Abstract
Exposure to cigarette smoke (CS) is the primary factor associated with the development of chronic obstructive pulmonary disease (COPD). CS increases the level of oxidants in the lungs, resulting in a depletion of antioxidants, which promotes oxidative stress and the destruction of alveolar tissue. In response to CS, pulmonary epithelial cells counteract increased levels of oxidants by activating Nrf2-dependent pathways to augment the expression of detoxification and antioxidant enzymes, thereby protecting the lung from injury. We hypothesize that increasing the pathways activated by Nrf2 will afford protection against CS-induced lung damage. To this end we have developed a novel mouse model in which the cytosolic inhibitor of Nrf2, Keap1, is genetically deleted in Clara cells, which predominate in the upper airways in mice. Deletion of Keap1 in Clara cells resulted in increased expression of Nrf2-dependent genes, such as Nqo1 and Gclm, as determined by microarray analysis and quantitative PCR. Deletion of Keap1 in airway epithelium decreased Keap1 protein levels and significantly increased the total level of glutathione in the lungs. Increased Nrf2 activation protected Clara cells against oxidative stress ex vivo and attenuated oxidative stress and CS-induced inflammation in vivo. Expression of KEAP1 was also decreased in human epithelial cells through siRNA transfection, which increased the expression of Nrf2-dependent genes and attenuated oxidative stress. In conclusion, activating Nrf2 pathways in tissue-specific Keap1 knockout mice represents an important genetic approach against oxidant-induced lung damage.
Keywords: cigarette smoke, Nrf2, Keap1, inflammation, oxidative stress
CLINICAL RELEVANCE.
The current study uses a novel conditional knockout mouse model in which the cytosolic inhibitor of Nrf2, Keap1, is genetically deleted in airway epithelial cells. The authors hypothesize that sustained activation of Nrf2 may provide new therapeutic strategies for lung diseases such as emphysema in which oxidative stress and inflammation are implicated.
Chronic obstructive pulmonary disease (COPD) is currently the fifth leading cause of death worldwide and affects more than 210 million people (1, 2). This debilitating disease is characterized by irreversible airflow limitation and abnormal inflammation in the lung that is attributed to pulmonary emphysema and chronic bronchitis. The primary risk factor for developing COPD is exposure to cigarette smoke (CS) (2).
CS contains greater than 1014 free radicals per puff that include reactive oxygen species (ROS) and reactive nitrogen species (RNS) (3). Inhaled oxidants from CS generate cellular damage by directly targeting proteins, lipids, and nucleic acids and deplete the level of antioxidants in the lung, thereby overwhelming the oxidant/antioxidant balance of the lung, leading to increased oxidative stress (4). Indeed, oxidative stress in the lungs has been strongly implicated in COPD severity (5, 6). Oxidative stress due to CS also drives the inflammation of macrophages and neutrophils to the lung and induces apoptosis of epithelial and endothelial cells (7). These factors all contribute to the destruction of alveolar structure leading to airspace enlargement, loss of elastic recoil, and ultimately reduced airflow.
Nuclear erythroid 2 p45-related factor–2 (Nrf2) is a basic leucine zipper transcription factor that regulates the expression of multiple genes involved in antioxidant and detoxification pathways (8). Under unstressed conditions, Nrf2 is maintained at low basal levels by binding to its cytosolic inhibitor Kelch-like ECH associating protein 1 (Keap1) (9). Keap1 is associated with the actin cytoskeleton, where it targets Nrf2 for ubiquitination through the Cul3/Roc-1 ubiquitin ligase complex, leading to the constitutive proteasomal degradation of Nrf2 (10–12). However, upon exposure to oxidative stress Nrf2 is released from Keap1 and translocates to the nucleus (13–15). In the nucleus, Nrf2 forms heterodimers with small Maf proteins and binds to the antioxidant response element (ARE) to promote the expression of detoxification enzymes such as NADPH quinone oxidoreductase-1 (Nqo1) and enzymes that are important in glutathione (GSH) biosynthesis such as glutamate cysteine ligase modulatory subunit (Gclm) (16, 17).
Chronic exposure to CS in Nrf2-knockout mice increases oxidative stress and inflammation in the lung, which results in earlier onset and more severe emphysema (18, 19). In addition, patients with COPD have a reduction in Nrf2 pathways that is associated with increased oxidative stress and alveolar destruction in the lungs (20–22). Interestingly, the lungs of patients with COPD have decreased expression of Nrf2 target genes, which correlate with increased lung damage and COPD severity (21). The responsiveness of the Nrf2 pathway may act as a major determinant of susceptibility to cigarette smoke–induced emphysema by up-regulating antioxidant defenses and decreasing lung inflammation. In fact, enhancing Nrf2-dependent antioxidant and cytoprotective pathways through a small molecular approach decreases oxidative stress in the lungs of mice and protects against CS-induced pulmonary emphysema (23). We hypothesize that sustained activation of Nrf2 may provide new therapeutic strategies for lung diseases such as emphysema in which oxidative stress and inflammation are implicated.
Genetic ablation of Keap1 causes the constitutive activation of Nrf2, leading to an increase in antioxidant and detoxification pathways. However, Keap1 knockout mice die postnatally due to malnutrition as a result of abnormal squamous metaplasia and progressive keratinization of the esophagus and forestomach (24). Lethality due to the loss of Keap1 can be overcome by generating tissue-specific knockout mice (25). For example, hepatocyte-specific disruption of Keap1 leads to increased expression of Nrf2-regulated genes in the liver and increases resistance to acetaminophen-induced toxicity (26). Keap1 tissue-specific knockouts represent an important genetic model in which to test whether increasing Nrf2 pathways in selected organs can protect against injury due to oxidative stress.
Clara cells are nonciliated secretory epithelial cells that line the primary bronchioles of the lungs localizing predominantly to the upper airways (27). Clara cells maintain lung homeostasis by secreting a variety of factors, including Clara cell secretory protein (CCSP) and a component of the lung surfactant (28, 29), and are enriched in detoxifying enzymes, such as cytochrome P450 (30). Since oxidative stress plays an important role in CS-induced emphysema, we used Clara cell–specific Keap1 knockout mice (Keap1Δ2–3/Δ2–3;CctCre+) to determine whether increasing the activity of Nrf2 pathways are protective against acute CS-induced inflammation and oxidative stress.
MATERIALS AND METHODS
Lineage-Specific Deletion of Keap1 Gene in Lungs
A targeting vector was constructed by inserting LoxP sites flanking exons 2 and 3 of the Keap1 gene and a neomycin cassette flanked by FRT sites for the selection of positive clones. Exons 2 and 3 of the Keap1 gene were selected for targeting because these exons are small in size, separated by a small intron and the intronic sequences flanking exon 2 and 3 are devoid of repetitive sequences. The Keap1 transcript lacking exons 2 and 3 will code for a truncated nonfunctional Keap1 protein. The mutant Keap1 protein contains the N-terminal BTB domain essential for Keap1 dimerization, but lacks the redox-sensitive IVR domain and Nrf2-binding Kelch domains. The vector was linearized by Not1 and transfected by electroporation into C57BL/6J embryonic stem cells. After selection with neomycin, surviving clones were expanded for PCR analysis to identify ES clones with homologous recombination at Keap1 locus. Two recombinant clones were identified and used for injection into recipient female mice (see Figure E1A in the online supplement). Chimeras with the targeted allele were backcrossed with C57BL/6 mice to generate Keap1flox/flox;Neo+ mice (Figure E1B). Keap1flox/flox;Neo+ mice were subsequently crossed with mice carrying the FLP recombinase (FLP+ mice on the C57BL/6 background), which targets excision of the FRT sequence flanking the neomycin cassette to generate Keap1flox/wt;FLP+ mice. The presence of a neomycin resistance cassette in an intron can result in an alteration of gene function and produce unwanted or lethal phenotypes (31); therefore, the FLP transgene was segregated by backcrossing with C57/BL6J mice to generate the Keap1flox/wt;FLP- mice. Confirmation of the floxed allele without the FLP transgene was identified through PCR analysis (Figure E1C). Keap1flox/wt;FLP- mice were further inbred to generate Keap1flox/flox mice, which are aphenotypic. Lung epithelium–specific Keap1-deficient (Keap1Δ2–3/Δ2–3;CctCre+) mice were generated by breeding Keap1flox/flox mice with CCtCre+ mice, which express Cre only in Clara cells (32). The CCtCre+ transgenic mice were bred into Keap1flox/flox mice to generate Keap1Δ2–3/wt mice;CctCre+ mice and backcrossed to generate Keap1Δ2–3/Δ2–3;CctCre+ mice. Initial experiments revealed no reduction in survival or fertility of conditionally targeted mice.
CS Exposure
Eight- to 10-week-old mice were housed under controlled conditions for temperature and humidity, using a 12-hour light/dark cycle. At 8 weeks of age, mice were exposed to CS for 5 hours using a TE-10 smoking machine (Teague Enterprises, Davis, CA) and 3R4F reference cigarettes (University of Kentucky, Tobacco Research Institute, Lexington, KY). Chamber atmosphere was monitored for total suspended particles and carbon monoxide with an average concentration of 150 mg/m3 and 875 ppm, respectively. All experimental protocols were performed in accordance with the standards established by the U.S. Animal Welfare Acts, as set forth in NIH guidelines and in the Policy and Procedures Manual of the Johns Hopkins University Animal Care and Use Committee.
DNA Isolation
Tissue frozen in liquid nitrogen was crushed to fine pieces with a mortar and pestle and incubated in DNA digestion buffer (50 mM Tris-HCL, pH 8.0, 100 mM EDTA, pH 8.0, 100 mM NaCl, 1% SDS, and 0.5 mg/ml proteinase K) overnight at 50°C. DNA was extracted through a phenol/chloroform/isoamyl alcohol extraction and precipitated with sodium acetate and isopropanol. The pellet was washed with 70% ethanol, dried, and resuspended in 10 mM Tris.
PCR Conditions
A 2,954-bp fragment was amplified from DNA derived from Keap1flox/flox mice and a 288-bp fragment from Keap1Δ2–3/Δ2–3;CctCre+ mice from different organs to identify the deletion of Keap1 in genomic DNA. The oligonucleotides used were Keap14F (5′-GAGTCCACAGTGTGTGGCC-3′) and NeoI3R (5′-GAGTCACCGTAAGCCTGGTC-3′). The PCR conditions were as follows: 5 minutes at 95°C followed by 35 cycles of 95°C for 60 seconds, 70°C for 30 seconds, and 72°C for 5 minutes 30 seconds, followed by 10 minutes of extension at 72°C.
RNA Isolation
RNA from tissue was isolated from the upper left lobe of the lung to enrich the proportion of Clara cells in the preparation in both control air–exposed mice and mice immediately after CS exposure using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's recommended protocol. RNA from cell culture experiments was isolated using the RNeasy Mini Kit according to the manufacturer's recommended protocol (Qiagen, Inc., Valencia, CA).
Gene Expression
Total RNA was isolated from the upper left lobe of the lung immediately after CS exposure, using Trizol reagent (Invitrogen), and cDNA was generated using Multiscribe reverse transcriptase (Applied Biosystems, Foster City, CA). Gene expression was measured using assays on demand probe sets (Applied Biosystems), and reactions were analyzed using the ABI 7000 Taqman system. Actin was used for normalization. The cycle threshold (CT) value indicates the number of PCR cycles that are necessary for the detection of a fluorescence signal exceeding a fixed threshold. The fold change (FC) was calculated using the following formulas: ΔCT = CT (target gene) – CT (Actin) and FC = 2 −(ΔCT2−ΔCT1), in which ΔCT1 represents the average ΔCT value among control samples and ΔCT2 represents the value of a particular sample. Control samples for the in vivo studies included air exposed Keap1flox/flox mice. Control samples for the in vitro studies included untreated control BEAS-2B cells transfected with SS siRNA. Results are expressed as mean ± SEM.
Transcriptional Profiling Using Microarray Analysis
Quantitation of RNA was performed using a NanoDrop spectrophotometer. Quality assessment was determined by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100. Processing of templates for GeneChip Analysis was in accordance with methods described in the Affymetrix GeneChip Expression Analysis Technical Manual, Revision five. Double-stranded cDNA was synthesized from 5 μg total RNA using the GeneChip Expression 3′ amplification reagents one-cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA) and subsequently column-purified using the GeneChip Sample Cleanup Module. Biotinylated cRNA was synthesized from the double-stranded cDNA by in vitro transcription (IVT) using the GeneChip Expression 3′ amplification reagents for IVT labeling (Affymetrix) according to the manufacturer's recommended protocol. Resultant cRNAs were purified by column purification with the GeneChip Sample Cleanup Module (Affymetrix) and quantified. Fifteen micrograms of cRNA were fragmented by metal-induced hydrolysis in fragmentation buffer (250 mM Tris acetate, pH 8.1, 150 mM MgOAc, 500 mM KOAc) at 94°C for 35 minutes. Quality of pre- and post-fragmentation cRNAs was assessed by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100. Hybridization cocktails were prepared as recommended for arrays of “Standard” format, including incubation at 94°C for 5 minutes and 45°C for 5 minutes, and centrifugation at maximum speed for 5 minutes before pipetting into the GeneChips (Affymetrix Mouse Genome 430 2.0). Hybridization was performed at 45°C for 16 hours at 60 rpm in the Affymetrix rotisserie hybridization oven. The signal amplification protocol for washing and staining of eukaryotic targets was performed in an automated fluidics station (Affymetrix FS450) using protocol FS450_0001. The arrays were scanned in a GeneChip 3000 7G laser scanner with autoloader (Affymetrix) at an emission wavelength of 570 nm and 2.5 μm resolution. Intensity of hybridization for each probe pair was computed by Command Console software.
Data Analysis
Raw data CEL files were imported into Genomic Suite Software (Partek, St. Louis, MO). Gene expression data was Robust Multichip Analysis (RMA) background corrected, adjusted for GC content, quantile normalized, and the median polish was used for probe summarization. Principal component analysis on overall gene expression was performed on the analyzed data and two samples were corrected for batch effect. Differentially expressed genes were detected between Keap1Δ2–3/Δ2–3;CctCre+ mice and Keap1flox/flox mice. Genes differentially expressed were defined as significant with P ≤ 0.05, showed a change in at least six out of nine comparisons, and a fold change greater than 1.5-fold. Differentially expressed genes were then subjected to pathway analysis using Ingenuity pathway analysis tool using default settings. (Ingenuity Systems, Redwood City, CA).
Immunoblotting
Tissue was lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton, 1% Sodium deoxycholate, 0.1% SDS, complete protease-inhibitor cocktail; Sigma-Aldrich, St. Louis, MO). Lysates were homogenized and the total protein content was determined by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). Equal amounts of total protein (50 μg) were resolved by SDS-PAGE and were transferred to nitrocellulose membranes (Millipore, Marlborough, MA). Nonspecific binding sites were blocked by incubation for 1 hour at room temperature with PBS containing 0.1% Tween-20 (PBST) and 5% nonfat dry milk. Membranes were exposed to either a goat anti-Keap1 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit anti-Keap1 polyclonal antibody (Santa Cruz) in PBST-0.5% milk overnight at 4°C followed by incubation with horseradish peroxidase–conjugated secondary antibodies (GE Healthcare UK Ltd, Buckinghamshire, UK). Immunoreactivity was visualized by chemiluminescence substrate according to the manufacturer's instructions (Amersham Biosciences, Piscataway, NJ). Denistometric analysis was performed with ImageJ software (National Institutes of Health, Bethesda, MD) by quantifying the intensity of the Keap1 and GAPDH proteins band and dividing the intensity of Keap1 by GAPDH and then multiplying by 100.
Nrf2 Immunohistochemistry
Formalin-fixed tissues were treated with an anti-Nrf2 antibody (H-300; Santa Cruz) at a dilution of 1:250 for 1 hour and developed using horseradish peroxidase (Dako, Glostrup, Denmark) as previously described (33).
Lung Morphometry
Lungs of mice were inflated with 0.6% agarose at a constant pressure of 25 cm H2O, as described (34). Lungs were fixed for 24 hours in 10% buffered formalin and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin. Slides were coded so as to minimize any bias involved in image acquisition. Ten to fifteen representative images were captured at ×100 magnification for each sample. MLI and S/V ratio were determined by using a macro designed with MetaMorph software (Molecular Devices, Sunnyvale, CA).
Clara Cell Isolation
Clara cells were purified as previously described (32). Briefly, mice were anesthetized with an overdose of ketamine and xylazine (n = 5 per group). The chest cavity was opened, exposing the lungs, thymus, and heart. The heart, thymus, and rib cage were resected, and the lungs were removed from the cavity. A majority of the lung parenchyma was removed, leaving major bronchi of the lungs, which was digested with 10 ml of a 0.15% pronase (Sigma-Aldrich) solution in F-12 media at 4°C for 18 hours. To remove the noncellular portion and to obtain a single-cell suspension, the protease solution was filtered through a 70-μm filter and the filtrate was centrifuged at 2,000 rpm for 10 minutes. The cellular pellet was resuspended in a 0.5 mg/ml DNase solution (Sigma-Aldrich) for 5 minutes on ice. The cellular filtrate was centrifuged at 2,000 rpm for 10 minutes and the pellet was resuspended in basic MTEC media with 10% FBS with antibiotics. Cells obtained from five mice of each genotype were pooled. Cells were plated for 3 hours in tissue culture plates to remove macrophages and fibroblasts. Based on staining with nitro blue tetrazolium (35), the purity of Clara cells was greater than 30% in the nonadherent cell population. Nonadherent cells were collected and stained with 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA; Invitrogen Corporation, Carlsbad, CA) for 1 hour at 37°C. Cells were washed once in phosphate-buffered saline, treated with or without 100 μM hydrogen peroxide for 2 hours, and submitted for flow cytometric analysis.
In Vitro Studies
Human lung epithelial cells, BEAS-2B cells obtained from the American Type Culture Collection (Manassas, VA) were cultured under recommended conditions. BEAS-2B cells were stained with DCFDA as described above. The plate was incubated in a 37°C and 5% CO2 incubator prior to reading on a fluorescent plate reader. Cells without dye were used to subtract background fluorescence. Readings for fluorescence intensity were measured using a SpectraMax fluorescent plate reader set at 485-nm excitation and 530-nm emissions (Molecular Devices, Sunnyvale, CA).
Transfection of siRNA Duplexes
KEAP1 siRNA corresponds to the coding region nucleotides 1545–1563 (5′-GGGCGTGGCTGTCCTCAAT-3′) in KEAP1 transcript variant 2. The KEAP1 siRNA duplex with the following sense and antisense sequences was used: 5′-GGGCGUGGCUGUCCUCAAUdUdU-3′ (sense) and 3′-dUdUCCCGCACCGACAGGAGUUA-5′ (antisense). To confirm the specificity of the inhibition, the siCONTROL non-targeting siRNA 1 (SS siRNA; 5′-UAGCGACUAAACACAUCAAUU-3′) with microarray-defined signature was used as a negative control. The siRNA duplexes were synthesized by Dharmacon Research (Lafayette, CO) (33). BEAS-2B cells in the exponential growth phase were plated at a density of 0.2 × 106 cells/ml, grown for 12 hours, and transfected with 50 nM siRNA duplexes using Lipofectamine 2000 and OPTI-MEM reduced serum medium (Invitrogen) according to the manufacturer's recommendations.
Flow Cytometry Analysis
A FACS Calibur flow cytometer (Becton Dickinson, Bedford, MA) equipped with a 488-nm argon laser was used for the flow cytometric analysis. Forward and side scatters were used to establish size gates and exclude cellular debris from the analysis. The excitation wavelength was set at 488 nm. In each measurement, a minimum of 5,000 cells was analyzed. Data were acquired and analyzed using the Cell Quest software (Becton Dickinson). Results are expressed as mean ± SEM.
Glutathione Quantification
For determination of glutathione concentration, lungs were harvested from mice immediately after a 5-hour CS exposure. Glutathione levels in lungs were quantified as previously described (36). Briefly, lungs were lysed in buffer containing 0.25 M sucrose, 10 mM Tris-HCL, and 1 mM EDTA. Protein was precipitated by adding sulfosalicylic acid to a final concentration of 6.5%, followed by incubation on ice for 10 minutes and centrifugation at 2,000 × g for 15 minutes. Lung supernatants were transferred to a 96-well plate for measurement of glutathione levels. Total intracellular glutathione was measured using the glutathione reductase-DTNB recycling assay, comparing the rate of color formation at 412 nm of the unknowns to a standard curve. Total intracellular glutathione levels were determined by quantifying the intracellular glutathione levels then dividing by the protein concentration. The data are expressed as nmoles of glutathione per milligram (mg) protein.
BAL and Phenotyping
Mice were anesthetized with an overdose of ketamine and xylazine. The lungs were aspirated twice with 1 ml of sterile PBS to collect BAL fluid. Cells were counted by using a hemocytometer, and differential cell counts were performed on 200 cells from BAL fluid with Wright-Giemsa stain (Baxter, McGaw Park, IL).
Protein Determination
Protein concentrations were determined with the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions.
Statistical Analysis
Data are given as mean ± SEM. Analyses were done using the software package GraphPad Prism 3.03 (GraphPad, San Diego, CA). One-way ANOVA was used to compare groups with one independent variable. A Dunnett's posttest was used to compare different treatments. Data comparing two group means were analyzed by independent samples t test. Significance was noted at P < 0.05. Outliers were detected through Grubb's test from GraphPad Software.
RESULTS
Generation of Airway Epithelium–Specific Keap1 Conditional Knockout Mice
Mice homozygous for Keap1 with a pair of loxP sites flanking exons 2 and 3 of the Keap1 gene (floxed allele) were crossed with CctCre+ mice, which express Cre recombinase under the direction of the CC10 promoter (32) to generate Keap1Δ2–3/Δ2–3;CctCre+ mice (Figure 1A; Figure E1B). Deleting exons 2 and 3 of the Keap1 gene resulted in the loss of a majority of the IVR domain and four out of six of the Kelch domains in the truncated Keap1 protein. The predicted size of the truncated Keap1 protein was 25 kD. Mice with Keap1 deleted specifically in airway epithelial cells were initially genotyped using isolated DNA obtained from tail biopsies (Figure E1C).
Figure 1.
Schematic of the gene targeting vector, breeding strategy, and analysis of genomic and messenger RNA of Keap1Δ2–3/Δ2–3;CctCre+ mice. (A) The breeding strategy to generate Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice is illustrated. Genomic organization of the wild-type Keap1 gene, the Keap1-targeting vector, the floxed Keap1 allele, and the truncated knockout Keap1 allele after Cre-mediated recombination are shown. Coding exons of the mouse Keap1 gene are shown with open boxes, the FRT sites are indicated as gray arrowheads, and the loxP sites are indicated as black arrowheads. The red arrows indicate the forward and reverse primers used for PCR analysis of genomic DNA with the predicted bp amplicon size of all PCR products. (B) PCR analysis showing the genomic deletion of exons 2 and 3 of the Keap1 gene in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice. Genomic DNA was purified from the lung and liver of Keap1wt/wt, Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ mice. PCR amplification using primers Keap1–4F and NeoI-3R generated either a 2,954-bp amplicon from the floxed Keap1 allele or a 288-bp amplicon from the Keap1 knockout allele. The deleted Keap1 gene was only apparent in the lung DNA preparation. (C) Representative light microscopy picture of Clara cells isolated from Keap1Δ2–3/Δ2–3;CctCre+ mice ex vivo. Cells were stained with nitro blue tetrazolium as previously described (35). Clara cells are indicated by arrows. (D) PCR analysis showing the genomic deletion Keap1 in airway epithelial cells from Keap1Δ2–3/Δ2–3;CctCre+ mice. Genomic DNA was purified and PCR amplification using primers Keap14F and NeoI3R generated either a 2,954-bp amplicon from the floxed Keap1 allele or a 288-bp amplicon from the Keap1 knockout allele. (E) Messenger RNA levels of Nqo1 in airway epithelium isolated from Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice (n = 8). (F) Messenger RNA levels of Keap1 in airway epithelium isolated from Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice (n = 8). Clara cells were isolated as described in Materials and Methods from 8-week-old mice. The fold change was calculated using the formula described in Materials and Methods. Data represented are mean fold change ± SEM. Asterisks indicate a significant difference compared with Keap1flox/flox mice (P < 0.05).
Genotypes and successful tissue-specific deletion was confirmed by analyzing DNA isolated from the lung. DNA from the liver and other organs were used as negative controls. PCR analysis revealed that exons 2 and 3 of Keap1 gene were selectively deleted in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice resulting in the amplification of a 288-bp product along with the larger 2,954-bp product from Keap1 floxed gene found in other cell types (Figure 1B). Since Clara cells are present only in more distal bronchioles of the murine airway and represent only 10% of the total lung volume (37, 38), the presence of the larger 2,954-bp product is most likely due to other cell types. Cre-mediated deletion in Keap1Δ2–3/Δ2–3;CctCre+ mice occurred only in the lungs and not in other organs such as the liver. No deletion of Keap1 was observed in the lungs of either Keap1wt/wt or Keap1flox/flox mice (Figure 1B). Keap1Δ2–3/Δ2–3;CctCre+ mice were born at Mendelian frequencies and exhibited no developmental abnormalities or morbidity.
To confirm that Keap1 was deleted in Clara cells of Keap1Δ2–3/Δ2–3;CctCre+ mice, airway epithelial cells were isolated ex vivo as previously described (32). This preparation enables the isolation of airway epithelial cells and based on staining with nitro blue tetrazolium (35), the percentage of Clara cells was greater than 30% in all preparations (Figure 1C). PCR analysis of genomic DNA isolated from airway epithelial cells of both Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice revealed that Keap1 was deleted only in airway epithelial cells from Keap1Δ2–3/Δ2–3;CctCre+ mice and not in Keap1flox/flox mice (Figure 1D). The expression levels of Nqo1 mRNA were significantly up-regulated in Keap1Δ2–3/Δ2–3;CctCre+ mice by 19.1-fold (P < 0.05) (Figure 1E). Expression levels of the Keap1 transcript were significantly decreased in Keap1Δ2–3/Δ2–3;CctCre+ mice to approximately 60% of control levels (P < 0.05) (Figure 1F). In addition, airway epithelium from Keap1Δ2–3/Δ2–3;CctCre+ mice had increased protein levels of Nrf2 compared with Keap1flox/flox mice as determined by immunohistochemistry (Figure E2). These results confirm the successful creation of lung specific Keap1 conditional knockout mice.
To characterize the effect of deleting Keap1 in airway epithelium in lung tissue, mRNA expression of Nrf2 target genes, Nqo1 and Gclm, as well as Keap1, was quantified through real-time PCR from lungs of Keap1wt/wt mice, Keap1flox/flox mice, and Keap1Δ2–3/Δ2–3;CctCre+ mice. Expression levels of Nqo1 and Gclm mRNA were significantly up-regulated in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice compared with Keap1flox/flox mice, 5.5- and 1.8-fold increase, respectively (P < 0.05) (Figure 2A). In addition, expression levels of the Keap1 transcript were significantly decreased in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice compared with Keap1flox/flox mice to approximately 55% of control levels (P < 0.05) (Figure 2B). No difference in Nrf2 expression was observed between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice (data not shown). No differences in Keap1 or Nrf2 target gene expression were observed between Keap1flox/flox mice and Keap1wt/wt mice indicating that the insertion of the floxed allele had no effect on gene expression (data not shown). In addition, genetic deletion of Keap1 in airway epithelial cells decreased the protein level of the full-length Keap1 protein in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice compared with Keap1flox/flox mice to approximately 50% of control levels as determined through immunoblotting (Figures 2C and 2D). No truncated Keap1 protein could be detected.
Figure 2.
Increased mRNA expression of Nrf2-dependent genes and decreased Keap1 transcript and protein levels in the lungs from Keap1Δ2–3/Δ2–3;CctCre+ mice. (A) Messenger RNA levels of Nqo1 and Gclm in the lungs of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice. Lungs were isolated from 8-week-old mice. The fold change was calculated using the formula described in Materials and Methods (n = 3). (B) Messenger RNA levels of Keap1 in the lungs of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice. Data represented are mean fold change ± SEM (n = 3). Asterisks indicate a significant difference compared with Keap1flox/flox mice (P < 0.05). (C) Lung lysates from Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ mice, and cell lysates from human cancer cells (positive control). Tissue and cell lysates were prepared and analyzed through immunoblotting using an anti-Keap1 goat polyclonal that detects the full-length protein. The blot was stripped and probed with an anti-GAPDH antibody to ensure equal protein loading between samples (n = 1). (D) Densitometric analysis of Keap1 protein expression normalized to GAPDH protein expression for experiment shown in C. Protein band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Keap1 Deletion in the Lung Induces Greater Expression of Nrf2 Target Antioxidant Genes
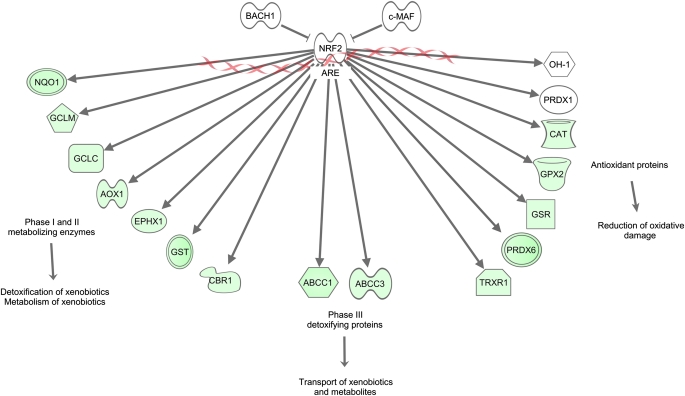
To examine the response of the Nrf2–ARE pathways in response to Keap1 deletion in the lung, we comprehensively investigated the gene expression profile of Keap1Δ2–3/Δ2–3;CctCre+ mice by a microarray approach. Microarray analysis was performed using total RNA from the lungs of Keap1Δ2–3/Δ2–3;CctCre+ and Keap1flox/flox mice. Expression levels of more than 50 genes were increased more than 2-fold in the Keap1Δ2–3/Δ2–3;CctCre+ lungs compared with those in the Keap1flox/flox lungs. Table 1 summarizes the antioxidant and detoxification enzymes significantly increased in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice. Many of these antioxidant and detoxification enzymes are well-characterized Nrf2–ARE target genes, such as Nqo1, Gsr1, Srxn1, and Gpx2 (39–42). In addition, carbonyl reductase 1 (Cbr1) and carbonyl reductase 3 (Cbr3) were up-regulated in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice. Cbr1 and Cbr3 are important detoxification enzymes that catalyze the reduction of xenobiotic compounds, including smoke-derived carcinogens, in a NADPH-dependent manner. However, not all Nrf2-dependent genes were increased in expression in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice. For example, the expression of heme oxygenase-1 (OH-1) and peroxiredoxin 1 (Prdx1) was not increased in Keap1 conditional knockout mice (Figure 3). A limited number of antioxidant and detoxification genes were significantly down-regulated in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice (Table 2). Many of these genes were associated with innate immune function such as lactotransferrin (Ltf), cytochrome b-245 (Cybb), and granzyme A (Gzma). None of the down-regulated genes are known to be involved in Nrf2-dependent cytoprotection and detoxification pathways.
TABLE 1.
GENES EXPRESSED CONSTITUTIVELY HIGHER IN THE LUNGS OF Keap1Δ2−3/Δ2−3;CctCre+ COMPARED TO Keap1flox/flox MICE
| Functional Classification and Entrez Gene Identification no. | Gene (Gene Symbol) | Fold Change |
|---|---|---|
| Antioxidant-associated enzymes | ||
| 233016 | Biliverdin reductase B (Blvrb) | 4.1 |
| 76650 | Sulfiredoxin 1 (Sxrn1)* | 3.6 |
| 14776 | Glutathione peroxidase 2 (Gpx2)* | 3.5 |
| 14629 | Glutamate-cysteine ligase, catalytic subunit (Gclc)* | 2.8 |
| 11758 | Peroxiredoxin 6 (Prdx6)* | 2.7 |
| 50493 | Thioredoxin reductase 1 (Txnrd1)* | 2.4 |
| 14630 | Glutamate-cysteine ligase, modifier subunit (Gclm)* | 2.1 |
| 14854 | Glutathione synthetase (Gss) | 2.0 |
| 14782 | Glutathione reductase 1 (Gsr1)* | 1.8 |
| 14380 | Glucose-6-phosphate dehydrogenase 2 | 1.7 |
| Phase II detoxification enzymes | ||
| 14857 | Glutathione S-transferase, α1 (Gsta1) | 9.8 |
| 14858 | Glutathione S-transferase, α2 (Gsta2)* | 7.1 |
| 18104 | NADPH dehydrogenase, quinone 1 (Nqo1)* | 6.4 |
| 14862 | Glutathione S-transferase, μ1 (Gstm1) | 4.3 |
| 109857 | Carbonyl reductase 3 (Cbr3) | 3.2 |
| 14864 | Glutathione S-transferase, μ3 (Gstm3)* | 3.0 |
| 14859 | Glutathione S-transferase, α3 (Gsta3) | 2.4 |
| 12408 | Carbonyl reductase 1 (Cbr1) | 2.1 |
| 12359 | Catalase (Cat) | 1.8 |
Genes that have already been reported to have AREs and to be regulated by Nrf2.
Figure 3.
Schematic representing the significant changes in gene expression in the lung of Keap1Δ2–3/Δ2–3;CctCre+ mice compared with Keap1flox/flox mice. Gene interactions were identified and described by Ingenuity Pathway Analysis (Ingenuity Systems). Genes indicated in green were significantly increased in expression in Keap1Δ2–3/Δ2–3;CctCre+ mice (fold change > 1.5). Genes indicated in red were significantly decreased in expression (fold change < 1.5). Genes indicated in white were not different between groups (n = 3). The X symbols represent genomic DNA. Abbreviations are as follows. ABCC1: ATP-binding cassette, sub-family C (CFTR/MRP) member 1; AOX1: aldehyde oxidase 1; ARE: antioxidant response element; BACH1: BTB and CNC homology 1; CAT: catalase; CBR1: carbonyl reductase 1; EPHX1: epoxide hydrolase 1; GCLC: glutamate-cysteine ligase catalytic subunit; GCLM: glutamate-cysteine ligase modifier subunit; GPX2: glutathione peroxidase 2; GSR: glutathione reductase; GST: glutathione S-transferase; OH-1: heme oxygenase-1; NQO1: NAD(P)H dehydrogenase, quinone 1; NRF2: nuclear erythroid 2 p45 related factor-2; PRDX6: peroxiredoxin 6; TXNRD1: thioredoxin reductase 1.
TABLE 2.
GENES EXPRESSED CONSTITUTIVELY LOWER IN THE LUNGS OF Keap1Δ2−3/Δ2−3;CctCre+ COMPARED TO Keap1flox/flox MICE
| Functional Classification and Entrez Gene Identification no. | Gene (Gene Symbol) | Fold Change |
|---|---|---|
| Cytoprotective-associated proteins | ||
| 29818 | Heat shock protein family, member 7 (Hspb7) | −1.9 |
| 22289 | Ubiquitously transcribed tetratricopeptide repeat gene, X chromosome (Utx) | −1.8 |
| 21452 | Transcobalamin 2 (Tcn2) | −1.6 |
| 68460 | Dehydrogenase/reductase (SDR) family member 7C (Dhrs7c) | −1.6 |
| 53945 | Solute carrier family 40 (Slc40a1) | −1.6 |
| Detoxification-associated proteins | ||
| 17002 | Lactotransferrin (Ltf) | −2.3 |
| 13058 | Cytochrome b-245, beta polypeptide (Cybb) | −1.9 |
| 11865 | Aryl hydrocarbon receptor nuclear translocator-like (Arntl) | −1.7 |
| 14938 | Granzyme A (Gzma) | −1.6 |
| 17132 | Avian musculoaponeurotic fibrosarcoma (V-maf) | −1.6 |
No Structural Changes Observable in the Lungs of Keap1Δ2–3/Δ2–3;CctCre+ Mice
Gross anatomy and histologic organization of the Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ lungs were normal and similar to Keap1wt/wt C57BL/6J mice. No pathologies or tumors were observed at least 6 months after birth. Morphologic measurements were conducted on the lungs of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice to determine whether any significant structural changes occurred as a result of the deletion of Keap1 during lung development. Airspace quantification was assessed by measuring the mean linear intercept (MLI), which is an indicator of alveolar size, and by measuring the surface to volume ratio, which is a determinant of alveolar lung destruction, using computer-assisted stereologic measurements. The MLI of normal air-exposed Keap1flox/flox mice was 50.1 ± 0.45 μm. The MLI of Keap1Δ2–3/Δ2–3;CctCre+ mice was 50.7 ± 1.02. Surface-to-volume ratios were also similar between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice (Table 3). There were no significant differences in MLI or surface-to-volume ratios between the groups. Therefore, deletion of Keap1 in airway epithelium does not alter the lung structure of mice less than 3 months of age.
TABLE 3.
COMPARISON OF LUNG STRUCTURE BETWEEN Keap1flox/flox AND Keap1Δ2-3/Δ2−3;CctCre+ MICE USING MORPHOMETRY
| Mice | Mean linear intercept, μm | Surface to volume ratio (x1,000) |
|---|---|---|
| Keap1flox/flox | 50.1 ± 0.45 | 35.1 ± 0.48 |
| Keap1Δ2−3/Δ2−3;CctCre+ | 50.7 ± 1.02 | 35.5 ± 1.89 |
N = 6–7 per group.
Clara Cells Isolated from Keap1 Conditional Knockout Mice Have a Significantly Lower Level of ROS after Exposure to Hydrogen Peroxide
To determine whether epithelial cells from Keap1Δ2–3/Δ2–3;CctCre+ mice are resistant to oxidative stress as a result of Keap1 deletion in murine airway epithelium, Clara cells were isolated from Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice and exposed to hydrogen peroxide. The endogenous levels of ROS were quantified through DCFDA staining followed by flow cytometry analysis. No difference in fluorescence (intracellular ROS) was observed in Clara cells between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice under normal conditions (Figures 4A and 4C, respectively). However, after exposure to 100 μM hydrogen peroxide the levels of ROS were significantly decreased in Keap1Δ2–3/Δ2–3;CctCre+ mice compared with Keap1flox/flox mice (Figures 4B and 4D, respectively), indicating that epithelial cells in Keap1Δ2–3/Δ2–3;CctCre+ mice are more resistant to oxidative stress compared with Keap1flox/flox mice (Figure 4E). These data indicate that deletion of Keap1 in Clara cells decreases ROS levels and protects against the redox imbalance due to oxidative stress in airway epithelial cells.
Figure 4.
Clara cells from Keap1Δ2–3/Δ2–3;CctCre+ mice have reduced ROS levels as a result of oxidative stress due to hydrogen peroxide. Clara cells were isolated from Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ mice as described in Materials and Methods. Clara cells were stained with DCFDA and treated with 100 μM hydrogen peroxide for 2 hours. Clara cells were analyzed through flow cytometry. (A) Representative histogram of untreated Clara cells isolated from Keap1flox/flox mice. (B) Representative histogram of Clara cells treated with hydrogen peroxide isolated from Keap1flox/flox mice. (C) Representative histogram of untreated Clara cells isolated from Keap1Δ2–3/Δ2–3;CctCre+ mice. (D) Representative histogram of Clara cells treated with hydrogen peroxide isolated from Keap1Δ2–3/Δ2–3;CctCre+ mice. (E) Quantification of Mean DCFDA fluorescence from flow cytometry. Data represented are mean values ± SEM (n = 3–4). Asterisk indicates a significant difference between Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ Clara cells treated with H2O2 (P < 0.05).
Constitutive Nrf2 Activity in Airway Epithelial Cells Leads to an Attenuation of CS-Induced Oxidative Stress and Inflammation in the Lung
To determine whether increased expression of Nrf2 pathway genes is protective against CS-induced oxidative stress and inflammation, Keap1Δ2–3/Δ2–3;CctCre+ and Keap1flox/flox mice were exposed to 5 hours of CS, and markers of oxidative stress and inflammation were quantified. No differences were observed in the mRNA expression levels of Nrf2 target genes through real-time PCR immediately after CS exposure between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice (Figures 5A and 5B). However, expression of Nrf2 target genes in the lungs remained up-regulated in Keap1Δ2–3/Δ2–3;CctCre+ mice the day after CS exposure (Figures 5A and 5B).
Figure 5.
Keap1Δ2–3/Δ2–3;CctCre+ mice have increased levels of GSH in the lung but no significant differences in Nrf2-dependent gene expression after acute exposure to CS. (A) Messenger RNA levels of Nqo1 in the lungs of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice. Lungs were isolated from 8-week-old mice immediately after CS exposure (CS) or the next day (Post-CS). The fold change was calculated using the formula described in Materials and Methods (n = 3). (B) Messenger RNA levels of Gclm in the lungs of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice. Lungs were isolated from 8-week-old mice immediately after CS exposure (CS) or the next day (Post-CS). The fold change was calculated using the formula described in Materials and Methods (n = 3). (C) Concentration of total glutathione in lungs of Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ mice before and immediately after exposure to CS. Data represented are mean values ± SEM (n = 3). Asterisks indicate a significant difference compared with air-exposed Keap1flox/flox mice (P < 0.05). Dagger indicates a significant difference compared with CS-exposed Keap1flox/flox mice (P < 0.05). NS indicates no significant difference compared with CS-exposed Keap1flox/flox mice.
The levels of the major antioxidant tripeptide, glutathione, were quantified in the lungs of mice immediately after CS. Keap1Δ2–3/Δ2–3;CctCre+ mice had significantly higher glutathione levels compared with Keap1flox/flox mice under normal conditions (P < 0.05) (Figure 5C). After CS exposure, both Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice had a significant decrease in GSH levels of 61% and 67%, respectively. The relative GSH levels between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice were significantly different after CS exposure (P < 0.05). These results indicate that genetic ablation of Keap1 in the lung provides greater antioxidant capacity compared with Keap1flox/flox mice.
Exposure to CS generated a predominantly macrophage-driven inflammation in the lungs of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice, which supports our previous findings using this exposure model (18). The greatest numbers of infiltrating inflammatory cells were recovered in the BAL fluid of Keap1flox/flox mice, which was significantly increased compared with air-exposed mice (P < 0.05) (Figure 6A). The number of macrophages was significantly increased in the lavage fluid of Keap1flox/flox mice after CS exposure (P < 0.05). The observed increase in the number of inflammatory cells recovered in the BAL of Keap1Δ2–3/Δ2–3;CctCre+ mice exposed to CS, however, was not significant. No difference was observed in the number of inflammatory cells between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice exposed to CS. Proinflammatory cytokine expression levels were also measured the day after CS exposure, which correlated with the results from the BAL. Monocyte chemotactic protein-1 (MCP-1) expression levels were significantly increased in Keap1flox/flox mice compared with air-exposed mice (P < 0.05), but not in Keap1Δ2–3/Δ2–3;CctCre+ mice (Figure 6B). No difference was observed in inflammation after 5 days of CS exposure between Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice, indicating that the attenuation of inflammation was transient in this model.
Figure 6.
Deletion of Keap1 in Keap1Δ2–3/Δ2–3;CctCre+ mice attenuates CS-induced inflammation. (A) Total and differential inflammatory cell populations in the BAL fluid of Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ mice before and after exposure to CS. (n = 3–12). Data represented are mean number of cells ± SEM. (B) Messenger RNA expression levels of monocyte chemotactic protein-1 (MCP-1) in the lungs of Keap1flox/flox and Keap1Δ2–3/Δ2–3;CctCre+ mice after exposure to CS. The fold change was calculated using the formula described in Materials and Methods. Data presented is mean fold change ± SEM (n = 3). Asterisks indicate a significant difference compared with air exposed Keap1flox/flox mice (P < 0.05). NS indicates no significant difference compared with air-exposed Keap1Δ2–3/Δ2–3;CctCre+ mice.
Genetic Knockdown of KEAP1 through siRNA Transfection in Human Epithelial Cells Decreases the Levels of ROS during Oxidative Stress
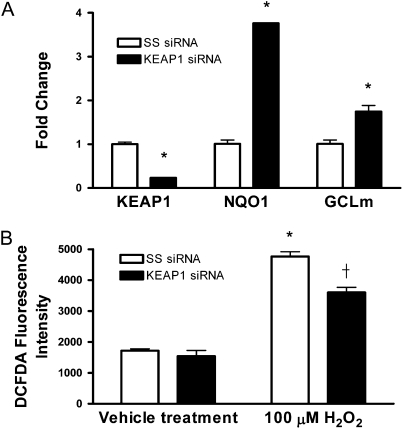
To determine whether activation of Nrf2 reduces the levels of ROS in human epithelial cells during exposure to oxidative stress, BEAS-2B cells were transfected with a siRNA duplex targeting KEAP1 mRNA, and the expression of KEAP1 and Nrf2-dependent genes were quantified through real-time PCR (33). Transfection of siRNA against KEAP1 significantly reduced the expression of KEAP1 mRNA greater than 75% after 48 hours (P < 0.05) (Figure 7A). To confirm the specificity of the inhibitory effect of siRNA targeting KEAP1 on Nrf2 downstream pathway, the transcript level of Nrf2-dependent genes, NQO1 and GCLm, were quantified. Both NQO1 and GCLm were significantly up-regulated compared with control cells transfected with a nontargeting siRNA (SS siRNA), 3.8- and 1.75-fold increase, respectively (P < 0.05) (Figure 7A). No difference in Nrf2 expression was observed between cells transfected with KEAP1 siRNA and cells transfected with SS siRNA (data not shown).
Figure 7.
Transfection of siRNA against KEAP1 mRNA in human epithelial cells leads to increased expression of Nrf2-dependent genes and decreased ROS levels during oxidative stress. (A) Relative mRNA levels of NQO1 and GCLm in BEAS-2B cells 48 hours after siRNA transfection. The fold change was calculated using the formula described in Materials and Methods (n = 3). (B) Quantification of DCFDA fluorescence in human epithelial cells. Data represented are mean values ± SEM (n = 6). Asterisks indicate a significant difference compared with SS siRNA control cells (P < 0.05). Dagger indicates a significant difference compared with SS siRNA cells treated with 100 μM H2O2 (P < 0.05).
Human lung epithelial cells transfected with or without KEAP1 siRNA were then exposed to hydrogen peroxide and the endogenous levels of ROS were quantified through DCFDA staining. Similar to primary Clara cells isolated from Keap1 conditional knockout mice, no difference in fluorescence was observed in human epithelial cells transfected with KEAP1 or nontargeting control siRNA under unstressed conditions (Figure 7B). However, after exposure to 100 μM hydrogen peroxide the levels of ROS were significantly decreased in epithelial cells transfected with KEAP1 siRNA compared with the nontargeting controls, indicating that Nrf2 activation in human epithelial cells leads to an attenuation of oxidative stress (Figure 7B). These data provide additional support in human epithelial cells that the deletion of KEAP1 decreases ROS levels and protects against the redox imbalance due to oxidative stress.
DISCUSSION
Exposure to CS is the primary factor associated with the development of COPD. Although the exact mechanism leading to the development of COPD has not been fully explained, mounting evidence indicates that oxidative stress plays a significant role (5). Since airways are continuously exposed to high levels of environmental oxidants, they must maintain the proper balance between oxidants and antioxidants to prevent cellular damage. Environmental toxicants, such as CS, that alter the cellular redox balance in the lung promote oxidative stress and over time lead to pulmonary injury and tissue damage.
CS creates an oxidant imbalance through two mechanisms. First, a burning cigarette releases millions of free radicals and increases the level of ROS and RNS in the lung. The release of these oxidants into the lungs of smokers overwhelms the normal antioxidant balance, which results in a subsequent decrease in antioxidant levels, thereby promoting oxidative stress. Xenobiotics are naturally detoxified through Phase I and Phase II enzymes, which modify xenobiotic toxicants through either oxidation or reduction of compounds (Phase I) and then promote the conjugation of phase I products with various hydrophilic moieties, including glutathione, to be safely secreted (Phase II) (43). Numerous Phase I and Phase II detoxification enzymes as well as enzymes associated with antioxidant metabolism are induced in the lung after exposure to CS through the master transcriptional regulator, Nrf2 (18). Indeed, chronic exposure to CS increases the expression of many Nrf2-dependent antioxidant and detoxification enzymes in airway epithelium (44). Therefore, to combat oxidative stress generated by CS, increasing antioxidant and detoxification Nrf2-dependent pathways may have therapeutic benefits.
Nrf2-dependent pathways can be activated either pharmacologically with the use of small triterpenoid molecules such as CDDO-Im (23, 45, 46) or genetically with tissue-specific Keap1 knockout mice as described in this and other studies (26). CDDO-Im is a potent synthetic triterpenoid, which is able to increase cytoprotective gene expression in the liver, lung, and other organs (45). This small molecule has been shown to be protective against LPS-induced inflammation and mortality by increasing antioxidant gene expression and decreasing ROS production and proinflammatory cytokine expression (46). More recently, CDDO-Im has been shown to be protective against both pulmonary CS-induced emphysema and cardiac dysfunction by decreasing oxidative stress and cellular apoptosis in the lung in an Nrf2-dependent manner (23). However, it is unclear whether selective targeting of Keap1 and enhancement of Nrf2 activation in the lungs will be sufficient to protect against the CS-induced oxidative stress and inflammation.
The present study is the first to report the creation of lung cell–specific Keap1 knockout mice in which Nrf2-dependent pathways are activated and more than 50 antioxidant and detoxification enzymes are increased in expression. Similar to the hepatocyte-specific Keap1 knockout mice, HO-1 and Prdx1, which are well-known Nrf2-dependent genes, were not significantly up-regulated in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice (Figure 3). The inability to induce the expression of these genes in airway epithelium may be due to the lack of other signaling events required to induce the expression of certain Nrf2-dependent genes, such as the recruitment of co-activators to the ARE (26). Alternatively, the inability to up-regulate HO-1 may be due to the presence of a repressor protein such as Bach-1 that binds to the OH-1 promoter and inhibits Nrf2-dependent transcription (47–49). A significant difference between the current study and the hepatocyte-specific Keap1 knockout mouse model is that Gclm was significantly up-regulated in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice but was not induced in the liver. This may indicate that cellular factors within different cell types influence which Nrf2-dependent genes are expressed in cells lacking a functional Keap1 protein.
Interestingly, a subset of cytoprotective and detoxification genes were significantly decreased in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice. Many of these genes were associated with the innate or adaptive immune response such as Cybb and Gzma, respectively. Since Keap1 is deleted from airway epithelial cells only and the microarray used tissue from the whole lung, it may be possible that certain genes are down-regulated as a compensatory mechanism to maintain homeostasis in the lung. Indeed, no difference existed between the lung structure of Keap1flox/flox mice and Keap1Δ2–3/Δ2–3;CctCre+ mice. Therefore, increasing Nrf2-dependent gene expression in Clara cells may have downstream effects in other pulmonary cell types.
Corresponding to the increase in antioxidant gene expression, Clara cells isolated from Keap1Δ2–3/Δ2–3;CctCre+ mice had decreased levels of intracellular ROS after the induction of oxidative stress (e.g., hydrogen peroxide). Keap1Δ2–3/Δ2–3;CctCre+ mice also had significantly increased levels of total GSH in the lungs during normal unstressed conditions as well as immediately after CS exposure. Recruitment of proinflammatory macrophages as a result of CS was blunted in Keap1Δ2–3/Δ2–3;CctCre+ mice, indicating that the increases in antioxidant and detoxification enzymes in the lung are beneficial to suppress inflammation.
The deletion of exons 2 and 3 of the Keap1 gene and elimination of the IVR domain and Kelch domains 1 through 4 in this novel mouse model led to a decrease in Keap1 gene expression and protein levels and an increase in Nrf2-dependent gene expression. No truncated Keap1 protein was detected in the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice. Therefore, we hypothesize that the truncated Keap1 protein in the conditional knockout mice is rapidly degraded through the proteasomal pathway. This hypothesis is supported by studies in which mutations in KEAP1 have been identified in a significant percentage of primary human lung cancer tumors and cell lines (33). Mutations in functionally important domains of KEAP1 in human non–small cell lung cancer cell lines decrease the protein levels of KEAP1 compared with nonmalignant cells (33). The loss of function in Keap1 and gain of Nrf2 function is not always beneficial and may promote tumor progression. However, the lungs of Keap1Δ2–3/Δ2–3;CctCre+ mice were analyzed at 8 weeks of age and have MLI and surface to volume ratios similar to the floxed control mice, indicating that the disruption of Keap1 in airway epithelial cells does not alter lung structure. In addition, Keap1Δ2–3/Δ2–3;CctCre+ mice greater than 6 months of age have no observable tumors or pathologies in the lung, suggesting that other mutations in addition to the deletion of Keap1 is necessary to trigger the development of lung cancer.
In summary, a novel Clara cell–specific Keap1 knockout mouse model was generated that had a significant increase in many Nrf2-dependent antioxidant and phase II detoxification enzymes in the lung. Clara cells from Keap1Δ2–3/Δ2–3;CctCre+ mice were protected against ROS in response to oxidative stress as a result of increased expression of Nrf2 pathways. In the lungs deletion of Keap1 led to a significantly increased level of total GSH that remained elevated after exposure to CS. The increased antioxidant levels correlated with an attenuation of inflammatory macrophages as well as dampened expression of MCP-1 after acute CS exposure. Increased expression of Nrf2 pathways in Clara cells may therefore protect against lung destruction due to chronic exposure to CS exposure.
Supplementary Material
Acknowledgments
The authors thank Anne Jedlicka and the Johns Hopkins University Malaria Research Institute for assistance with the Affymetrix GeneChip experiment and analyses.
This work was supported by National Institutes of Health grant F32HL094018 (D.J.B.), R01HL081205 (S.B.), COPD SCCOR grant P50HL084945 (S.B., R.M.T.), NIEHS center grant P30ES00381, Flight Attendant Medical Research Institute Clinical Innovator Award (S.B.), and Young Clinical Scientist Award (A.S.).
This article contains an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2009-0054OC on June 11, 2009
Conflict of Interest Statement: S.B. has received consultancy fees from MerckFrost for $5,001 to $10,000, lecture fees from Novartis for less than $1,000, a grant from Quark Pharmaceuticals for more than $100,001, and a sponsored grant from the National Institutes of Health (NIH) for more than $100,001. D.J.B. has received a sponsored grant from the National Heart, Lung, and Blood Institute (NHLBI) for $50,001 to 100,000. R.M.T. has received sponsored grants from NHLBI for more than $100,001 and Alpha 1 Foundation for $50,001 to $100,000. E.G. has received advisory board fees from Cardium Therapeutics for $10,001 to $50,000, expert witness fees from Bowman and Brooke Law Firm for $5,001 to 10,000, a research grant from Kanglaite Co. for $50,001 to $100,000, and two sponsored grants (one his own and one his spouse's) from National Cancer Institute for more than $100,001. He has stock ownership in Cardium for $10,001 to $50,000. T.J.M. has received sponsored grants from NIH for more than $100,001 and FAMRI for more than $100,001. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Viegi G, Pistelli F, Sherrill DL, Maio S, Baldacci S, Carrozzi L. Definition, epidemiology and natural history of copd. Eur Respir J 2007;30:993–1013. [DOI] [PubMed] [Google Scholar]
- 2.Chronic obstructive pulmonary disease. World Health Organization fact sheet 315. 2009.
- 3.Pryor WA, Prier DG, Church DF. Electron-spin resonance study of mainstream and sidestream cigarette smoke: nature of the free radicals in gas-phase smoke and in cigarette tar. Environ Health Perspect 1983;47:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacNee W. Oxidants/antioxidants and COPD. Chest 2000;117:303S–317S. [DOI] [PubMed] [Google Scholar]
- 5.Rahman I, MacNee W. Role of oxidants/antioxidants in smoking-induced lung diseases. Free Radic Biol Med 1996;21:669–681. [DOI] [PubMed] [Google Scholar]
- 6.Barnes PJ. Chronic obstructive pulmonary disease. N Engl J Med 2000;343:269–280. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida T, Tuder RM. Pathobiology of cigarette smoke-induced chronic obstructive pulmonary disease. Physiol Rev 2007;87:1047–1082. [DOI] [PubMed] [Google Scholar]
- 8.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-are pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116. [DOI] [PubMed] [Google Scholar]
- 9.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal neh2 domain. Genes Dev 1999;13:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa M, Xiong Y. Btb protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the cullin 3-roc1 ligase. Mol Cell Biol 2005;25:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for cul3-based e3 ligase to regulate proteasomal degradation of nrf2. Mol Cell Biol 2004;24:7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a cul3-dependent ubiquitin ligase complex. Mol Cell Biol 2004;24:10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinkai Y, Sumi D, Fukami I, Ishii T, Kumagai Y. Sulforaphane, an activator of Nrf2, suppresses cellular accumulation of arsenic and its cytotoxicity in primary mouse hepatocytes. FEBS Lett 2006;580:1771–1774. [DOI] [PubMed] [Google Scholar]
- 14.Kraft AD, Johnson DA, Johnson JA. Nuclear factor e2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci 2004;24:1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 2003;8:379–391. [DOI] [PubMed] [Google Scholar]
- 16.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 2002;62:5196–5203. [PubMed] [Google Scholar]
- 17.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997;236:313–322. [DOI] [PubMed] [Google Scholar]
- 18.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest 2004;114:1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka T, Ishii Y, Itoh K, Kiwamoto T, Kimura T, Matsuno Y, Morishima Y, Hegab AE, Homma S, Nomura A, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells 2005;10:1113–1125. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki M, Betsuyaku T, Ito Y, Nagai K, Nasuhara Y, Kaga K, Kondo S, Nishimura M. Down-regulated nf-e2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2008;39:673–682. [DOI] [PubMed] [Google Scholar]
- 21.Malhotra D, Thimmulappa R, Navas-Acien A, Sandford A, Elliott M, Singh A, Chen L, Zhuang X, Hogg J, Pare P, et al. Decline in Nrf2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, dj-1. Am J Respir Crit Care Med 2008;178:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Amara N, Crestani B, Fournier M, Leseche G, Soler P, Boczkowski J, et al. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax 2008;63:916–924. [DOI] [PubMed] [Google Scholar]
- 23.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, Yates MS, Kombairaju P, Yamamoto M, Liby KT, et al. Targeting Nrf2 with the triterpenoid cddo-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci USA 2009;106:250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 2003;35:238–245. [DOI] [PubMed] [Google Scholar]
- 25.Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods 2001;24:71–80. [DOI] [PubMed] [Google Scholar]
- 26.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the Keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun 2006;339:79–88. [DOI] [PubMed] [Google Scholar]
- 27.Widdicombe JG, Pack RJ. The Clara cell. Eur J Respir Dis 1982;63:202–220. [PubMed] [Google Scholar]
- 28.Patton SE, Gupta RP, Nishio S, Eddy EM, Jetten AM, Plopper CG, Nettesheim P, Hook GE. Ultrastructural immunohistochemical localization of clara cell secretory protein in pulmonary epithelium of rabbits. Environ Health Perspect 1991;93:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cutz E, Conen PE. Ultrastructure and cytochemistry of Clara cells. Am J Pathol 1971;62:127–141. [PMC free article] [PubMed] [Google Scholar]
- 30.Walker SR, Hale S, Malkinson AM, Mason RJ. Properties of isolated nonciliated bronchiolar cells from mouse lung. Exp Lung Res 1989;15:553–573. [DOI] [PubMed] [Google Scholar]
- 31.Scacheri PC, Crabtree JS, Novotny EA, Garrett-Beal L, Chen A, Edgemon KA, Marx SJ, Spiegel AM, Chandrasekharappa SC, Collins FS. Bidirectional transcriptional activity of pgk-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis 2001;30:259–263. [DOI] [PubMed] [Google Scholar]
- 32.Simon DM, Arikan MC, Srisuma S, Bhattacharya S, Tsai LW, Ingenito EP, Gonzalez F, Shapiro SD, Mariani TJ. Epithelial cell PPAR[gamma] contributes to normal lung maturation. FASEB J 2006;20:1507–1509. [DOI] [PubMed] [Google Scholar]
- 33.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional Keap1-Nrf2 interaction in non-small-cell lung cancer. PLoS Med 2006;3:e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasahara Y, Tuder RM, Taraseviciene-Stewart L, Le Cras TD, Abman S, Hirth PK, Waltenberger J, Voelkel NF. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J Clin Invest 2000;106:1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devereux TR, Fouts JR. Isolation and identification of Clara cells from rabbit lung. In Vitro 1980;16:958–968. [DOI] [PubMed] [Google Scholar]
- 36.Rahman I, Kode A, Biswas SK. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc 2006;1:3159–3165. [DOI] [PubMed] [Google Scholar]
- 37.Plopper CG. Comparative morphologic features of bronchiolar epithelial cells: the Clara cell. Am Rev Respir Dis 1983;128:S37–S41. [DOI] [PubMed] [Google Scholar]
- 38.Plopper CG, Mariassy AT, Hill LH. Ultrastructure of the nonciliated bronchiolar epithelial (Clara) cell of mammalian lung: I. A comparison of rabbit, guinea pig, rat, hamster, and mouse. Exp Lung Res 1980;1:139–154. [DOI] [PubMed] [Google Scholar]
- 39.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-fos and fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:Quinone oxidoreductase1 gene. Proc Natl Acad Sci USA 1996;93:14960–14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh A, Rangasamy T, Thimmulappa RK, Lee H, Osburn WO, Brigelius-Flohe R, Kensler TW, Yamamoto M, Biswal S. Glutathione peroxidase 2, the major cigarette smoke-inducible isoform of gpx in lungs, is regulated by Nrf2. Am J Respir Cell Mol Biol 2006;35:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A, Ling G, Suhasini AN, Zhang P, Yamamoto M, Navas-Acien A, Cosgrove G, Tuder RM, Kensler TW, Watson WH, et al. Nrf2-dependent sulfiredoxin-1 expression protects against cigarette smoke-induced oxidative stress in lungs. Free Radic Biol Med 2008;46:376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvey CJ, Thimmulappa RK, Singh A, Blake DJ, Ling G, Wakabayashi N, Fujii J, Myers A, Biswal S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic Biol Med 2008;46:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motohashi H, Yamamoto M. Nrf2-keap1 defines a physiologically important stress response mechanism. Trends Mol Med 2004;10:549–557. [DOI] [PubMed] [Google Scholar]
- 44.Adair-Kirk TL, Atkinson JJ, Griffin GL, Watson MA, Kelley DG, DeMello D, Senior RM, Betsuyaku T. Distal airways in mice exposed to cigarette smoke: Nrf2-regulated genes are increased in Clara cells. Am J Respir Cell Mol Biol 2008;39:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, Gribble GW, Johnson DA, Johnson JA, Burton NC, et al. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther 2007;6:154–162. [DOI] [PubMed] [Google Scholar]
- 46.Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, Sporn MB, Yamamoto M, Kensler TW, Biswal S. Nrf2-dependent protection from LPS induced inflammatory response and mortality by cddo-imidazolide. Biochem Biophys Res Commun 2006;351:883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J 2002;21:5216–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K. Heme regulates the dynamic exchange of Bach1 and Nf-e2-related factors in the MAF transcription factor network. Proc Natl Acad Sci USA 2004;101:1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:Quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem 2005;280:16891–16900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.