Abstract
Rationale: Much effort is being made to discover noninvasive biomarkers of chronic airway disease that might enable better management, predict prognosis, and provide new therapeutic targets.
Objectives: To undertake a comprehensive, unbiased proteomic analysis of induced sputum and identify novel noninvasive biomarkers for chronic obstructive pulmonary disease (COPD).
Methods: Induced sputum was obtained from patients with COPD with a spectrum of disease severity and from control subjects. Two-dimensional gel electrophoresis and mass spectrometric identification of differentially expressed proteins were first applied to induced sputum from patients with GOLD stage 2 COPD and healthy smoker control subjects. Initial results thus obtained were validated by a combination of immunoassays (Western blotting and ELISA) applied to a large subject cohort. The biomarkers were localized to bronchial mucosa by immunohistochemistry.
Measurements and Main Results: Of 1,325 individual protein spots identified, 37 were quantitatively and 3 qualitatively different between the two groups (P < 0.05%). Forty protein spots were subjected to tandem mass spectrometry, which identified 15 separate protein species. Seven of these were further quantified in induced sputum from 97 individuals. Using this sequential approach, two of these potential biomarkers (apolipoprotein A1 and lipocalin-1) were found to be significantly reduced in patients with COPD when compared with healthy smokers. Their levels correlated with FEV1/FVC, indicating their relationship to disease severity.
Conclusions: A potential role for apolipoprotein A1 and lipocalin-1 in innate defense has been postulated previously; our discovery of their reduction in COPD indicates a deficient innate defense system in airway disease that could explain increased susceptibility to infectious exacerbations.
Keywords: two-dimensional polyacrylamide gel electrophoresis, induced sputum, proteome, biomarkers, chronic obstructive pulmonary disease
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Studies to date have identified numerous clinical and biological variables associated with chronic obstructive pulmonary disease (COPD); however, there is a need for specific biomarkers of COPD that can be sampled noninvasively and that correlate with disease severity.
What This Study Adds to the Field
We have identified two biomarkers in induced sputum that correlate with the severity of COPD. Their known role in innate defense and their reduction in disease may provide additional insight into the pathobiology of COPD.
Chronic obstructive pulmonary disease (COPD) is characterized by progressive airflow limitation that results from an abnormal inflammatory and tissue remodeling response in the lungs to noxious particles or gases (www.gold.org), the most common of which is cigarette smoke (1–3). A major question relevant to disease pathogenesis and identification of “at risk” smoking individuals concerns why all smokers do not develop disease. It is also unclear why lung function of some patients with COPD continues to deteriorate even though they stop smoking (4, 5).
As with other diseases, much hope has been placed in the discovery of biomarkers that would help understand the mechanisms of COPD (6–10). It is also hoped that some of these could speed up drug discovery by serving as surrogate markers that respond to novel drugs within a shorter time span than spirometric measurements of lung function, currently the main clinical outcome in drug trials. Although studies in patients with COPD have identified several biomarkers, most of these have not been fully validated and their prognostic value remains unclear (6, 11–15). Many of the markers have been identified in blood and, although it is recognized that there are nonpulmonary consequences of COPD, some of which can be viewed as systemic (16, 17), biomarkers measured and/or generated in the lungs are likely to be most informative.
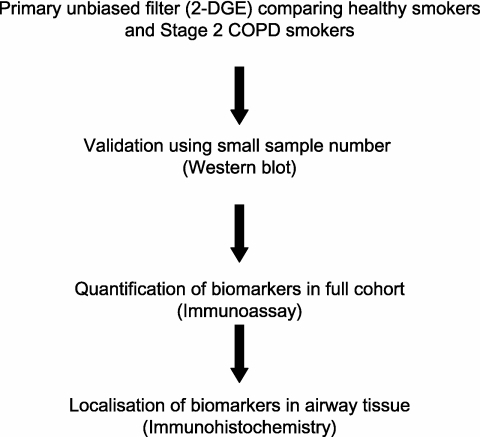
We report the results of a staged COPD biomarker discovery program (Figure 1) in which proteomics and immunoassays were applied to compare the proteins in the epithelial lining fluid sampled by sputum induction, a technique widely used to study airway diseases, including COPD (18). We hypothesized that smoking results in altered expression of proteins in smokers who develop COPD, that is, different from that in smokers whose lung function remains normal. Initially, we used two-dimensional gel electrophoresis (2-DGE) as the primary protein fractionation step and identified differentially expressed proteins (DEPs) by tandem mass spectrometry (MS). This provided an unbiased filter for more than 1,000 proteins that could be identified by 2-DGE in sputum, and resulted in 40 DEPs. This phase was followed by a focused step in which the DEPs were validated as “candidate biomarkers” of COPD by applying Western blot assays to the same samples used in the 2-DGE analysis. Further validation of the candidate biomarkers was performed by using ELISA and Western blots on a large collection of sputum samples from healthy smokers, smokers with various levels of COPD severity, and healthy nonsmoking control subjects and checking for disease specificity by analyzing samples from subjects with asthma. Finally, the validated biomarkers were localized within the bronchial mucosa by immunohistochemistry, using bronchoscopic biopsies from the same subject groups.
Figure 1.
Schematic diagram of study design for biomarker discovery. 2-DGE = two-dimensional gel electrophoresis; COPD = chronic obstructive pulmonary disease.
METHODS
Subjects and Sample Collection
Ninety-seven subjects were recruited for the study. These were classified as current smokers with mild, moderate, and severe COPD (Global Initiative for Chronic Obstructive Lung Disease [GOLD] stage 1 to 3); smokers with chronic bronchitis (previously classified as GOLD stage 0); smokers with low transfer factor of the lung for carbon monoxide (TlCO, % predicted) and a control subject group consisting of healthy smokers, nonsmokers (NS), and nonsmoking subjects with asthma (Tables 1 and 2). No ex-smokers were included. The groups were similar with respect to age and sex except for the asthmatic group, where the mean age was significantly lower. Smoking history was similar in HS and subjects with COPD. Full history was taken and the St. George Quality of Life Questionnaire was completed. The smokers underwent histamine challenge and assessment of carbon monoxide transfer (DlCO) and bronchodilator reversibility. Healthy controls also had high-resolution computed tomography (HRCT) of the lungs (19) to exclude emphysema. Any subjects with COPD with acute exacerbation/chest infection within 6 weeks of the study, positive sputum culture, decompensated cor pulmonale, and treatment with inhaled corticosteroids were excluded. All patients were steroid naive, had no comorbidities, and were not taking any other medication at the time of the study other than bronchodilators. Of these 97 subjects, 15 were excluded from the healthy smoker category used for 2-DGE and subsequent ELISA despite normal spirometry, 8 due to symptoms of chronic bronchitis (stage 0 COPD according to previous GOLD criteria) and 7 due to low measurements of TlCO (likely to be indicative of emphysema) (Table 2).
TABLE 1.
SUBJECT CHARACTERISTICS FOR SPUTUM AND SERUM DONORS WITH STAGES I–III COPD AND HEALTHY SMOKER CONTROL SUBJECTS
| Healthy Smokers | COPD Stage 1 | COPD Stage 2 | COPD Stage 3 | |
|---|---|---|---|---|
| No. of subjects | 20 | 16 | 25 | 3 |
| F/M | 14/6 | 5/11 | 7/18 | 1/2 |
| Height, cm | 169.4 (161–174) | 173 (165.0–178.0) | 173 (170.1–177.9) | 165.1 (149.0–174.5) |
| Age, yr | 52.9 (47.3–56.4) | 57.9 (52.4–63.4) | 60.3 (52.6–67.7) | 61.2 (57.6–70.8) |
| Smoking, pack-years | 33.6 (28.8–51.8) | 52.5 (38.7–85.0) | 43.8 (33.5–50.5) | 67.5 (36.0–79.5) |
| FEV1 % predicted (postbronchodilator) | 100 (96–107) | 89 (84–97) | 73 (63–80) | 45 (38–46) |
| FEV1/FVC % (postbronchodilator) | 77 (73–80) | 66 (62–71) | 57.4 (54–62) | 48 (40–48) |
| TlCO % predicted | 94 (84–103) | 85 (68–96) | 83 (67–91) | nd |
| Total SGRQ score | 13.5 (8.7–18.9) | 22.9 (10.6–36.8) | 25.1 (11.7–40.7) | 50.4 (5.2–55.9) |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; F/M = females/males; nd = not done, SGRQ = St. George Quality of life Questionnaire; TlCO = transfer factor for carbon monoxide. Values shown represent medians (interquartile range in parentheses).
TABLE 2.
SUBJECT CHARACTERISTICS FOR SPUTUM AND SERUM DONORS WITH CHRONIC BRONCHITIS, LOW TlCO, ASTHMA AND NONSMOKER CONTROL SUBJECTS
| Healthy Nonsmokers | Normal Spirometry, Symptoms of Chronic Bronchitis | Normal Spirometry, low TlCO | Subjects with Asthma | |
|---|---|---|---|---|
| No. of subjects | 7 | 8 | 7 | 11 |
| F/M | 3/4 | 6/2 | 7/0 | 6/5 |
| Height, cm | 170.2 (139.7–185.0) | 163 (161–178) | 159 (158–164) | n/d |
| Age, yr | 51.1 (35.5–69.3) | 49 (42–60) | 54 (44–57) | 49.6 (22–78) |
| Smoking, pack-years | 0 (0–0) | 42 (25–51) | 35 (18–37) | 0 (0–0) |
| FEV1 % predicted (postbronchodilator) | 113 (96–118) | 94 (85–103) | 97 (84–118) | 84 (57–90)* |
| FEV1/FVC % (postbronchodilator) | 83 (71–84) | 94.5 (87–97) | 93 (90–99) | 69.46 (49–87)* |
| TlCO % predicted | nd | 85 (54–90) | 68 (62–75) | nd |
| Total SGRQ score | 10.4 (1.5–21.0) | 23.2 (10.4–37.2) | 14.2 (5.7–26.7) | n/a |
Definition of abbreviations: F/M = female/male; nd = not done; n/a = not applicable; SGRQ = St. George Quality of Life Questionnaire; TlCO = transfer factor for carbon monoxide. Values shown represent medians (interquartile range in parentheses).
Prebronchodilator values are shown for the subjects with asthma.
Fifty-six NS, HS, and subjects with COPD (Table E1 in the online supplement), the majority from within the main cohort, also underwent bronchoscopy (19).
All subjects gave written informed consent and the study was approved by the Southampton and South West Hampshire Ethics Committee.
Sampling and Processing
A standard protocol for sputum induction and our previously reported (with minor modifications) sputum-processing protocol (20) were applied. None of the samples was reported as significantly positive for pathogenic bacteria, as determined according to standard microbiological assessment procedures. Endobronchial biopsies were processed as previously reported (21).
Initial Biomarker Screen for DEP Spots by 2-DGE
Sputum samples from a subset of 15 subjects with stage 2 COPD and 18 HS were selected for this first unbiased stage of DEP identification. High molecular weight proteins and contaminants were first removed, using a combination of chaotropic agent (urea), reducing agent (dithioerythritol, DTE), and sequential molecular weight filtration steps (see the online supplement for details). Proteins were visualized by SYPRO Ruby protein staining and digital image capture. Gels were restained overnight with colloidal Coomassie stain.
Gel Image Analysis, Differential Protein Expression Assessment, and Protein Identification
PDQuest software (Bio-Rad, Hemel Hempstead, UK) was applied for manual matching of spots and for subsequent image analysis to determine spot quantity changes between the two groups. Data were normalized for total valid spot pixel density before quantification. Six images were rejected because of insufficient quality (due to running quality or faintness of image). Only gels having more than 500 detectable spots (based on samples from 13 healthy smoker and 14 smokers with stage 2 COPD) were included for further analysis. The characteristics of the analyzed subjects are shown in Table E2 in the online supplement.
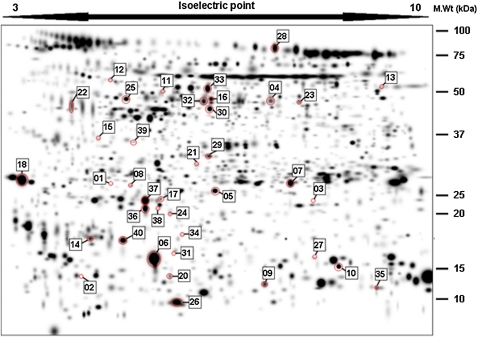
A total of 1,325 individual spots was found (Figure 2; and Table E3 in the online supplement). Differences in spot quantities between healthy smokers and subjects with stage 2 COPD were determined either quantitatively by Mann-Whitney U test (if present in ≥80% of all gels in each group) or qualitatively (if present in >80% of one and <20% of the other group) (annotated PDQuest files of gel images available on request).
Figure 2.
Distribution of differentially expressed protein spots (marked by red circles) displayed on a master gel image generated from 33 individual patient gel images. Each spot number relates to data shown in Table 3.
The protein spots were digested with trypsin (22) and the resulting peptides were separated by nano–reversed-phase liquid chromatography and electrosprayed into a quadrupole time-of-flight tandem mass spectrometer. All data were acquired by Q-tof Global Ultima (Waters Ltd, Milford, MA) fitted with a NanoLockSpray source to achieve greater than 10 ppm mass accuracy for the precursor ions. Tandem mass spectrometry (MS/MS) spectra were automatically processed by MassLynx 4.0 (Waters Ltd) and searched against the NCBI nonredundant database, using ProteinLynx Global Server 2.05, and proteins were identified on the basis of previously described criteria (23).
Initial Biomarker Validation by Western Blotting
A subset of seven samples from the two subject categories used for 2-DGE (HS and stage 2 COPD) were probed on Western blots with antibodies specific for the nine biomarkers discovered by 2-DGE. Images were analyzed by densitometry, using QuantityOne software (Bio-Rad, Hercules, CA) and results were obtained as pixel density (pdu) per millimeter squared. The two groups were compared by Mann-Whitney U test.
Validation of Candidate Biomarkers in a Wider Cohort
Samples from all 97 subjects who provided sputum (main cohort) were finally analyzed by ELISA and Western blotting. Albumin, transthyretin, α2-HS (Heremans–Schmid) glycoprotein, and apolipoprotein A1 were quantified in duplicate DTE-treated sputum samples by commercial ELISA after optimization of sputum dilutions. Spiking with recombinant protein showed mean percent recoveries of 85% for α2-HS glycoprotein, 102% for transthyretin, 95% for apolipoprotein A1, and 72% for albumin. In the absence of commercially available ELISA, lipocalin-1, PSP94 (prostate secretory protein of 94 amino acids), and PLUNC (palate, lung, and nasal epithelium clone protein) were quantified by Western immunoblotting of samples fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Band densities were normalized with a standard protein mixture fractionated on every gel. The quantified biomarkers were compared as absolute values (weight of protein per volume [ml] of sputum), or as values normalized for protein content using relative protein content determined from fluorescent blot stains (expressed as weight of protein per pixel density unit) (see the online supplement for details).
Tissue Localization of Identified Biomarkers by Immunohistochemistry
Bronchial biopsy samples from 56 subjects (Table E1) were embedded in GMA resin as previously described (21), and 2-μm sections were stained for lipocalin-1 and apolipoprotein A1 and subjected to image analysis. Staining was expressed as the percentage of total epithelial surface area (24).
Quantification of Apolipoprotein A1 in Plasma Samples
Apolipoprotein A1 was quantified in citrated plasma samples from the cohort described in Table E1, using a commercial ELISA kit (Alerchek, Portland, OR) according to the manufacturer's instructions (see the online supplement for details).
Statistical Analyses
All data were analyzed with SPSS software, version 13 (SPSS, Chicago, IL). Data were compared for statistical significance, using parametric and nonparametric tests as appropriate. Clinical variables from the two main test groups (healthy smokers and smokers with stage 2 COPD) were compared by t test and results were confirmed by Mann-Whitney U test. The first comparison of protein expression, using 2-DGE spot volume data from both groups, was based on Mann-Whitney U tests (exact, two-tailed, corrected for ties) as the spot volumes did not show a Gaussian distribution. The primary outcome of the study was the comparison between the ELISA/Western blot data from HS with normal DlCO and no HRCT evidence of emphysema and all subjects with stage 2 COPD, also using Mann-Whitney U tests. This was followed by an exploratory analysis comparing by Mann-Whitney U test all subject groups from the entire cohort of 97 subjects (Tables 1 and 2). Because the primary outcome analysis was restricted to a single comparison, no adjustments were made for these exploratory multiple comparisons. Data from the primary analysis groups (HS and subjects with stage 2 COPD) were then examined by Kendall's tau b test for correlations between each biomarker and clinical outcomes such as FEV1/FVC and FEV1 (% predicted). Principal components analysis was performed on the significantly differentially expressed 2-DGE spots as a data reduction technique to extract the main features of the spot volumes and represent them as a set of uncorrelated components. The first two principal components were plotted against each other to look for clusters in the data.
RESULTS
2-DGE Group Descriptors
When examined using t tests and confirmed by Mann-Whitney U test, no differences at the 5% level were found between the two test groups (healthy smokers and smokers with stage 2 COPD) used for 2-DGE analysis in terms of height, weight, smoking pack-years, age, sputum volume, sputum lymphocytes, or sputum epithelial cells (Table E2). Significant differences between the two groups were found in terms of sputum neutrophils (P < 0.01), macrophages (P < 0.01), and squamous cells (P < 0.05).
Primary (Unbiased) Screen for Proteins Differentially Expressed in Patients with GOLD Stage 2 COPD Compared with Healthy Smokers
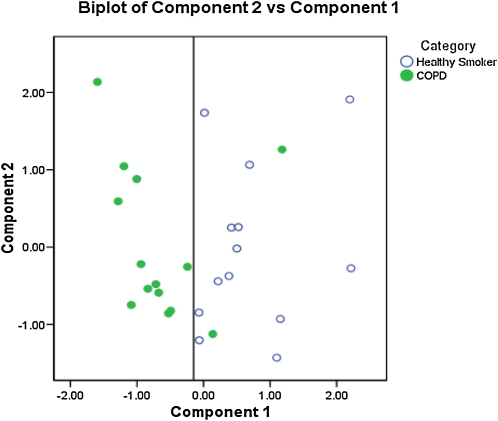
A total of 1,325 individual valid spots (those matched between 2 gels or more) (Table E3 and Figure 2) were identified as present across 27 gels (from 13 HS and 14 subjects with stage 2 COPD; details of subjects are presented in Table E2). Three of these spots fulfilled the criteria of qualitative differences between healthy smoker and COPD stage 2 populations. Of the 363 spots that fulfilled the criteria of being present in at least 80% of gels in both groups (Table E4 in the online supplement), 37 spots had a significantly different median volume when comparing the two groups (P < 0.05). Between 0.22- and 2.83-fold changes (stage 2 COPD/HS) were observed in these differentially expressed protein spots (DEPs) (Table 3). No effect of sex was found in the distribution of these spots (results not shown). The distribution of both quantitatively and qualitatively different spots (DEPs) in the 2D profile can be seen on a master image generated from the spots identified on all 33 gels initially analyzed (Figure 2; and Figure E1 in the online supplement). Principal component analysis of these DEPs indicated that the combined data from these spots segregated the smokers with and without COPD (Figure 3).
TABLE 3.
TANDEM MASS SPECTROMETRIC ANALYSIS OF DIFFERENTIALLY EXPRESSED PROTEIN SPOTS
| Original SSP | ID No. | P Value | Experimental kDa | Experimental pI | Mean Spot Volume, HS | Mean Spot Volume, COPD Stage 2 | Mean Fold Change | Protein ID |
|---|---|---|---|---|---|---|---|---|
| 2304 | 1 | — | 26.6 | 4.8 | — | — | −7* | Apolipoprotein A1 (5) |
| 1019 | 2 | — | — | — | — | — | +9* | Not detected |
| 7203 | 3 | — | — | — | — | — | +9* | Not detected |
| 6606 | 4 | 0.002 | 46.8 | 5.5 | 1,085.8 | 2,469.9 | 2.3 | Hypothetical protein (1) |
| 5201 | 5 | 0.002 | 25.2 | 5.4 | 9,076.8 | 4,386.1 | 0.5 | Albumin (4) |
| 3107 | 6 | 0.004 | 16.7 | 4.9 | 11,974.6 | 17,568.2 | 1.5 | PSP94 (2) |
| 6317 | 7 | 0.004 | 26.8 | 5.6 | 8,872.1 | 5,009.3 | 0.6 | Albumin (8) |
| 2308 | 8 | 0.006 | 26.3 | 4.9 | 2,301.3 | 498.6 | 0.2 | Apolipoprotein A1 (11) |
| 6002 | 9 | 0.006 | 13.0 | 5.5 | 964.3 | 1,935.3 | 2.0 | Apolipoprotein A1 (4) |
| 7112 | 10 | 0.006 | 15.7 | 5.9 | 24,410.6 | 12,301.0 | 0.5 | Albumin (6) |
| 3713 | 11 | 0.006 | 49.3 | 5.0 | 757.1 | 381.2 | 0.5 | Albumin (12) |
| 2709 | 12 | 0.007 | 54.5 | 4.8 | 796.3 | 428.9 | 0.5 | Albumin (3) |
| 8721 | 13 | 0.008 | — | — | 2,432.0 | 1,204.8 | 0.5 | Not detected |
| 1122 | 14 | 0.013 | 18.1 | 4.7 | 5,176.9 | 2,891.5 | 0.6 | Lipocalin-1 (3) |
| 2506 | 15 | 0.013 | 35.6 | 4.7 | 453.3 | 1,284.3 | 2.8 | Albumin (3) |
| 4626 | 16 | 0.013 | 47.3 | 5.4 | 1,178.9 | 2,154.3 | 1.8 | Lactoferrin (6) |
| 3220 | 17 | 0.014 | 23.7 | 5.1 | 698.8 | 430.3 | 0.6 | Apolipoprotein A1 (1) |
| 302 | 18 | 0.015 | 27.4 | 0 | 10,628.1 | 15,246.1 | 1.4 | IgJ (2)PLUNC (1) |
| 2009 | 19 | 0.017 | 12.5 | 4.8 | 1,136.9 | 514.1 | 0.5 | Albumin (2) |
| 3013 | 20 | 0.017 | 13.2 | 5.0 | 2,354.6 | 1,536.6 | 0.7 | Albumin (2) |
| 4414 | 21 | 0.017 | 30.2 | 5.3 | 2,253.6 | 1,195.0 | 0.5 | Zinc-α2-glycoprotein (2) |
| 1606 | 22 | 0.02 | 45.5 | 0 | 1,587.5 | 2,785.3 | 1.8 | Albumin (4) |
| 6619 | 23 | 0.022 | 46.5 | 5.7 | 670.4 | 1,356.1 | 2.0 | Albumin (7) |
| 4202 | 24 | 0.023 | 20.5 | 5.1 | 1,126.6 | 514.8 | 0.5 | α2-HS glycoprotein (2)Transferrin (2)α1-Antitrypsin (4)Albumin (16) |
| 3607 | 25 | 0.026 | 47.7 | 4.9 | 1,530.9 | 2,553.0 | 1.7 | α2-HS glycoprotein (2)Hemoglobin (1) |
| 4002 | 26 | 0.026 | — | — | 3,642.9 | 6,634.8 | 1.8 | Not detected |
| 7104 | 27 | 0.026 | 16.7 | 5.8 | 4,960.4 | 1,830.9 | 0.4 | Lactoferrin (4)Albumin (2) |
| 5905 | 28 | 0.028 | 75.0 | 5.5 | 1,359.8 | 2,653.9 | 1.9 | Albumin (7) |
| 4418 | 29 | 0.029 | 31.8 | 5.4 | 2,512.3 | 1,587.5 | 0.6 | Albumin (4) |
| 4625 | 30 | 0.029 | 44.5 | 5.4 | 4,071.2 | 2,420.9 | 0.6 | Transthyretin (3)IgA (1) |
| 4101 | 31 | 0.029 | 17.1 | 5.1 | 793.6 | 411.3 | 0.5 | Transthyretin (3)IgA (1) |
| 4623 | 32 | 0.031 | 46.7 | 5.3 | 2,710.0 | 4,431.2 | 1.6 | Albumin (3)Unnamed protein (2)Lactoferrin (1) |
| 4713 | 33 | 0.033 | 49.9 | 5.3 | 5,027.5 | 2,735.4 | 0.5 | Albumin (9) |
| 4104 | 34 | 0.035 | — | — | 329.3 | 562.5 | 1.7 | Not detected |
| 8008 | 35 | 0.038 | 12.6 | 0 | 1,971.7 | 2,566.3 | 1.3 | SAP (1) |
| 3206 | 36 | 0.042 | 21.5 | 4.9 | 7,407.0 | 4,276.0 | 0.6 | Albumin (3) |
| 3208 | 37 | 0.042 | 23.5 | 4.9 | 12,879.1 | 6,916.0 | 0.5 | Albumin (4) |
| 3212 | 38 | 0.043 | — | — | 630.6 | 290.7 | 0.5 | Not detected |
| 2521 | 39 | 0.049 | — | — | 339.9 | 608.6 | 1.8 | Not detected |
| 2111 | 40 | 0.05 | 18.0 | 4.8 | 10,164.1 | 5,889.4 | 0.6 | Lipocalin-1 (4) |
Definition of abbreviations: α2-HS glycoprotein = α2-Heremans–Schmid glycoprotein; 2-DGE = two-dimensional gel electrophoresis; COPD = chronic obstructive pulmonary disease; HS = healthy smokers; pI = isoelectric point; PLUNC = palate, lung, and nasal epithelium clone protein; PSP94 = prostate secretory protein of 94 amino acids; SAP = salivary acidic protein; SSP = sample spot protein.
Protein spots determined as significant (P ≤ 0.05) from comparison of healthy smokers and smokers with stage 2 COPD.
Sputum 2-DGE spot profiles were excised from gels and subjected to tandem mass spectrometry to determine protein identifications. Values in parentheses denote the number of peptides detected per protein.
For qualitatively detected protein spots, spot differences are expressed as the difference in the number of gels containing that protein spot between the two groups, according to the principle stated in Methods.
Figure 3.
Principal component analysis of differentially expressed protein spot volumes, identifying a component capable of segregating healthy smokers from smokers with stage 2 chronic obstructive pulmonary disease (COPD).
Four fresh 2-DGE gels from representative healthy smokers and patients with stage 2 COPD were prepared for protein identification by peptide-mass fingerprinting (results of this analysis are shown in Table 3). Of the 40 DEP spots, 15 individual protein identities were obtained (lactoferrin, albumin, transthyretin, PLUNC, lipocalin-1, cystatin, apolipoprotein A1, hemoglobin, IgJ chain, hypothetical protein, zinc-α2-glycoprotein, α1-antitrypsin, α2-HS glycoprotein, PSP94, and IgA).
Preliminary Analysis to Identify Candidate Biomarkers
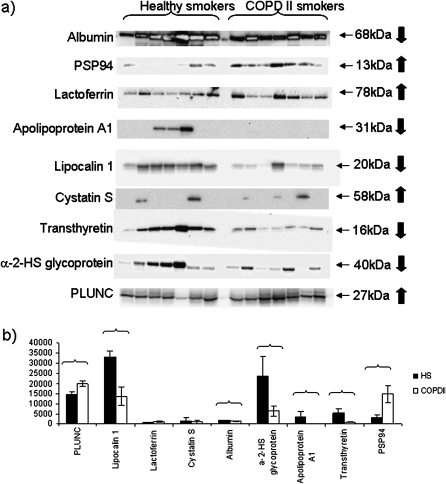
Sputum samples from seven healthy smokers and seven smokers with stage 2 COPD were selected at random from the full cohort and analyzed by Western blot for nine of the biomarkers identified by 2-DGE/peptide/mass fingerprinting analysis (Figure 4). Comparison of band intensities by Mann-Whitney U test confirmed that, in these samples, all biomarkers were altered in accordance with the trend predicted from 2-DGE spot volume analysis for each individual protein except cystatin S, which showed a few positive samples in each group, and lactoferrin, which also showed no robust change in overall direction. Seven of the potential “candidate biomarkers” were statistically significantly different between groups, namely albumin, α2-HS glycoprotein, transthyretin, PSP94, apolipoprotein A1, lipocalin-1, and PLUNC (Figure 4).
Figure 4.
(a) Western blot analysis of biomarkers in induced sputum samples from seven healthy smokers and seven smokers with stage 2 chronic obstructive pulmonary disease (COPD II). Experimental molecular masses of the major band are shown. Arrows indicate predicted direction of change in patients with COPD from two-dimensional gel electrophoresis data. α-2-HS glycoprotein = α2-Heremans–Schmid glycoprotein; HS = healthy smokers; PLUNC = palate, lung, and nasal epithelium clone protein; PSP94 = prostate secretory protein of 94 amino acids. (b) Densitometry analysis of results from the Western blot, indicating significance of change between the two groups compared by Student t test (denoted by brackets: P < 0.05).
Validation of Biomarkers in the Whole Cohort
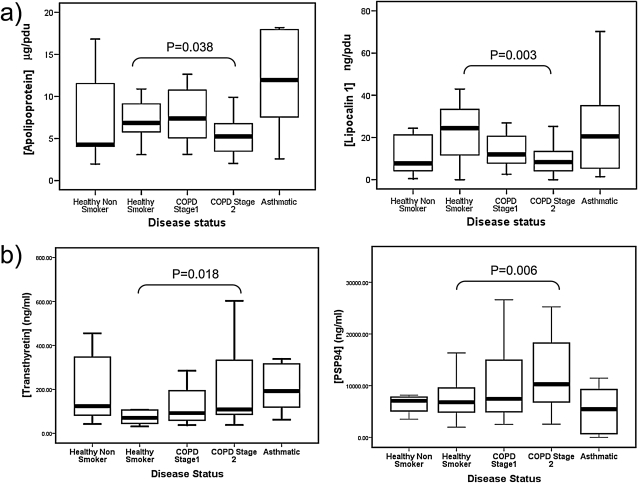
To identify proteins that were differentially expressed in smokers with COPD (the primary outcome of this study) both absolute data or data normalized for protein content were used to compare HS (n = 20) and subjects with GOLD stage 2 COPD (n = 25), using Mann-Whitney U tests (Table 4). When normalized data were used, two biomarkers had significantly reduced median values in the COPD stage 2 group when compared with HS (apolipoprotein A1, P = 0.038; and lipocalin-1, P = 0.003) (Table 4 and Figure 5a). When raw data (i.e., concentrations of protein per volume [ml] of sputum supernatant not normalized for protein content) from the same subjects were compared, two biomarkers (transthyretin and PSP94) were significantly different between the groups, and lipocalin-1 showed borderline significance, whereas apolipoprotein A1 was no longer significantly different (Figure 5b).
TABLE 4.
STATISTICAL ANALYSIS COMPARING BIOMARKER QUANTITIES BETWEEN HEALTHY SMOKERS AND SUBJECTS WITH STAGE 2 CHRONIC OBSTRUCTIVE PULMONARY DISEASE, USING BOTH NORMALIZED AND NONNORMALIZED (RAW) DATA FROM SAMPLES FROM THE WHOLE COHORT
| Whole Cohort |
|||
|---|---|---|---|
| HS (n = 20) | COPD 2 (n = 25) | Exact Significance | |
| Normalized data | |||
| Apolipoprotein A1, μg/pdu | 6.87 (5.72–9.33) | 5.22 (3.34–7.09) | 0.038* |
| Albumin, μg/pdu | 0.592 (0.431–0.816) | 0.502 (0.351–0.809) | 0.465 |
| α2-HS glycoprotein, μg/pdu | 0.0035 (0.0023–0.0056) | 0.0028 (0.0017–0.0041) | 0.185 |
| Transthyretin, ng/pdu | 0.8873 (0.6502–1.2045) | 1.2617 (0.6973–1.8306) | 0.193 |
| Lipocalin-1, ng/pdu | 24.44 (11.17–34.57) | 8.28 (4.15–16.41) | 0.003† |
| PLUNC, ng/pdu | 18.97 (10.72–25.26) | 16.06 (5.05–53.82) | 0.945 |
| PSP94, ng/pdu | 69.26 (46.35–143.43) | 86.67 (53.69–131.44) | 0.479 |
| Nonnormalized data | |||
| Apolipoprotein A1, μg/ml | 597.8 (390.1–1,051.4) | 703.6 (417.4–1,045 3) | 0.537 |
| Albumin, μg/ml | 54.5 (31.1–77.1) | 69.4 (45.3–133.5) | 0.171 |
| α2-HS glycoprotein, μg/ml | 0.30 (0.16–0.46) | 0.34 (0.15–0.83) | 0.698 |
| Transthyretin, ng/ml | 70.3 (43.5–106.9) | 108.6 (78.0–410.7) | 0.018† |
| Lipocalin-1, ng/ml | 1,776.2 (1,069.0–3,006.4) | 1,075.9 (639.6–1,824.7) | 0.052 |
| PLUNC, ng/ml | 1,614.6 (1,128.7–2,252.0) | 2,184.5 (673.7–6,254.6) | 0.361 |
| PSP94, ng/ml | 6,788.3 (4,713.7–9,649.8) | 10,283.5 (6,821.3–18,393 3) | 0.006† |
Definition of abbreviations: α2-HS glycoprotein = α2-Heremans–Schmid glycoprotein; COPD 2 = stage 2 chronic obstructive pulmonary disease; HS = healthy smokers; pdu = pixel density unit; PLUNC = palate, lung, and nasal epithelium clone protein; PSP94 = prostate secretory protein of 94 amino acids.
Data were compared by Mann-Whitney U test; data values shown are medians (interquartile range in brackets). P values < 0.05 were considered significant.
P < 0.05.
P < 0.01.
Figure 5.
Biomarker distribution in nonsmokers, healthy smokers (HS), and smokers with increasing severity of chronic obstructive pulmonary disease (COPD), showing biomarker relationship to disease severity using (a) absolute biomarker values (weight/ml) and (b) values normalized for protein content (weight/pixel density unit [pdu]). Asthmatic nonsmokers are included as a chronic airway disease control. Only the P values of the primary comparison (see Statistical Analyses) between HS and subjects with stage 2 COPD. Lines represent the median values, boxes the standard deviation, and bars the range, excluding outliers. PSP94 = prostate secretory protein of 94 amino acids.
To check the robustness of our proteomic approach, the normalized data were used to compare by Mann-Whitney U test the biomarker quantities from the smaller set of samples that was used for the 2-DGE analysis (n = 9 HS and n = 12 smokers with stage 2 COPD (Table 5); 6 samples did not have sufficient remaining quantity to be analyzed in this manner). Despite reduced sample size, apolipoprotein A1 levels remained significantly different between the two groups (P < 0.01) whereas lipocalin-1 showed borderline significance (P = 0.082). A further protein (albumin) was found to be significantly different between the two groups (P < 0.05), and α2-HS glycoprotein showed borderline significance (P = 0.058) (Table 5). No significant biomarker quantity differences were observed in the samples used for 2-DGE analysis when raw data (i.e., data not normalized for protein content) were used.
TABLE 5.
STATISTICAL ANALYSIS COMPARING BIOMARKER QUANTITIES BETWEEN HEALTHY SMOKERS AND SUBJECTS WITH STAGE 2 CHRONIC OBSTRUCTIVE PULMONARY DISEASE, USING BOTH NORMALIZED AND NONNORMALIZED (RAW) DATA FROM THE SUBSET OF PATIENTS WHOSE SAMPLES WERE USED FOR 2-DGE
| 2-DGE Subset |
|||
|---|---|---|---|
| HS (n = 9) | COPD 2 (n = 12) | Exact Significance | |
| Normalized data | |||
| Apolipoprotein A1, μg/pdu | 8.19 (6.33–25.05) | 5.01 (3.15–6.53) | 0.002* |
| Albumin, μg/pdu | 0.691 (0.524–0.930) | 0.452 (0.349–0.699) | 0.023 |
| α2-HS glycoprotein, μg/pdu | 0.0046 (0.0034–0.0107) | 0.0031 (0.0019–0.004) | 0.058 |
| Transthyretin, ng/pdu | 1.075 (0.936–1.265) | 0.786 (0.525–1.360) | 0.382 |
| Lipocalin-1, ng/pdu | 12.89 (6.77–25.03) | 6.77 (3.61–17.71) | 0.082 |
| PLUNC, ng/pdu | 18.99 (8.15–24.96) | 39.35 (5.46–66.02) | 0.345 |
| PSP94, ng/pdu | 65.65 (40.11–128.75) | 85.46 (58.58–182.73) | 0.277 |
| Nonnormalized data | |||
| Apolipoprotein A1, μg/ml | 750.4 (505.4–2660.9) | 653.9 (394.9–784.6) | 0.193 |
| Albumin, μg/ml | 72.7 (52.3–104.2) | 58.9 (40.3–79.5) | 0.277 |
| α2-HS glycoprotein, μg/ml | 0.44 (0.29–1.66) | 0.36 (0.19–0.85) | 0.464 |
| Transthyretin, ng/ml | 95.5 (77.6–158.8) | 99.2 (59.4–143.7) | 0.917 |
| Lipocalin-1, ng/ml | 1,238.6 (907.3–2,720.4) | 1,068.4 (464.0–1,542.5) | 0.310 |
| PLUNC, ng/ml | 1,974.5 (1,144.8–2,283.9) | 3,789.8 (758.5–9,080.0) | 0.310 |
| PSP94, ng/ml | 7,442.1 (5,319.7–9,850.5) | 9,560 (6,708.7–20,609.3) | 0.148 |
Definition of abbreviations: α2-HS glycoprotein = α2-Heremans–Schmid glycoprotein; 2-DGE = two-dimensional gel electrophoresis; COPD 2 = stage 2 chronic obstructive pulmonary disease; HS = healthy smokers; pdu = pixel density unit; PLUNC = palate, lung, and nasal epithelium clone protein; PSP94 = prostate secretory protein of 94 amino acids.
Data were compared by Mann-Whitney U test; data values shown are medians (interquartile range in brackets). P values < 0.05 were considered significant.
P < 0.05.
P < 0.01.
When post-hoc (exploratory) statistical analysis using the Wilcoxon rank-signed test was applied to the other groups in the cohort (healthy nonsmokers, subjects with COPD stages 1 and 3, and subjects with asthma), the expression of lipocalin-1 was observed to be raised in healthy smokers, when compared with nonsmokers, with borderline significance (P = 0.053), and this trend was reversed in smokers with COPD (P = 0.003) (Figure 5). Apolipoprotein A1 was observed to be decreased solely in COPD when compared with HS, with no difference observed in nonsmokers or subjects with asthma when compared with HS. When analyzed by nonparametric correlation using samples from healthy smokers and smokers with stage 2 COPD only (Kendall's tau b; Table 6), lipocalin-1 concentration positively correlated with FEV1/FVC (τb = 0.31, P = 0.01), and apolipoprotein A1 showed a trend toward correlation (τb = 0.21, P = 0.067) using normalized data. When nonnormalized data were analyzed for association with lung function, both transthyretin and lipocalin-1 were significantly correlated with FEV1/FVC, and α2-HS glycoprotein and PSP94 showed borderline significance; however, apolipoprotein A1 showed no significant correlation. No significant correlations were observed using either normalized or nonnormalized data between biomarkers and FEV1 % predicted values.
TABLE 6.
POST-HOC CORRELATION ANALYSIS TO DETERMINE RELATIONSHIP BETWEEN BIOMARKERS IN SPUTUM AND SPIROMETRIC MEASUREMENTS IN HEALTHY SMOKERS AND SMOKERS WITH STAGE II CHRONIC OBSTRUCTIVE PULMONARY DISEASE
| FEV1, % Predicted |
FEV1/FVC Ratio |
|||
|---|---|---|---|---|
| Biomarker | Nonnormalized | Normalized | Nonnormalized | Normalized |
| Apolipoprotein A1 | −0.042 (0.73) | 0.17 (0.17) | −0.07 (0.54) | 0.21 (0.067) |
| Albumin | −0.057 (0.64) | 0.19 (0.13) | −0.18 (0.11) | 0.001 (0.99) |
| α2-HS glycoprotein | −0.061 (0.62) | 0.068 (0.58) | −0.2 (0.08) | −0.069 (0.54) |
| Transthyretin | −0.141 (0.25) | −0.04 (0.73) | −0.26 (0.02)* | −0.18 (0.11) |
| Lipocalin-1 | 0.07 (0.57) | 0.16 (0.34) | 0.29 (0.01)* | 0.31 (0.01)* |
| PLUNC | −0.13 (0.28) | −0.07 (0.56) | −0.08 (0.48) | −0.04 (0.71) |
| PSP94 | −0.17 (0.16) | −0.05 (0.71) | −0.21 (0.057) | −0.56 (0.62) |
Definition of abbreviations: α2-HS glycoprotein = α2-Heremans–Schmid glycoprotein; COPD 2 = stage II chronic obstructive pulmonary disease; PLUNC = palate, lung, and nasal epithelium clone protein; PSP94 = prostate secretory protein of 94 amino acids.
Analysis was performed using Kendall's tau b, comparing the quantity of each biomarker measured in sputum from healthy smokers and smokers with stage 2 COPD, with their corresponding FEV1 and FEV1/FVC values. Prebronchodilator lung function data were used for the comparison. Biomarker quantities were compared using raw values and also those normalized for protein content. Kendall's tau b values are shown, with P values in parentheses.
P < 0.05 (considered significant).
Bronchial Mucosal Localization of Lipocalin-1 and Apolipoprotein A1
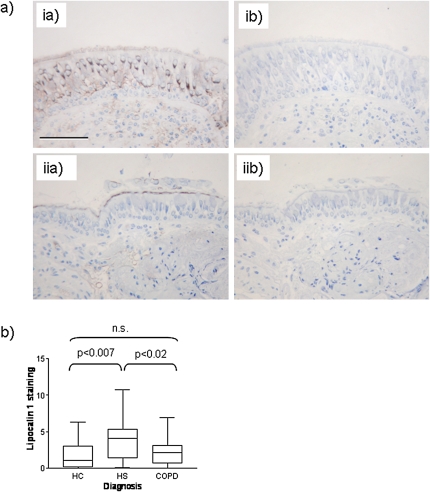
Lipocalin-1 was seen mainly in a perinuclear location within bronchial epithelial cells (Figure 6a). Apolipoprotein A1 staining was located only in the airway and endothelial lumen, suggesting a systemic origin. An increase in lipocalin-1–specific staining was observed in HS individuals when compared with NS (Figure 6b); in accordance with the biomarker quantification in sputum, a reduction in staining was observed in smokers with COPD.
Figure 6.
(a) Immunohistochemical staining for lipocalin-1 and apolipoprotein A1 in bronchial tissue. Panel ia: lipocalin-1 staining by specific antibody is observed in the airway epithelium. Panel ib: IgG control for lipocalin. Panel iia: Apolipoprotein staining by specific antibody is observed in the airway lumen and endothelium. Panel iib: IgG control for apolipoprotein A1. In all cases, specific staining is seen in brown; hematoxylin counterstain is in blue. Scale bar: 100 μm. (b) Comparison of staining intensity for lipocalin-1 in the epithelium of nonsmokers, healthy smokers, and smokers with chronic obstructive pulmonary disease (COPD) determined by image analysis and shown as percentage staining of the epithelium. HC = healthy control subjects; HS = healthy smokers; n.s. = not significant.
Quantification of Plasma Levels of Apolipoprotein A1
No significant differences were seen in apolipoprotein A1 levels in the plasma of healthy smokers and smokers with COPD (data not shown), indicating that the sputum levels of this protein are under independent control of the systemic amounts.
DISCUSSION
This study provides the first large-scale unbiased proteomic analysis of bronchial secretions from patients with COPD. The term “unbiased” in this study refers to the lack of a priori selection for study of specific known proteins and does not exclude a degree of bias because of the proteomic platform used. As the first study of its kind in any airway disease it provides a proof of principle for future biomarker discovery. We have demonstrated that induced sputum samples can be compared by 2-DGE to obtain biomarkers that can be shown subsequently to be differentially expressed in diseased individuals. This was achieved through a combination of a novel approach to sputum processing for 2-DGE analysis and sample normalization, coupled with careful matching of protein spots in gels before statistical analysis. Two main biomarkers were identified: apolipoprotein A1 and lipocalin-1. Their concentrations in sputum were found to be reduced when compared with control smoking subjects who had no evidence of COPD as shown by detailed lung function tests and HRCT. Furthermore, the reduction in sputum concentrations of these two biomarkers, especially, apolipoprotein A1, correlated with the extent of airflow limitation. The value of lipocalin-1 as a predictive biomarker in COPD was confirmed by the finding that its expression in the epithelium, quantified by immunohistochemistry and image analysis, followed the same pattern of differential expression as in sputum when comparing nonsmokers, healthy smokers, and patients with COPD. The lack of difference in the same biomarkers in blood samples confirms the value of using lung-derived samples to identify biomarkers of airway disease.
The results of this study justify a staged approach to biomarker discovery, with each stage enabling narrowing down of DEPs and their validation. The first phase of the program, which used 2-DGE as an unbiased filter, identified 15 potential biomarkers. However, commercially available antibodies were available only for nine of these and it was beyond the scope of this program to develop new antibodies. Of these, only seven showed significant differences between HS and smokers with stage 2 COPD, following preliminary analysis by Western blot, to warrant further quantification by ELISA and Western blot analysis in the full cohort of smokers with or without COPD, age-matched healthy nonsmokers, and patients with asthma. This final analysis confirmed that the concentrations of two biomarkers, lipocalin-1 and apolipoprotein A1, were significantly reduced in the sputum of smoking patients with COPD when compared with healthy smoking control subjects. A further protein (PSP94) showed an association with smoking as it was raised in healthy smokers and smokers with COPD when compared with nonsmokers, and nonsmoking subjects with asthma.
The effect of normalization of proteomic data from induced sputum samples using protein content has not been well examined to date. In this study we chose to use normalized data for comparison, following the same principle used to compare sputum inflammatory cell counts set out by the European Respiratory Society Task Force for Induced Sputum (25). Studies have conclusively shown that only the differential cell counts, whereby the counts of a cell type are expressed as a percentage of total inflammatory cell counts, give reproducible results (25), most probably because absolute cell counts depend on the dilution of the cell fraction by the inhaled saline and sputum viscosity combined with the inevitable variability in duration of effective induction between repeat procedures. Nevertheless, for the sake of completeness, we also analyzed the nonnormalized data and this showed two potential biomarkers (transthyretin and PSP94) that were distinct from those identified on the basis of normalized data. Furthermore, like the biomarkers identified from normalized data, these biomarkers were also correlated with FEV1/FVC ratio, indicating their potential utility as biomarkers providing information about the pathophysiology of COPD in relation to airflow obstruction.
Of the 15 DEPs initially identified, most belong to three major groups: acute-phase proteins (lactoferrin, albumin, and transthyretin), innate defense proteins (PLUNC, lipocalin-1, α1-antitrypsin, and cystatin S), and the transport proteins (apolipoprotein A1, hemoglobin, α2-HS glycoprotein, and PSP94), in addition to IgA, an abundant immune protein present in mucosal secretions. This combination of biomarkers could be seen as representing acute responses to an initial stimulus (smoking), preclinical risk factors for hypertension and dyslipidemia which might lead to comorbidities in the future, and innate defense deficiencies.
Apolipoprotein A1 is known to have a systemic origin, playing a role in cholesterol transport to the liver. Consistent with this, no local origin of this protein was seen by immunohistochemical analysis of bronchial biopsies. Of note, analysis of plasma samples did not indicate a significant difference in apolipoprotein A1 levels in the systemic circulation (data not shown), although a trend was observed toward a reduction in the COPD group. Previous studies have shown a reduction in Lp(a), a lipid measure closely related to apolipoprotein A1, in patients with COPD compared with age-matched healthy smoker control subjects (26); however, no study has examined the relationship between apolipoprotein A1 and COPD in depth or found any causal relationship.
The reduction of a systemically derived protein in diseased airways is counterintuitive to current dogma, which asserts that increased “leakiness” of tissue fluid in emphysema and bronchitis accounts for increased systemic protein load in the sputum. Our results for all systemic proteins show a reverse trend for proteins of systemic origin when using data normalized for protein content, indicating that plasma proteins might in fact have a reduced tendency to appear in induced sputum. We speculate that this occurs because of such factors as increased degradation or consumption. Other members of the apolipoprotein family have been found to be altered in COPD: for example, plasma levels of apolipoprotein E were found to be increased in COPD in one large biomarker study (27) and apolipoprotein A1 and high-density lipoprotein levels were positively correlated with FEV1 (28). In a mouse model, apolipoprotein A1 has been shown to be protective against proinflammatory stimulus and sepsis (29), possibly through sequestration of LPS, and in pigs plasma levels are reduced during sepsis (30).
Consistent with past studies (31–33), lipocalin-1 was shown to be expressed in the bronchial epithelium, in keeping with local production. This localization served to further confirm that staining was specific for lipocalin-1 rather than the neutrophil-derived lipocalin-2 (also known as neutrophil lipocalin), which is a well-recognized biomarker of neutrophilia in a number of diseases including COPD. Its expression showed the same pattern in disease in both sputum and bronchial tissue samples, that is, an increase in HS relative to NS and a reduction in COPD, suggesting up-regulation as a consequence of cigarette smoke exposure and a reduction in diseased airways. Lipocalin-1 is an innate defense molecule and is the archetypal form of the lipocalin superfamily. It has been demonstrated to be a biomarker in tear fluid for a number of conditions including exposure to cigarette smoke (34), with a putative role in antimicrobial defense. Since it was discovered to be identical to Von Ebner's gland protein, a protein abundant in lingual saliva, roles for this protein in taste acuity (35) and protease inhibition (36) have also been proposed. However, because of its structural similarity to other antimicrobial proteins in this superfamily and widespread distribution in the bronchial mucosa, its primary role is thought to be in epithelial defense in the respiratory tract. This is supported by the finding of increased concentrations of this protein in bronchial secretions of patients with cystic fibrosis (31). An increase in healthy smokers, when compared with healthy nonsmoking subjects, was therefore not surprising, and the mechanism leading to this change could involve either direct effects of smoking or an increased susceptibility of smokers for bacterial infections. In contrast, the loss of this protein seen in COPD in the present study was unexpected and may reflect deficiencies in the innate epithelial defense, which could have implications for microbial susceptibility in the epithelium of patients with COPD. Intriguingly, many lipocalins (including but not exclusive to retinol-binding protein) are capable of binding and transporting retinols to their nuclear receptors (37), raising the possibility that the lack of lipocalin-1 may contribute to, or be a result of, lack of retinol stimulation of the epithelium, a known cause of squamous metaplasia in COPD. Although little is known about the modulation of lipocalin-1 expression, its lack of up-regulation in the subjects with asthma in this study suggests that it is not a general marker of inflammatory airway disease.
Two other markers were considered to be of potential relevance from analysis of the nonnormalized data: PSP94 and transthyretin were found to be elevated in smokers with stage 2 COPD when compared with healthy smoker controls. Because PSP94 has been shown to be localized in glandular tissue of the bronchial tissue (38), one might speculate that it is a biomarker of increased glandular activity in any type of airway disease, possibly increasing in expression as a result of smoking. Previous studies have implicated PSP94 (immunoglobulin binding factor, IgBF) in chronic airway disease (39), although no mechanism has been identified. It has been proposed as an immunomodulatory factor important in areas of exposure of epithelium to the external environment.
Transthyretin is thought to have an important role in protein transport as it has the capability to bind to thyroxine, and also to retinols through association with retinol-binding proteins. It has also been used as an indicator of nutritional status (40), and this role may explain in part its elevation in patients with COPD.
Sputum has long been viewed as a source of cells and soluble mediators that might serve as biomarkers of airway disease. However, no study to date has been able to validate a single biomarker that would help to differentiate between healthy smokers and smokers with COPD. An elegant study by Casado and colleagues (41), applying nonquantitative mass spectrometric, non-gel–based techniques (capillary liquid chromatography, CapLC), identified a number of potential biomarkers, some of which were found in the current study, but none of these were clearly shown to be quantitatively different in COPD. Similarly, Gray and colleagues (42) used mass spectrometric techniques to identify several sputum biomarkers altered in several airway diseases; however, smoking was not controlled for in that study. Previous attempts to use induced sputum in 2-DGE proteomic studies have suffered from difficulties in resolving sputum proteins from the induced sputum sample type because of high salt content (due to saline being used to induce expectoration), large amounts of viscous mucus glycoprotein, contamination with salivary proteins, and large amounts of cellular debris and microbial cells. Through our previous work in identifying the global proteome using so-called “shotgun” techniques (1D and 2D Gel-Liquid chromatography-tandem mass spectrometry) (23), we have optimized the methods for handling sputum samples for polyacrylamide gel electrophoresis fractionation and this has enabled us to identify a number of potentially interesting protein species. In particular, we have optimized the processing and 2-DGE fractionation procedures for induced sputum to such an extent that we can confidently obtain good-quality 2-DGE profiles for more than 80% of sputum samples.
This study has a number of limitations. COPD is a syndrome representing a spectrum of diseases composed of varying degrees of emphysema and chronic bronchitis. Considerable effort has gone into finding associations between the morphologic changes in the lungs and lung function, but many gaps in our understanding remain. There have been two main explanations for the decline in lung function: destruction of the parenchyma and changes in the small airways. Although we have identified changes in protein biomarkers with disease, we have not correlated these with changes in the two compartments to determine whether they arise from changes in the alveolar or bronchial tissue, or both. Furthermore, this study has not included ex-smokers because we chose to study current smokers with and without disease, thus keeping smoking as a constant factor.
Contrary to our expectations, this study has not identified a large number of differentially expressed proteins. The reasons for this are not clear but could include the fact that COPD is a heterogeneous disease, with each subphenotype having a different protein fingerprint. Clearly, a study that would address the subphenotypes of COPD was beyond the scope of this project, not least because of the expense and complexity of the 2-DGE approach. Future studies, using high-throughput non-gel–based mass spectrometric methods that pick up low molecular weight proteins, should make this possible. It is also likely that differences in individual biomarkers, which on their own only tend toward significance, are far more important when analyzed by multidimensional, system biology approaches. The observed power of the principal component analysis, which took all the differential expressed proteins spots into account, distinguished between HS and subjects with COPD and this would argue in favor of this approach. Finally, as with any proteomics study of this kind, when throughput is limited by the technology and the desire for stringency in the analysis, the number of data points obtained far exceeds the patient and clinical variable number, leading inevitably to problems with multivariate statistical analysis and multiple testing issues. To deal with this issue, we have validated the potential biomarkers arising from the 2-DGE proteomic findings, using alternative quantitative methods. Further validation in a much larger independent cohort would be the next, highly desirable step to determine the validity of these biomarkers in a clinical setting.
Supplementary Material
Supported by the National Institutes for Health (US): RO1-HL72356–01.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200906-0857OC on January 28, 2010
Conflict of Interest Statement: B.L.N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; P.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; S.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; D.S. received $,001–$5,000 from GlaxoSmithKline, $10,001–$50,000 from Chiesi Pharmaceuticals, $1,001–$5,000 from AstraZeneca, $1,001–$5,000 from CILPA, and $5,001–$10,000 from Allmiral in consultancy fees, $1,001–$5,000 from AstraZeneca, $1,001–$5,000 from MSD, and $1,001–$5,000 from Forest in advisory board fees, $1,001–$5,000 from GlaxoSmithKline, up to $1,000 from Boehringer Ingelheim, and $10,001–$50,000 from AstraZeneca in lecture fees, more than $100,001 from Chiesi Pharmaceuticals, more than $100,001 from GlaxoSmithKline, and more than $100,001 from UCB, more than $100,001 from AstraZeneca, and more than $100,001 from Novartis in industry-sponsored grants; D.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; R.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; G.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; J.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; B.G.-N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; S.W. received up to $1,000 from Kyowa Hakko in institutional consultancy fees, more than $100,001 from GlaxoSmithKline, and more than $100,001 from Novartis in institutional industry-sponsored grants; P.H. received up to $1,000 from Functional Therapeutics for serving on an advisory board, $1,001–$5,000 from GlaxoSmithKline in lecture fees, and $10,001–$50,000 from Schering Plough and $50,001–$100,000 from Shionogi in industry-sponsored grants (clinical trials institutional); D.E.D. received $10,001–$50,000 from Synairgen Research Ltd in consultancy fees, holds a patent from the University of Southampton for interferon-ß for virus-induced exacerbations of asthma and COPD, and holds $50,001–$100,000 in stock ownership or options from Synairgen plc; S.R. received $1,001–$5,000 from Able Associates, up to $1,000 from Adelphi Research, $10,001–$50,000 from Almirall/Prescott, $1,001–$5,000 from APT Pharma/Britnall, $1,001–$5,000 from Aradigm, $1,001–$5,000 from AstraZeneca, $5,001–$10,000 from Boehringer Ingelheim, up to $1,000 from Chiesi, up to $1,000 from Common Health, up to $1,000 from Consult Complete, $1,001–$5,000 from COPDForum, up to $1,000 from Data Monitor, up to $1,000 from Decision Resources, up to $1,000 from Defined Health, $1,001–$5,000 from Dey, up to $1,000 from Dunn Group, up to $1,000 from Eaton Associates, up to $1,000 from Equinox, up to $1,000 from Gerson, $10,001–$50,000 from GlaxoSmithKline, up to $1,000 from Infomed, up to $1,000 from KOL Connection, up to $1,000 from M. Pankove, up to $1,000 from MedaCorp, up to $1,000 from MDRx Financial, up to $1,000 from Mpex, $10,001–$50,000 from Novartis, $10,001–$50,000 from Nycomed, $1,001–$5,000 from Oriel Therapeutics, $1,001–$5,000 from Otsuka, up to $1,000 from Pennside Partners, $5,001–$10,000 from Pfizer (Varenicline), up to $1,000 from PharmaVentures, $1,001–$5,000 from Pharmaxis, up to $1,000 from Price Waterhouse, up to $1,000 from Propagate, up to $1,000 from Pulmatrix, up to $1,000 from Reckner Associates, up to $1,000 from Recruiting Resources, $1,001–$5,000 from Roche, up to $1,000 from Schlesinger Medical, up to $1,000 from Scimed, up to $1,000 from Sudler and Hennessey, $1,001–$5,000 from TargeGen, $1,001–$5,000 from Theravance, $1,001–$5,000 from UBS, $1,001–$5,000 from Uptake Medical, and $5001–$10,000 from Vantage Point Mgmt in consultancy advisory board fees, $10,001–$50,000 from AstraZeneca, $5,001–$10,000 from Boehringer Ingelheim, $10,001–$50,000 from Creative Educational Concept, $5,001–$10,000 from the France Foundation, $1,001–$5,000 from Information TV, $1,001–$5,000 from the Network for Continuing Ed, $10,001–$50,000 from Novartis, $1,001–$5,000 from Pfizer, and $1,001–$5,000 from SOMA in lecture fees, $50,001–$100,000 from AstraZeneca, $50,001–$100,000 from Biomarck, $50,001–$100,000 from Centocor, $50,001–$100,000 from Mpex, $50,001–$100,000 from Nabi, $50,001–$100,000 from Novartis, and $50,001–$100,000 from Otsuka in industry-sponsored grants. S.R. received funding from RJ Reynolds to evaluate the effect of a harm reduction product in normal smokers (1996) and in subjects with chronic bronchitis (1999) and to assess the effect of smoking cessation on lower respiratory tract inflammation (2000); participated in a Philip Morris multicenter study to assess biomarkers of smoke exposure (2002); received funding for a clinical trial from the Institute for Science and Health (2005), which receives support from the tobacco industry, to evaluate biomarkers in exhaled breath associated with smoking cessation and reduction. This study was supplemented with funding from Lorillard and RJ Reynolds. S.R. received a grant from the Philip Morris External Research Program (2005) to assess the impact of cigarette smoking on circulating stem cells in the mouse. S.R. has consulted with RJ Reynolds on the topic of harm reduction until 2007, but did not receive personal remuneration for this. There are no active tobacco industry–funded projects. All ties with tobacco industry companies and entities supported by tobacco companies were terminated in 2007; C.D.O. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; G.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; R.D. received $10,001–$50,000 from Synairgen plc (and is the cofounder of this university spinout company), $10,001–$50,000 from Novartis in lecture fees, more than $100,001 from 3VBioSciences in industry-sponsored grants, has a patent filed for a research model developed by my research group from the University of Southampton, holds more than $100,001 from Synairgen plc, as a cofounder of Synairgen, a University of Southampton spinout company, and received $1,001–$5,000 from Boehringer Ingelheim for sponsorship of attendance of ATS and ERS annual meetings. R.D.'s institution holds more than $100,001 in stock ownership or options in Synairgen. The University of Southampton helped Synairgen to spin out from the research activity of its three founders, one of whom is R.D.
References
- 1.Burrows B, Knudson RJ, Camilli AE, Lyle SK, Lebowitz MD. The “horse-racing effect” and predicting decline in forced expiratory volume in one second from screening spirometry. Am Rev Respir Dis 1987;135:788–793. [DOI] [PubMed] [Google Scholar]
- 2.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 2001;163:1256–1276. [DOI] [PubMed] [Google Scholar]
- 3.Rennard SI. COPD: overview of definitions, epidemiology, and factors influencing its development. Chest 1998;113:235S–241S. [DOI] [PubMed] [Google Scholar]
- 4.Fletcher C, Peto R. The natural history of chronic airflow obstruction. BMJ 1977;1:1645–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthonisen NR, Connett JE, Kiley JP, Altose MD, Bailey WC, Buist AS, Conway WA Jr, Enright PL, Kanner RE, O'Hara P. Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1: the Lung Health Study. JAMA 1994;272:1497–1505. [PubMed] [Google Scholar]
- 6.Franciosi LG, Page CP, Celli BR, Cazzola M, Walker MJ, Danhof M, Rabe KF, la Pasqua OE. Markers of exacerbation severity in chronic obstructive pulmonary disease. Respir Res 2006;7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurst JR, Donaldson GC, Perera WR, Wilkinson TM, Bilello JA, Hagan GW, Vessey RS, Wedzicha JA. Use of plasma biomarkers at exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;174:867–874. [DOI] [PubMed] [Google Scholar]
- 8.Wielders PL, Dekhuijzen PN. Disease monitoring in chronic obstructive pulmonary disease: is there a role for biomarkers? Eur Respir J 1997;10:2443–2445. [DOI] [PubMed] [Google Scholar]
- 9.Aldonyte R, Eriksson S, Piitulainen E, Wallmark A, Janciauskiene S. Analysis of systemic biomarkers in COPD patients. COPD 2004;1:155–164. [DOI] [PubMed] [Google Scholar]
- 10.Casado B, Iadarola P, Luisetti M, Kussmann M. Proteomics-based diagnosis of chronic obstructive pulmonary disease: the hunt for new markers. Expert Rev Proteomics 2008;5:693–704. [DOI] [PubMed] [Google Scholar]
- 11.Barnes PJ, Chowdhury B, Kharitonov SA, Magnussen H, Page CP, Postma D, Saetta M. Pulmonary biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;174:6–14. [DOI] [PubMed] [Google Scholar]
- 12.Pinto-Plata V, Toso J, Lee K, Bilello J, Mullerova H, De SM, Vessey R, Celli B. Use of proteomic patterns of serum biomarkers in patients with chronic obstructive pulmonary disease: correlation with clinical parameters. Proc Am Thorac Soc 2006;3:465–466. [DOI] [PubMed] [Google Scholar]
- 13.Merkel D, Rist W, Seither P, Weith A, Lenter MC. Proteomic study of human bronchoalveolar lavage fluids from smokers with chronic obstructive pulmonary disease by combining surface-enhanced laser desorption/ionization-mass spectrometry profiling with mass spectrometric protein identification. Proteomics 2005;5:2972–2980. [DOI] [PubMed] [Google Scholar]
- 14.Tzortzaki EG, Lambiri I, Vlachaki E, Siafakas NM. Biomarkers in COPD. Curr Med Chem 2007;14:1037–1048. [DOI] [PubMed] [Google Scholar]
- 15.Bowler RP, Canham ME, Ellison MC. Surface enhanced laser desorption/ionization (SELDI) time-of-flight mass spectrometry to identify patients with chronic obstructive pulmonary disease. COPD 2006;3:41–50. [DOI] [PubMed] [Google Scholar]
- 16.Bowler RP, Ellison MC, Reisdorph N. Proteomics in pulmonary medicine. Chest 2006;130:567–574. [DOI] [PubMed] [Google Scholar]
- 17.Ashitani J, Mukae H, Arimura Y, Matsukura S. Elevated plasma procoagulant and fibrinolytic markers in patients with chronic obstructive pulmonary disease. Intern Med 2002;41:181–185. [DOI] [PubMed] [Google Scholar]
- 18.Joos GF, O'Connor B, Anderson SD, Chung F, Cockcroft DW, Dahlen B, DiMaria G, Foresi A, Hargreave FE, Holgate ST, et al. Indirect airway challenges. Eur Respir J 2003;21:1050–1068. [DOI] [PubMed] [Google Scholar]
- 19.O'Donnell RA, Peebles C, Ward JA, Daraker A, Angco G, Broberg P, Pierrou S, Lund J, Holgate ST, Davies DE, et al. Relationship between peripheral airway dysfunction, airway obstruction, and neutrophilic inflammation in COPD. Thorax 2004;59:837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadjicharalambous C, Dent G, May RD, Handy RL, Anderson IK, Davies DE, Djukanovic R. Measurement of eotaxin (CCL11) in induced sputum supernatants: validation and detection in asthma. J Allergy Clin Immunol 2004;113:657–662. [DOI] [PubMed] [Google Scholar]
- 21.Britten KM, Howarth PH, Roche WR. Immunohistochemistry on resin sections: a comparison of resin embedding techniques for small mucosal biopsies. Biotech Histochem 1993;68:271–280. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A, Chernushevich I, Ens W, Standing KG, Thomson B, Wilm M, Mann M. Rapid “de novo” peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom 1997;11:1015–1024. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas B, Skipp P, Mould R, Rennard S, Davies DE, O'Connor CD, Djukanovic R. Shotgun proteomic analysis of human-induced sputum. Proteomics 2006;6:4390–4401. [DOI] [PubMed] [Google Scholar]
- 24.Puddicombe SM, Polosa R, Richter A, Krishna MT, Howarth PH, Holgate ST, Davies DE. Involvement of the epidermal growth factor receptor in epithelial repair in asthma. FASEB J 2000;14:1362–1374. [DOI] [PubMed] [Google Scholar]
- 25.Djukanovic R, Sterk PJ, Fahy JV, Hargreave FE. Standardised methodology of sputum induction and processing. Eur Respir J 2002;37:1s–2s. [DOI] [PubMed] [Google Scholar]
- 26.Basili S, Ferroni P, Vieri M, Cardelli P, Ceci F, Paradiso M, Labbadia G, Gazzaniga PP, Cordova C, Alessandri C. Lipoprotein(a) serum levels in patients affected by chronic obstructive pulmonary disease. Atherosclerosis 1999;147:249–252. [DOI] [PubMed] [Google Scholar]
- 27.Bandow JE, Baker JD, Berth M, Painter C, Sepulveda OJ, Clark KA, Kilty I, VanBogelen RA. Improved image analysis workflow for 2-D gels enables large-scale 2-D gel-based proteomics studies: COPD Biomarker Discovery Study. Proteomics 2008;8:3030–3041. [DOI] [PubMed] [Google Scholar]
- 28.Cirillo DJ, Agrawal Y, Cassano PA. Lipids and pulmonary function in the Third National Health and Nutrition Examination Survey. Am J Epidemiol 2002;155:842–848. [DOI] [PubMed] [Google Scholar]
- 29.Jiao YL, Wu MP. Apolipoprotein A-I diminishes acute lung injury and sepsis in mice induced by lipoteichoic acid. Cytokine 2008;43:83–87. [DOI] [PubMed] [Google Scholar]
- 30.Carpintero R, Pineiro M, Andres M, Iturralde M, Alava MA, Heegaard PM, Jobert JL, Madec F, Lampreave F. The concentration of apolipoprotein A-I decreases during experimentally induced acute-phase processes in pigs. Infect Immun 2005;73:3184–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redl B, Wojnar P, Ellemunter H, Feichtinger H. Identification of a lipocalin in mucosal glands of the human tracheobronchial tree and its enhanced secretion in cystic fibrosis. Lab Invest 1998;78:1121–1129. [PubMed] [Google Scholar]
- 32.Leclair EE. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochem Soc Trans 2003;31:801–805. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl M, Stahlbom B, Tagesson C. Identification of a new potential airway irritation marker, palate lung nasal epithelial clone protein, in human nasal lavage fluid with two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight. Electrophoresis 2001;22:1795–1800. [DOI] [PubMed] [Google Scholar]
- 34.Xu S, Venge P. Lipocalins as biochemical markers of disease. Biochim Biophys Acta 2000;1482:298–307. [DOI] [PubMed] [Google Scholar]
- 35.Lacazette E, Gachon AM, Pitiot G. A novel human odorant-binding protein gene family resulting from genomic duplicons at 9q34: differential expression in the oral and genital spheres. Hum Mol Genet 2000;9:289–301. [DOI] [PubMed] [Google Scholar]
- 36.van't Hoff W, Blankenvoorde MF, Veerman EC, Amerongen AV. The salivary lipocalin von Ebner's gland protein is a cysteine proteinase inhibitor. J Biol Chem 1997;272:1837–1841. [DOI] [PubMed] [Google Scholar]
- 37.Bratt T. Lipocalins and cancer. Biochim Biophys Acta 2000;1482:318–326. [DOI] [PubMed] [Google Scholar]
- 38.Ulvsback M, Lindstrom C, Weiber H, Abrahamsson PA, Lilja H, Lundwall A. Molecular cloning of a small prostate protein, known as β-microsemenoprotein, PSP94 or β-inhibin, and demonstration of transcripts in non-genital tissues. Biochem Biophys Res Commun 1989;164:1310–1315. [DOI] [PubMed] [Google Scholar]
- 39.Ichikawa W, Ogushi F, Tani K, Maniwa K, Kamada M, Ohmoto Y, Sakatani M, Sone S. Characterization of immunoglobulin binding factor in sputum from patients with chronic airway diseases. Respirology 1999;4:375–381. [DOI] [PubMed] [Google Scholar]
- 40.Shenkin A. Serum prealbumin: is it a marker of nutritional status or of risk of malnutrition? Clin Chem 2006;52:2177–2179. [DOI] [PubMed] [Google Scholar]
- 41.Casado B, Iadarola P, Pannell LK, Luisetti M, Corsico A, Ansaldo E, Ferrarotti I, Boschetto P, Baraniuk JN. Protein expression in sputum of smokers and chronic obstructive pulmonary disease patients: a pilot study by CapLC-ESI-Q-TOF. J Proteome Res 2007;6:4615–4623. [DOI] [PubMed] [Google Scholar]
- 42.Gray RD, MacGregor G, Noble D, Imrie M, Dewar M, Boyd AC, Innes JA, Porteous DJ, Greening AP. Sputum proteomics in inflammatory and suppurative respiratory diseases. Am J Respir Crit Care Med 2008;178:444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.