clone II had the same sequence type (ST2) (2,2-2,2-2,2-2), including isolate 585, which had a distinct but related ST in the first analysis. Among isolates that belonged to European clone I, two sequence types were determined: ST20 (3–1-1,1-5–1-1) (Libya, United Arab Emirates, Bahrain) and ST1 (1,1-1,1-5–1-1) (United Arab Emirates, Reunion, Egypt, Belgium, Algeria). Isolates 910 (Reunion), 861 (Egypt), and BEL (Belgium) were included in ST1. These isolates had a distinct ST according to methods of Bartual et al. (19). The 2 most recent isolates were classified into 2 STs, a new ST (3–2-2,2-5–4-8) for isolate DOS (France) and ST15 (6,6-8–2-3–5-4) for isolate 877 (Brazil) (Table).
Location and Transferability of the blaOXA-23 Gene
Location of the blaOXA-23 gene was evaluated by using the I-CeuI method. Eleven isolates had the blaOXA-23 gene on the chromosome, with a hybridization signal for an ≈40-kb band for isolate AS1 and an ≈200-kb band for 10 isolates (Table). Nine isolates carried the blaOXA-23 gene on a plasmid and 1 isolate had 2 copies of the blaOXA-23 gene, 1 on the chromosome and 1 on a 7–kb plasmid (Table).
To examine the copy number of the blaOXA-23 gene in different A. baumannii genomes, we performed Southern blot hybridization on EcoRI-digested DNA fragments using a 589-bp DNA probe specific for the blaOXA-23 gene. Sixteen isolates showed only 1 copy of the blaOXA-23 gene. Isolates BEL, Ab14, and DOS had 2 copies of the blaOXA-23 gene on different plasmids, and Ab13 had 1 copy on the chromosome and 1 copy on a plasmid according to results of the I-Ceu1 technique.
Mating-out assays were performed by using the 10 plasmid-positive strains as donor strains and rifampin-resistant A. baumannii BM4547 as the recipient strain. Five transconjugants were obtained; all had a 130-kb plasmid that did not provide additional antimicrobial drug resistance to the A. baumannii recipient strain, except in 1 case (co-resistance to kanamycin and amikacin on a blaOXA-23–carrying plasmid that originated from isolate 1190). Plasmids carrying the blaOXA-23 gene in isolates Ab14, DOS, BEL, and 877 were not self-transferable (Table) (24).
Variability of Genetic Structures Flanking the blaOXA-23 Gene
The 10 isolates that belonged to European clone II had a blaOXA-23 gene that was part of Tn2006. The 9-bp direct repeat (DR) that corresponded to duplication of the Tn2006 target site, which was consistent with a transposition event, was identified in the 9 ST22/ST2 isolates. Tn2006 was inserted in different locations on the chromosomes of those isolates (Table). For isolates 240, 512, 810, 859, 883, and Aus, the insertion occurred between 2 genes encoding hypothetical proteins (DR: GTCATTTAA) (Figure 1). In isolate 761, transposon Tn2006 was located between a gene encoding a hypothetical protein and a gene encoding an isoleucyl tRNA synthase (DR: ATTCGCGGG). In isolate 863, Tn2006 was identified between a gene encoding a cytochrome D terminale oxidase and a putative transposase (DR: ATAATTATT). In isolate 585, Tn2006 was located between a gene encoding a hypothetical protein and a sul1 gene (DR: ATTCGCGGG). The plasmid-borne blaOXA-23 gene identified in isolate Ab13 was also part of Tn2006 but was inserted into the sul gene that encoded a putative sulfonamide resistance determinant (DR: ATTCGCGGG).
Isolates that belonged to European clone I had diverse genetic structures at the origin of blaOXA-23 acquisition. Two isolates had transposon Tn2006: one on the chromosome (AS1) and 1 on a plasmid (910). Transposon Tn2007 was identified in 3 isolates; it was specific for the same open reading frame in 2 isolates (BEL and Ab14) (Figure 2). Only 1 copy of ISAba1 was identified upstream of the blaOXA-23 gene in isolates AS3, 1190, 861, and 877. Transposon Tn2008 was identified only in isolate 614 (Figure 1). Sequences of these specific genetic structures have been deposited in Genbank (accession nos. EF127491, EF059914, GQ861438, and GQ861439).
Discussion
This study was conducted to define which features may explain the worldwide dissemination of the blaOXA-23 gene in A. baumannii. Isolates were from the Middle East, Europe, and Asia; there were no isolates from North America. Except for 2 isolates, the isolates investigated in this study belonged to European clones I or II. Clustering of A. baumannii isolates was determined by MLST and PFGE; our collection was composed of 13 PFGE types corresponding to 9 STs. Eight STs were identified among the OXA-23–producing A. baumannii; the most common STs were ST22/ST2 found in France (n = 2), Vietnam, New Caledonia, Thailand, Australia, Reunion, South Africa, and Tahiti. Spread of blaOXA-23–positive A. baumannii isolates that belong to clone ST22 has been demonstrated in South Korea (25). Analysis of the target site of blaOXA-23 acquisition showed that in the same clone, such as ST22, acquisition of the Tn2006 composite transposon had occurred at different positions in the A. baumannii genome, which suggested that Tn2006-mediated acquisition of blaOXA-23 may occur as independent events, or that Tn2006 is a structure that is mobile in a given genome. A single clone could have different genetic structures at the origin of the blaOXA-23 acquisition.
We showed that the blaOXA-23 gene associated with Tn2006 could be located on the chromosome or a plasmid. This result agrees with our recent findings, which showed that Tn2006 is capable of transposition (13). We have also observed that 5 isolates with different sequence types (STNew1, ST25) harbored a similar 130-kb plasmid. The same strains with the same genetic structure were identified in 8 countries in different parts of the world.
In conclusion, the current worldwide dissemination of the blaOXA-23 gene is driven by >7 MLST types associated with different genetic structures and plasmids. We have identified complex and dynamic spreading of blaOXA-23 that will be difficult to control because this spread is not associated with a single entity.
Acknowledgment
We thank Rémy Bonnin for technical assistance.
This study was supported by a grant from the Ministère de la Recherche, Université Paris XI, Paris, France; grants from the European Community (DRESP2, LSHM-CT-2005-01705, and TROCAR HEALTH-F3-2008-223031); and the Institut National de la Santé et de la Recherche Médicale, France.
Biography
Ms Mugnier is a doctoral student at Hôpital de Bicêtre, Institut National et de la Santé et de la Recherche Médicale, Unité, South-Paris Medical School, University Paris-XI in Le Kremlin-Bicêtre, France. Her research interests are genetic plasticity and drug-resistance mechanisms in A. baumannii.
Footnotes
Suggested citation for this article: Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis [serial on the Internet]. 2010 Jan [date cited]. Available from http://www.cdc.gov/EID/content/16/1/35.htm
References
- 1.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84. 10.1128/AAC.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanism and epidemiology. Clin Microbiol Infect. 2006;12:826–36. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- 4.Scaife W, Young HK, Paton RH, Amyes GB. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36:585–7. 10.1093/jac/36.3.585 [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob Agents Chemother. 2008;52:1252–6. 10.1128/AAC.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoeva T, Higgins PG, Bojkova K, Seifert H. Clonal spread of carbapenem-resistant OXA-23 positive Acinetobacter baumannii in a Bulgarian university hospital. Clin Microbiol Infect. 2008;14:723–7. 10.1111/j.1469-0691.2008.02018.x [DOI] [PubMed] [Google Scholar]
- 7.Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, Li LJ, et al. Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1 blaOXA-23 gene in a Chinese hospital. J Med Microbiol. 2007;56:1076–80. 10.1099/jmm.0.47206-0 [DOI] [PubMed] [Google Scholar]
- 8.Carvalho KR, Carvalho-Assef AP, Peirano G, Santos LC, Pereira MJ, Asensi MD. Dissemination of multidrug-resistant Acinetobacter baumannii genotypes carrying blaOXA-23 collected from hospitals in Rio de Janeiro, Brazil. Int J Antimicrob Agents. 2009;34:25–8. 10.1016/j.ijantimicag.2008.12.009 [DOI] [PubMed] [Google Scholar]
- 9.Calhoun JH, Murray CK, Manring MM. Multidrug-resistant organisms in military wounds from Iraq an Afghanistan. Clin Orthop Relat Res. 2008;466:1356–62. 10.1007/s11999-008-0212-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naas T, Levy M, Hirschauer C, Marchandin H, Nordmann P. Outbreak of carbapenem OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J Clin Microbiol. 2005;43:4826–9. 10.1128/JCM.43.9.4826-4829.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corvec S, Poirel L, Naas T, Drugeon H, Nordmann P. Genetics and expression of the carbapenems-hydrolysing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:1530–3. 10.1128/AAC.01132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams-Haduch JM, Paterson DL, Sidjabat HE, Pasculle AW, Potoski BA, Muto CA, et al. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob Agents Chemother. 2008;52:3837–43. 10.1128/AAC.00570-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mugnier PD, Poirel L, Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol. 2009;191:2414–8. 10.1128/JB.01258-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mugnier P, Poirel L, Pitout M, Nordmann P. Carbapenem-resistant and OXA-23 producing Acinetobacter baumannii isolates in the United Arab Emirates. Clin Microbiol Infect. 2008;14:879–82. 10.1111/j.1469-0691.2008.02056.x [DOI] [PubMed] [Google Scholar]
- 15.Mugnier PD, Bindayna KM, Poirel L, Nordmann P. Diversity of plasmid-mediated carbapenem-hydrolysing oxacillinases among carbapenem-resistant Acinetobacter baumannii isolates from Kingdom of Bahrain. J Antimicrob Chemother. 2009;63:1071–3. 10.1093/jac/dkp052 [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. M100–S18. Wayne (PA): The Institute; 2008.
- 17.Dortet L, Legrand P, Soussy CJ, Cattoir V. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J Clin Microbiol. 2006;44:4471–8. 10.1128/JCM.01535-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13:807–25. 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 19.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382–90. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemec A, Krizova L, Maixnerova M, Diancourt L, Van der Reijden TJK, Brisse S, et al. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug resistant strains of European clone II. J Antimicrob Chemother. 2008;62:484–9. 10.1093/jac/dkn205 [DOI] [PubMed] [Google Scholar]
- 21.Liu SL, Hessel A, Sanderson KE. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A. 1993;90:6874–8. 10.1073/pnas.90.14.6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Guibert M, Bellais S, Naas T, Nordmann P. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin–clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype typhimurium DT104 from French patients. Antimicrob Agents Chemother. 1999;43:1098–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogaerts P, Cuzon G, Naas T, Bauraing C, Deplano A, Lissoir B, et al. Carbapenem-resistant Acinetobacter baumannii isolates expressing the blaOXA-23 gene associated with ISAba4 in Belgium. Antimicrob Agents Chemother. 2008;52:4205–6. 10.1128/AAC.01121-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park YK, Choi JY, Jung SI, Park KH, Lee H, Jung DS, et al. Two distinct clones of carbapenem-resistant Acinetobacter baumannii isolates from Korean hospitals. Diagn Microbiol Infect Dis. 2009;64:389–95. 10.1016/j.diagmicrobio.2009.03.029 [DOI] [PubMed] [Google Scholar]