Abstract
Epigenetic silencing of gluthathione-S-transferase pi (GSTP1) is recognized as being a molecular hallmark of human prostate cancer. We investigated the effects of green tea polyphenols (GTP) on GSTP1 re-expression and further elucidated its mechanism of action and long-term safety, compared with nucleoside-analog inhibitor of DNA methyltransferase (DNMT), 5-aza-2′deoxycitidine. Exposure of human prostate cancer LNCaP cells to 1–10μg/mL of GTP for 1–7 days caused a concentration- and time-dependent re-expression of GSTP1 which correlated with DNMT1 inhibition. Methyl-specific-PCR and sequencing revealed extensive demethylation in the proximal GSTP1 promoter and regions distal to the transcription factor binding sites. GTP exposure in a time-dependent fashion diminished the mRNA and protein levels of MBD1, MBD4 and MeCP2; HDAC 1–3 and increased the levels of acetylated histone H3 (LysH9/18) and H4. Chromatin immunoprecipitation assays demonstrated that cells treated with GTP have reduced MBD2 association with accessible Sp1 binding sites leading to increased binding and transcriptional activation of the GSTP1 gene. Exposure of cells to GTP did not result in global hypomethylation, as demonstrated by methyl-specific PCR for LINE-1 promoter; rather, GTP promotes maintenance of genomic integrity. Furthermore, exposure of cells to GTP did not cause activation of the pro-metaststic gene S100P, a reverse response noted after exposure of cells to 5-aza-2′deoxycitidine. Our results, for the first time, demonstrate that GTP has dual potential to alter DNA methylation and chromatin modeling, the two global epigenetic mechanisms of gene regulation, and their lack of toxicity makes them excellent candidates for the chemoprevention of prostate cancer.
Keywords: prostate cancer, GSTP1, chemoprevention, DNA methylation, histone modification, epigenetics
INTRODUCTION
Aberrant hypermethylation of CpG islands in the glutathione-S-transferase pi (GSTP1) gene promoter is increasingly being recognized as a precursor to the genesis of prostate cancer.1, 2 GSTP1 is a member of the glutathione S-transferase superfamily that catalyzes conjugation of the glutathione peptide with electrophilic compounds, including carcinogens, resulting in less toxic and more readily excretable metabolites.3, 4 A genetic variant of GSTP1 has been associated with cancer susceptibility5 and mice lacking GSTP1 exhibit increased skin tumorigenesis,6 findings that corroborate the notion that GSTP1 is a tumor suppressor. In contrast, over-expression of GSTP1 has been associated with the development of some types of cancer and has been noted to be associated with the acquisition of drug resistance in some neoplasms.7, 8 The 5′-untranslated region of GSTP1 contains GC-rich regions including CpG islands occupied by two putative Sp1 binding sites that play a central role in regulating basal levels of GSTP1 transcription.9 Detailed bisulfite sequencing analysis of the CpG islands spanning the core promoter region of the GSTP1 gene has demonstrated that methylation is extensive at essentially all CpG sites in androgen-responsive human prostate cancer LNCaP and MDA PCa 2b cells.10, 11 In clinical specimens, methylation of the GSTP1 promoter is the most frequently detected abnormality, demonstrable in over 90% of invasive cancers and in about 70% of prostatic intraepithelial neoplasia (PIN) lesions, but is rarely detected in normal prostate tissue or in benign hyperplastic tissue.12 It is also detected in a subset of proliferative inflammatory atrophy (PIA) lesions.13 Therefore, promoter methylation and loss of GSTP1 expression have been proposed as markers of prostate cancer.12–14 Furthermore, inhibition of aberrant methylation of GSTP1 may be effective in preventing the onset of the pathogenic process.
In recent years, DNA methylation has emerged as an attractive target in cancer therapeutics.15 DNA methylation is mediated by a family of three DNA methyltransferase enzymes: DNMT1, DNMT3a and DNMT3b.16 Protein and activity levels of DNMT1 have been shown to be elevated in human prostate cancer and in the autochonthous murine model of prostate cancer, TRAMP.17, 18 Hypothetically the use of a DNA methylation inhibitor, 5-aza-2′-deoxycytidine (5-Aza-dC) and zebularine could reverse the methylation process.19 Administration of 5-Aza-dC has been shown to inhibit prostate cancer progression in TRAMP mice.20 However, there are some concerns about the suitability of DNMT inhibitors in anticancer therapy, for two reasons. First, 5-Aza-dC, once incorporated into DNA in place of cytosine, can covalently trap DNA methyltransferase to DNA, causing irreversible inhibition of the enzyme. These covalent enzyme-DNA adducts have been linked to high cytotoxicity.21 This data suggests that, for suppression of tumor growth, inhibition of DNA methylation-independent activities of DNMT1 might be more critical than inhibition of its catalytic activity. Another reason for concern is that catalytic DNMT inhibitors cause ‘global’ demethylation which could trigger the expression of several pro-metastatic genes viz. S100A4, urokinase plasminogen activator (uPA) and S100P, thereby increasing the aggressiveness of the cancer.22–24 With these concerns in mind, it is preferable to target the regulatory protein-protein interactions of DNMT1 rather than just the catalytic activity by selecting effective and minimally toxic agents for DNMT inhibition.
Dietary polyphenols from green tea and its major constituent, epigallocatechin-3-gallate (EGCG) have been demonstrated to possess cancer preventive and therapeutic activity.25, 26 These effects have been attributed to antioxidant and anti-inflammatory properties and to induction of selective apoptosis in cancer cells.27 Recent studies have demonstrated that green tea polyphenols and EGCG inhibit DNA methyltransferase and may reactivate methylation-silenced genes.28 However, the molecular mechanisms underlying gene re-expression and demethylation remain unclear. It has not been ascertained whether regular consumption of green tea polyphenols cause global hypomethylation and genomic instability, hence facilitating cancer progression. The goal of this study is to elucidate the mechanism of action of green tea polyphenols in the process of GSTP1 reactivation, and to address safety issues related to their long-term consumption. Our results, for the first time, demonstrate that green tea polyphenols, at non-toxic doses, mediates dual action to remodel chromatin and alters DNA methylation patterns in the GSTP1 gene promoter.
MATERIALS AND METHODS
Cell Lines and Treatments
Human prostate cancer cell lines LNCaP, MDA PCa 2b and DU145 obtained from American Type Culture Collection (Manassas, VA) and normal human prostate epithelial cells (PrEC), obtained from Lonza Walkersville, Inc. were maintained in appropriate culture conditions. Cells received the following treatments: 5 or 10μM 5-aza-2′-deoxycytidine (Sigma, St. Louis, MO), 10nM Trichostatin A (Sigma), 5–20μM EGCG and 1–10μg/ml Polyphenon E® (Mitsui Norin, Japan) hereafter referred as green tea polyphenols (GTP) for indicated times. Concentrations of 1, 2.5, 5 and 10μg/mL Polyphenon E correspond to 1.4, 3.5, 7 and 14.0 μM EGCG. The details of the constituents present in Polyphenon E® are listed in Supplemental Table, S1.
DNA Methyltransferase Activity Assay
Nuclear extracts from untreated, 5-Aza-dC, EGCG and GTP treated LNCaP cells were prepared using the EpiQuik Nuclear Extraction Kit (Epigentek, New York, NY) as per the manufacturer’s protocol. Protein concentration was estimated and 20μg nuclear lysate was used to measure DNMT activity or inhibition with the EpiQuik DNA Methyltransferase Activity Assay Kit (Epigentek) as per vendor’s protocol.
ELISA Assay for GSTP1
Protein in whole cell lysate prepared from various treated and untreated LNCaP cells was estimated by Bradford Assay. GSTP1 was determined in cell lysate by a standard ELISA assay according to vendor’s protocol (Biotrin International). The substrate reaction was read spectrophotometrically at 450nm using the 96-well automated VersaMax Tunable Microplate Reader (Molecular Devices, Sunnyvale, CA).
HDAC activity assay
HDAC enzymatic activity was analyzed by using Colorimetric HDAC Activity Assay Kit (BioVision, Mountain View, CA) as per manufacturer’s protocol. Briefly, 50μg of whole cell lysate from untreated and treated LNCaP cells were incubated in 96-well plate with HDAC assay buffer and HDAC colorimetric substrate at 37°C for 1 h. The reactions were stopped by adding the Lysine Developer and further incubated at 37°C for 30 min. Nuclear extract from HeLa cells treated with and without TSA were used as negative and positive controls, respectively. Samples were read at 405 nm using a micro-plate reader.
Bisulfite Treatment and Methyl-specific PCR
Bisulfite modification of genomic DNA (2μg) was performed as per the protocol provided with the EZ DNA Methylation–Gold Kit (Zymo, Orange County, CA) followed by additional desalting and purification using the DNA Clean and Concentrator-5 Kit (Zymo). DNA was suspended in 10μlof H2O and stored at −20°C. Primers to perform MS-PCR on the GSTP1 promoter and LINE-1 were designed using the Methyl Primer Express® Software v1.0 (Applied Biosystems, Foster City, CA; Table S2). PCR products were resolved in 4–20% acrylamide-TBE gels and electrophoresed at 200V along with low DNA mass ladder (Invitrogen, Carlsbad, CA), visualized and quantified on the Kodak Image Station 2000R.
Reverse Transcriptase-PCR
First strand synthesis were performed using the High-Capacity cDNA Reverse Transcription Kit (ABI). Gene-specific primers were used to perform the second strand synthesis under the appropriate conditions using 1–2.5U Platinum Taq DNA Polymerase per reaction and 1.5mM MgCl2. GAPDH and TBP, a TATA box-binding protein primers were used as endogenous controls. PCR products were electrophoresed in 4–20% acrylamide-TBE gels along with low DNA mass marker (Invitrogen) and were analyzed as described above.
Genomic Bisulfite Sequencing of GSTP1 Promoter
Genomic DNA from untreated, 5-Aza-dC and GTP-treated cells was isolated using the protocol provided with DNA isolation kit (Roche, Indianapolis, IN) and subjected to PCR reaction for the proximal and distal GSTP1 promoter. PCR products from two independent MS-PCR reactions were resolved on agarose gel; the excised PCR fragments were purified using the ZymocleanGel DNA Recovery Kit (Zymo) and cloned into the TOPO-TA cloning vector (Invitrogen). At least fifteen individual clones for each treatment were sequenced using the T3 and T7 sequencing primers in the ABI Prism 3730 DNA Analyzer Sequencer (Applied Biosystems). The methylation status for each CpG site was determined and compared between untreated LNCaP cells; 5-Aza-dC and GTP treated clones.
Quantitative Real-Time Reverse Transcription-PCR
RNA was extracted and reverse transcribed as described above. Expression patterns of DNMT1, MBD1, MBD4, MeCP2, HDAC1, HDAC2 and HDAC3 were assayed with the appropriate Taq-man gene expression assay systems using the ABI Prism 7500 Real-Time PCR. Details of the expression assay kits used have been provided in Supplemental Table, S2. The mean expression levels are represented as the ratio between the different detectors and GAPDH expression.
Western Blot Analysis
Whole cell, cytoplasmic, nuclear and acid-extracted histone lysate were resolved in 4–20%Tris-glycine polyacrylamide gel, transferred onto nitrocellulose membrane, blocked in 5% nonfat dry milk and probed using appropriate primary antibodies in blocking buffer. The details of antibodies are provided in the Supplemental Table S2. The membrane was then incubated with appropriate secondary antibody conjugated with horse radish peroxidase followed by detection using an enhanced chemiluminescence kit (Amersham Life Sciences Inc.). To ensure equal protein loading, the membranes were stripped (Pierce) and reprobed with anti-Oct 1 or anti-β-actin antibodies (Santa Cruz Biotechnology) for nuclear and whole cell-cytoplasmic lysate, respectively.
Fluorescence Immunocytochemistry
For immunofluorescence studies, cells were grown on glass slides and fixed in ice-cold acetone for 30 min at 4°C, followed by sequential grading with cold ethanol and PBS and incubated with rabbit polyclonal anti-human GSTP1 antibody followed by incubation in polyclonal swine anti-rabbit FITC-tagged antibody (Dako Cytomation). Nuclear staining was done with DAPI, mounted with aqueous gel mount (Biomeda, Foster City, CA) and the fluorescence was detected by an Olympus microscope (Olympus, Model-DX51) with the appropriate filters. The photographs were taken by an Olympus cooled digital camera (Model DP70) and interpreted by MicroSuite Five software (Olympus Imaging Systems).
Acid Extraction of Histone Proteins
Histone proteins were extracted as per the protocol provided by Upstate-Millipore (Temecula, CA). Briefly, treated and untreated cells were suspended in lysis buffer (10mM HEPES, pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, 1.5mM PMSF). Hydrochloric acid was added to a final concentration of 0.2M and the lysates incubated on ice for 30 min followed by centrifugation at 11,000 × g for 10 min at 4°C. The supernatant fraction was collected and either stored at −80°C or 100μl of the fraction dialyzed on MF-Membrane filters (Millipore, Temecula, CA ) twice against 0.1M (0.1N) acetic acid for 30 min each, followed by three dialysis against H2O for 30 min each. The histone extracts were stored at −80°C and quantified by Bradford assay prior to use. 40μg lysate was immunoblotted to probe for total histone H3 and H4 and acetylated histone H3 (Lys 9/18) and H4 proteins as per the protocol provided with the respective antibodies.
Chromatin Immunoprecipitation (ChIP) assay for Sp1 and MBD2 on the Human GSTP1 Promoter
Cells were grown to 70–80% confluence, fixed in 1% formaldehyde, reaction stopped with glycine, washed twice with PBS and lysed in SDS lysis buffer (1%SDS, 10mM EDTA, 50mM Tris-HCl, pH 8.0). Briefly, chromatin was sheared by sonication on the Bioruptor 200 sonicator (Diagenode, Liège, Belgium) with 30 second pulses following 1 min break, followed by centrifugation at 15,000 × g. Diluted supernatants were precleared, blocked with Protein A/G agarose (Millipore, Temecula, CA) and the sonicated chromatin-DNA complex was precipitated overnight with an anti-Sp1 (Millipore) or anti-MBD2 (Imgenex) antibody. Bound DNA was eluted by incubating the beads in elution buffer (0.1 M NaHCO3 and 1% SDS) and reverse cross-linked was performed overnight, purified and amplified using primers flanking the Sp1 binding sites in the human GSTP1 proximal promoter.
Statistical Analysis
Statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey’s significant difference test with 95% confidence limits were used for comparing the effects of different treatment groups. Pearson’s correlation coefficient (r) and P value were also determined to ascertain association between concentration and efficacy. Data are presented in the figures as mean ± SD.
RESULTS
GTP inhibits DNA methyltransferase activity and expression in human prostate cancer cells
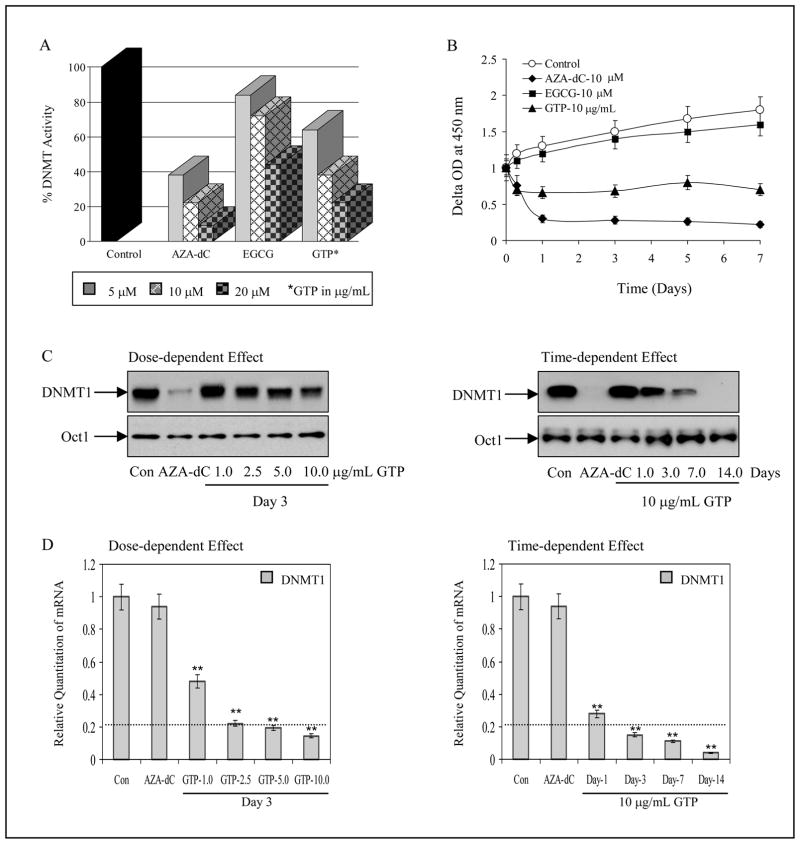
DNA methyltransferase (DNMT) activity was determined in nuclear extract of human prostate cancer LNCaP and MDA PCa 2b cells after various treatments. A dose-dependent inhibition of DNMT activity (16%, 28% and 56%) was observed in LNCaP cells after 7 days exposure of cells with 5, 10 and 20μM EGCG. Interestingly, higher inhibition in DNMT activity (36%, 62% and 78%) was observed after 5, 10 and 20 μg/mL GTP exposure, compared to untreated control. Exposure of cells with 5, 10 and 20μM 5-Aza-dC for 3 days resulted in 62%, 78% and 96% inhibition in DNMT activity (Fig. 1A). We also determined the time-course response of these agents in inhibition of DNMT activity. For time-dependent studies, maximum inhibition in DNMT activity was achieved within 24 h of exposure with these agents, with alterations in the order 5-Aza-dC>GTP>EGCG, which persisted till 7 days of treatment (Fig. 1B). Similar responses were noted in MDA PCa 2b cells after exposure with various agents (data not shown).
Figure 1.
Effect of green tea polyphenols (GTP), its major constituent, epigallocatechin-3-gallate (EGCG) and nucleoside DNA methyltransferase inhibitor, 5-aza-2′deoxycitidine (5-Aza-dC) on 5-cytosine DNA methyltransferase (DNMT) in human prostate cancer LNCaP cells. A, dose-dependent inhibition of DNMT activity by various concentrations of GTP 5–20μg/mL and 5–20μM of EGCG and 5-Aza-dC B, time-dependent inhibition of DNMT activity at 10μg/mL GTP and 10μM of EGCG and 5-Aza-dC for 1–7 days. The results are mean of 6–8 determinations and are analyzed by one way ANOVA, bars ± SD. C, dose- and time- dependent inhibition of DNMT1 protein expression after GTP treatment, anti-Oct-1 is used as loading control, and D, dose- and time- dependent inhibition of DNMT1 mRNA by GTP at indicated doses and times. The results of mRNA levels are mean ± SD of 6–8 determinations **P<0.001, compared to control. Dotted line in the graph represents DNMT1 mRNA levels expressed in normal human prostate epithelial cells. The details are described in ‘Materials and Methods’ section.
Next we determined whether decreased DNMT activity correlated with decreased DNMT at the protein and message levels. Exposure of cells to GTP resulted in a decrease in DNMT protein expression in the nuclear fraction in a dose-dependent fashion, whereas 5-Aza-dC completely inhibited DNMT protein expression. In time-dependent studies, exposure of cells to GTP up to 14 days demonstrated complete inhibition of DNMT protein, as had been noted with 3 days exposure to 5-Aza-dC (Fig. 1C). At the message level, untreated LNCaP cells exhibited the highest levels of DNMT mRNA, while cells treated with varying doses of GTP for 3 days resulted in a dose-dependent 0.5 to 0.85-fold decrease in the message levels of DNMT1 at doses ranging from 1–10μg/mL. Similar results were noted with cells exposed to 10μg/mL GTP from 1–14 days which resulted in 0.7 to 0.95-fold inhibition in the message levels of DNMT1 (Fig. 1D). No significant decrease in the mRNA levels of DNMT1 was observed in 5-Aza-dC treated cells (Fig. 1D). Since we noted that exposure of LNCaP cells to 10μg/mL GTP for 14 days caused growth arrest, we considered a maximum of 7 days of GTP treatment to these cells for further studies. We also determined the protein expression of DNMT3a and DNMT3b after GTP treatment. No significant changes in the levels of these proteins were observed after GTP treatment (data not shown).
GTP reactivates GSTP1 expression in human prostate cancer cells
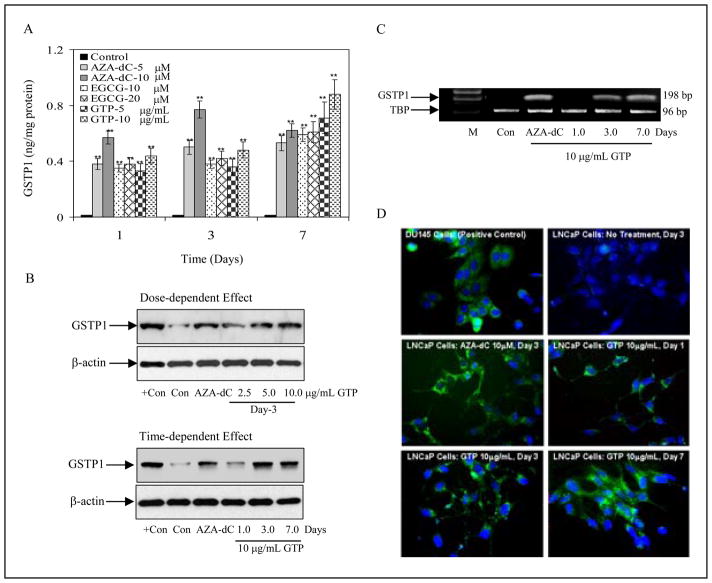
To ascertain whether GTP-induced inhibition of DNMT1 leads to re-expression of GSTP1, we determined protein levels of GSTP1 by ELISA assay after exposure of cells to 5-Aza-dC, EGCG and GTP. Exposure of cells to these agents for one day resulted in significantly increased GSTP1 levels in the following order: 5-Aza-dC (38–57%), GTP (33–44.5%), and EGCG (35–38.2%). The expression of GSTP1 induced by GTP and EGCG was consistently increased in cells exposed up to 7 days whereas the highest levels of GSTP1 peaked after 3 days of treatment with 5-Aza-dC. In summary, 7 days of exposure to GTP and EGCG caused induction of higher GSTP1 levels compared to 5-Aza-dC treatment. Since GTP treatment appeared to be more efficacious in inhibiting DNMT activity than EGCG, we elected to use GTP for further studies (Fig. 1A). We also confirmed induction of GSTP1 expression in LNCaP cells by Western Blot analysis after treatment with various doses of GTP for 3 days. Exposure of cells to GTP resulted in an increase in GSTP1 protein expression in a dose-dependent fashion, a response similarly observed after exposure of cells to 5-Aza-dC. In time-dependent studies, exposure of cells to 10μg/mL GTP for 7 days demonstrated significantly increased GSTP1 protein expression compared with 3 days exposure with 5-Aza-dC (Fig. 2B). We further determined whether increased GSTP1 protein levels correlated with increased message levels. No GSTP1 mRNA levels were detected in the untreated LNCaP cells. Exposure of LNCaP cells to 10μg/mL GTP caused re-expression of the message levels of GSTP1, which increased in a time-dependent fashion. As expected, treatment with 5-Aza-dC also resulted in re-expression of GSTP1 mRNA in these cells (Fig. 2C).
Figure 2.
Effect of green tea polyphenols (GTP), its major constituent, epigallocatechin-3-gallate (EGCG) and nucleoside DNA methyltransferase inhibitor, 5-aza-2′deoxycitidine (5-Aza-dC) on glutathione-S-transferase pi (GSTP1) re-expression in human prostate cancer LNCaP cells. A, dose- and time- dependent re-expression of GSTP1 protein by 5–10μg/mL GTP, 5–10μM 5-Aza-dC and 10–20μM of EGCG by using ELISA assay from Biotrin International. The results are mean of 6–8 determinations and are analyzed by one way ANOVA, bars ± SD and **P<0.001, compared to control. B, dose- and time-dependent re-expression of GSTP1 protein at indicated doses and times of treatment with GTP, positive control used is total lysate from DU145 cells. C, GSTP1 re-expression at message level using 10μg/mL GTP for 1–7 days and 10μM 5-Aza-dC for 3 days, M, marker; TBP is used as internal control. D, immunofluorescence staining for GSTP1 after GTP treatment. Constitutive cytoplasmic expression of GSTP1 in DU145 cells, positive control. Lack of detectable GSTP1 expression in either the cytoplasm or nucleus of naïve human prostate cancer LNCaP cells (control) as evident from the complete absence of green fluorescence in the cells. Re-expression of GSTP1 in LNCaP cells after 1, 3 and 7 days of treatment with 10μg/ml concentration with GTP and with 10μM 5-Aza-dC for 3 days as evident by the green fluorescence in the cytoplasm surrounding the blue, DAPI-stained nucleus.
We next evaluated whether the re-expressed GSTP1 after GTP treatment was optimally localized. In normal prostate epithelial cells GSTP1 is expressed in the cytoplasm. Immunofluorescence studies conducted in LNCaP cells demonstrated that the re-expressed GSTP1 was optimally localized in the cytoplasm and its expression increased in a temporal fashion, with low levels of GSTP1 detected after 1 day of treatment and with further increases after 3 and 7 days of treatment with GTP (Fig. 2D).
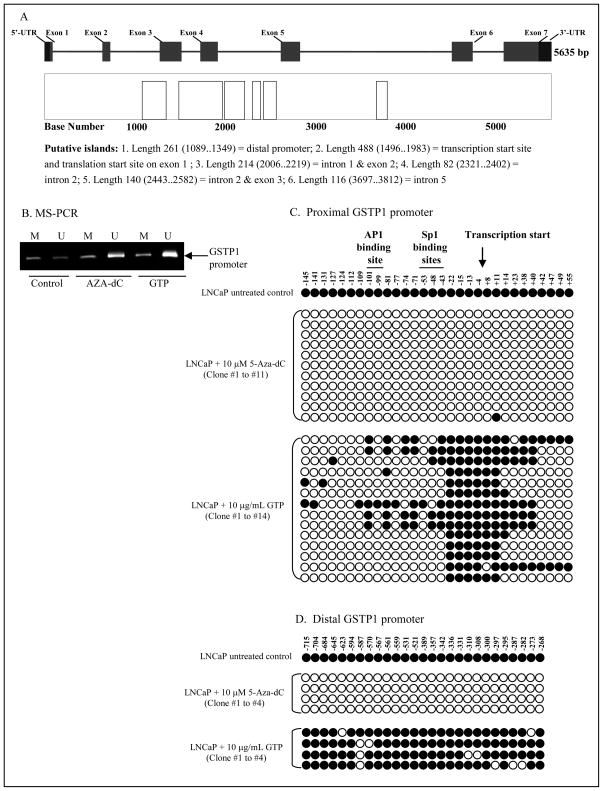
GTP causes demethylation of the GSTP1 promoter in human prostate cancer cells
The human GSTP1 gene is approximately 4kb in length and is comprised of 7-exons and 6-introns that encode a 715-base mRNA. The entire gene contains 6-putative CpG islands susceptible to hypermethylation in human prostate cancer (Fig. 3A). The 1.5kb promoter is present within the second CpG island and is hypermethylated early in prostate cancer leading to gene silencing.10 We evaluated whether the loss in DNMT1 expression and upregulation of GSTP1 expression after exposure of cells to GTP was an epigenetic effect. As shown in fig. 3B, methyl-specific PCR on the proximal GSTP1 promoter revealed that exposure of cells to 10μg/mL of GTP for 7 days resulted in promoter demethylation, potentially leading to GSTP1 reactivation. A significant increase in unmethylated sequences of the GSTP1 promoter was observed in GTP-treated cells, with higher levels of demethylation than were observed after 3 days of treatment with 5-Aza-dC.
Figure 3.
Effect of green tea polyphenols (GTP) on reversal of methylation in human prostate cancer LNCaP cells. A, Map of the GSTP1 gene and putative CpG islands. The relative position and sizes of the 7 exons (solid bars) of the GSTP1 are depicted. B, MS-PCR for GSTP1 promoter on genomic DNA isolated from cells after treatment with 10μg/mL GTP for 7 days and 10μM 5-Aza-dC for 3 days. Displayed are the products generated with primers specific for unmethylated GSTP1 CpG island alleles (U) and for hypermethylated GSTP1 CpG island alleles (M). C, methylation analysis of individual clones from proximal and D, distal GSTP1 promoter after GTP and 5-Aza-dC treatment. Reduction in GSTP1 CpG island hypermethylation was noted in LNCaP DNA by these treatments. At least 15 PCR clones from each treatment were sequenced; the CpG island DNA methylation pattern for selected clones are displayed. Closed circles indicate a methylated CpG and open circles indicate an unmethylated CpG. The details are described in ‘Materials and Methods’ section.
The proximal promoter in the human GSTP1 gene contains two Sp1 binding sites, a consensus AP-1 site and a TATAA box, and is separated from the distal promoter by a distinct (ATAAA)n sequence.29 We next studied whether the CpG islands in the proximal and distal promoters of the GSTP1 gene were similarly demethylated after treatment of LNCaP cells with GTP and 5-Aza-dC. For this, at least fifteen individual clones for each treatment were sequenced after DNA isolation and amplification for the for the proximal and distal GSTP1 promoter. As shown in fig. 3C&D, MS-sequencing performed on the proximal and distal promoters of the GSTP1 gene revealed that GTP caused a time-dependent demethylation of the GSTP1 promoter, with the proximal promoter being more susceptible to GTP-mediated demethylation than the distal promoter. The extent of demethylation varied from clone to clone, although in all clones of the proximal promoter, CpG islands at position −22 to +11 remained methylated. However, this methylation did not seem to inhibit transcription, since after 7 days of GTP exposure, we observed re-expression of GSTP1. During the same time period, no substantial demethylation of CpG dinucleotides was observed in the distal promoter, indicating that GTP exposure results in higher levels of demethylation of the proximal promoter, and this effect was sufficient to overcome epigenetic gene silencing.
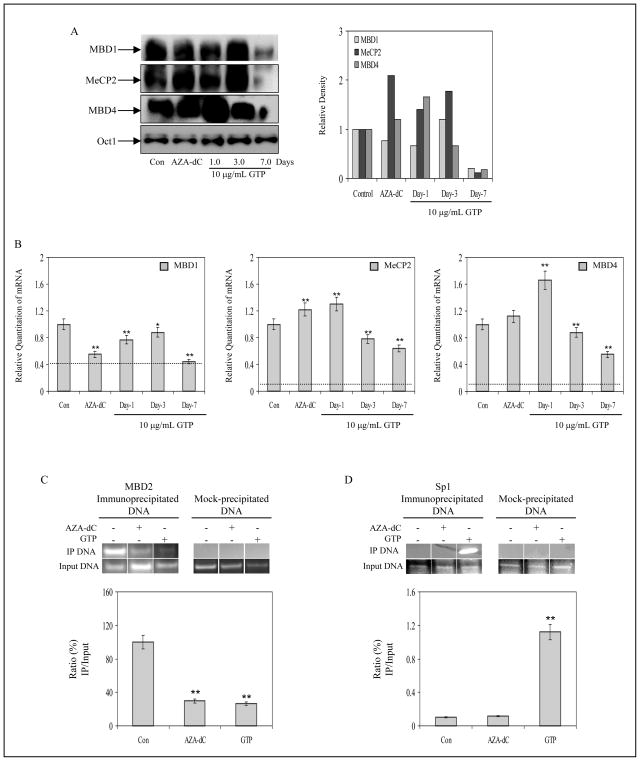
GTP downregulates methyl CpG-binding domain expression in human prostate cancer cells
Transcriptional inhibition on hypermethylated CpG islands is further assisted by the accumulation of the methyl-binding domain (MBD) proteins that are found in association with the 5′-methylated cytosine residue.30 Accumulation of MBDs on the methylated CpG dinucleotides some of which are contained within the consensus sequence of transcription factor binding sites, prevents docking and assembly of the transcriptional machinery. We determined the levels of these proteins by Western Blot analysis in the nuclear fractions and observed an initial increase in MBD1 and MeCP2 after 3 days of GTP exposure, followed by a decrease in their levels after 7 days of GTP treatment. MBD4 exhibited a steady decrease in protein levels after day 1 of GTP treatment (Fig. 4A). We also investigated whether treatment with GTP had any affect on the message levels of MBD proteins. As shown in fig. 4B, real-time PCR data revealed that exposure of LNCaP cells to GTP causes a time-dependent alteration of MBD1, MBD4 and MeCP2 mRNA levels in these cells. Exposure of cells to 10μg/mL GTP caused a 0.2, 0.1 and 0.6-fold decreases in the mRNA levels of MBD1 at 1, 3 and 7 days, respectively as compared to untreated LNCaP cells. GTP exposure initially led to a 1.3 and 1.6-fold increase in MeCP2 and MBD4 levels after 1 day. However, continued GTP treatment for 3 and 7 days resulted in 0.7-0.6 and 0.8-0.5-fold decreases in the transcript levels of MeCP2 and MBD4, respectively. In comparison, exposure of cells to 5-Aza-dC resulted in a 0.5-fold decrease in MBD1, and 1.2 and 1.1-fold increases in the transcript levels of MeCP2 and MBD4 were observed. Previous studies have demonstrated an increase association between MBD2 with the GSTP1 promoter in tumor cells.31, 32 Through chromatin immunoprecipitation (ChIP) assay, we next analyzed whether GTP could affect MBD2 association with the GSTP1 promoter. Compared to untreated control, 7 days of 10μg/mL GTP treatment of LNCaP cells resulted in 74% inhibition of MBD2 associated with the GSTP1 promoter. A 71% decrease in MBD2 association with GSTP1 promoter was observed after 3 day of exposure with 10μM 5-Aza-dC (Fig. 4C).
Figure 4.
Effect of green tea polyphenols (GTP) on methyl-CpG binding proteins and chromatin accessibility by transcription factor in human prostate cancer LNCaP cells. A, protein expression and densitometry of MBD1, MeCP2 and MBD4 in LNCaP cells after 1, 3 and 7 days of treatment with 10 μg/ml concentration with GTP and with 10μM 5-Aza-dC for 3 days. B, alterations in message expression of MBD1, MeCP2 and MBD4 after similar treatments. The results of mRNA levels are mean ± SD of 6–8 determinations **P<0.001, compared to control. Dotted line in the graph represents corresponding MBDs mRNA levels expressed in normal human prostate epithelial cells. C, ChIP assay for MBD2 association with GSTP1 promoter, D, ChIP assay for Sp1 promoter accessibility to the DNA by PCR analysis. GTP treatment to LNCaP cells caused decrease association with MBD2 and an increase in Sp1 binding to the GSTP1 proximal promoter, an effect not observed after treatment with 5-Aza-dC. The details are described in ‘Materials and Methods’ section.
GTP allows binding of the transcription factor Sp1 on the GSTP1 promoter to initiate transcription in human prostate cancer cells
The GSTP1 promoter has two Sp1 binding sites (−57 to −49 and −47 to −39) which are separated by a CpG dinucleotide. In addition, the distal Sp1 site contains another CpG dinucleotide.29 Binding of Sp1 to the GSTP1 promoter is absolutely required for optimal levels of GSTP1 transcription and the distal Sp1 recognition motif (−57 to −49) is the preferred binding site regulating its basal levels.9 Since GTP exposure causes a dose- and time- dependent increase in GSTP1 transcripts, with decreased levels of various repressor MBD proteins, we hypothesized that GTP treatment might lead to increased transcription factor binding and performed ChIP assays for Sp1 after GTP exposure. As shown in fig. 4D, untreated LNCaP cells demonstrated undetectable levels of Sp1 associated with the GSTP1 promoter. In contrast, exposure of LNCaP cells to GTP caused a 12-fold enhanced association of Sp1 with the GSTP1 promoter. We did not observe this phenomenon in 5-Aza-dC treated cells.
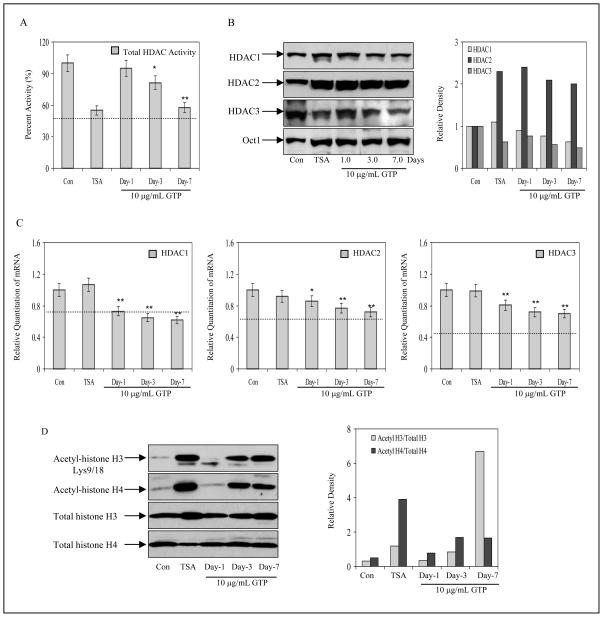
GTP causes inhibition of histone deacetyalses in human prostate cancer cells
For GSTP1 re-expression to occur after GTP treatment, it is reasonable to assume that chromatin re-modeling may follow CpG island demethylation. Chromatin modification is primarily regulated by histone deacetylases (HDACs), enzymes that are responsible for increased deacetylation of histone H3 and H4 proteins observed in epigenetically silenced genes.33 High HDAC activity has been previously reported in LNCaP cells, therefore, we probed whether GTP treatment inhibited HDAC activity.33 An assay representative of total HDAC activity revealed that GTP inhibited HDAC activity in a time-dependent manner. At a concentration of 10μg/mL GTP, a decline of 5%, 19% and 43% HDAC activity was observed in 1, 3 and 7 days. Exposure of cells to HDAC inhibitor tricostatin A (TSA) at 10nM caused 45% inhibition after 24 h of exposure (Fig. 5A). We also probed if inhibition in HDAC activity correlated with a decrease in HDAC protein levels. Exposure of cells with GTP showed a time-dependent decrease in HDAC1 (10% to 38%) and HDAC3 (33 to 51%) protein expression, whereas a 120% increase in HDAC2 was observed after GTP treatment with gradual time-course inhibition from 1–7 days. Similar results were noted after treatment of cells with 10nM TSA for 24 h (Fig. 5B). We also probed if GTP treatment decreased mRNA levels of the HDACs. Real-time PCR analysis revealed that GTP caused a 0.8 to 0.6-fold decrease for HDAC1, 0.8 to 0.7-fold decrease for HDAC2 and 0.8 to 0.7-fold decrease for HDAC3 mRNA levels after 1, 3 and 7 days of treatment, respectively. Treatment with TSA did not result in changes in the HDACs mRNA expression, indicating that while TSA can only inhibit the catalytic function of HDACs, GTP can down-regulate protein and mRNA levels of HDACs (Fig. 5C).
Figure 5.
Effect of green tea polyphenols (GTP) and histone deacetylase inhibitor, Trichostatin A (TSA) on histone deacetylases (HDACs) and histone modification in human prostate cancer LNCaP cells. A, time-dependent inhibition of total HDAC activity after 10μg/mL GTP treatment and 10nM TSA for 24 h. B, protein expression and densitometry of HDAC1, HDAC2 and HDAC3 in LNCaP cells after 1, 3 and 7 days of treatment with 10μg/ml concentration with GTP and with 10nM TSA for 24 h. C, alterations in messenger expression of HDAC1, HDAC2 and HDAC3 after similar treatments. The results of mRNA levels are mean ± SD of 6–8 determinations **P<0.001, compared to control. Dotted line in the graph represents corresponding HDACs mRNA levels expressed in normal human prostate epithelial cells. D, histone acetylation after GTP treatment. Increase in acetylation of H3 at lysine 9 and 18 and total H4 acetylation was observed with10μg/ml concentration with GTP and with 10nM trichostatin A, an inhibitor or histone deacetylase. The details are described in ‘Materials and Methods’ section.
GTP causes acetylation of the histone proteins in human prostate cancer LNCaP cells
We next evaluated whether decrease in HDAC expression by GTP has an effect on histone acetylation or deacetylation. As expected, TSA exposure increased histone H4 (7-fold) and H3 (3-fold) acetylation, particularly at lysine 9 and 18. Similar effects were noted with GTP exposure: 1.1, 3 and 22-fold increases in acetylation of histone H3 and 1.5, 2.5 and 2.2-fold H4 acetylation were observed at day 1, 3 and 7, respectively. These results demonstrate that in addition to altering DNA methylation, GTP exposure is also capable of altering the histone modification processes of epigenetic regulation (Fig. 5D).
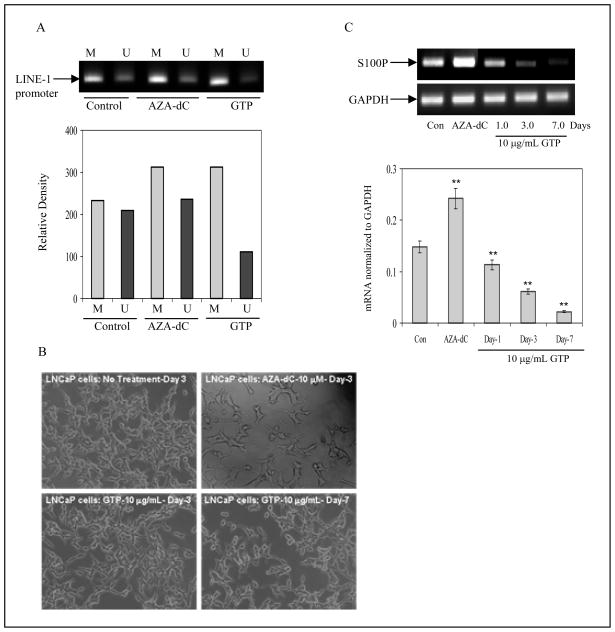
GTP causes alterations in LINE-1 expression in human prostate cancer LNCaP cells
Genome-wide changes in DNA methylation may, in particular, affect those repetitive DNA sequences that are comparatively rich in CpG dinucleotides.34 LINE-1 elements are the most important repetitive transposable elements or retrotransposons littered in the human genome.35 Usually, LINE-1 promoters are heavily methylated in normal cells, limiting activation of retro-elements and restricting their participation in gene recombination. We analyzed LINE-1 promoter sequences in tumor cells exposed to GTP. As shown in fig. 6A, MS-PCR for LINE-1 in LNCaP cells demonstrated a high degree of unmethylated sequence amplification, which further increased after 5-Aza-dC exposure. In sharp contrast, GTP exposure caused a significant decrease in unmethylated LINE-1 sequences. Compared to the findings in unexposed control cells, cells exposed to both 5-Aza-dC and GTP exhibited amplification of the methylated sequence in LINE-1 promoter. These results correlate well with the observation that exposure of LNCaP cells to 10μg/mL GTP for 3 and 7 days caused cell growth inhibition with no apparent signs of toxicity (although some damaged cells with cytoplasmic vesicles were observed), whereas exposure of cells to 10μM 5-Aza-dC for 3 days caused widespread severe cytotoxicity, as evidenced by altered cellular morphology (Fig. 6B).
Figure 6.
Effect of green tea polyphenols (GTP) on cellular toxicity. A, global hypomethylation in human prostate cancer LNCaP cells was assessed by the methylation status of LINE-1 promoter. MS-PCR analysis of CpG island within LINE-1 promoter after treatment of cells with10μg/ml concentration with GTP and with 10μM 5-Aza-dC for 3 days was assessed. A decrease in unmethylated sequences was observed after GTP treatment whereas 5-Aza-dC treatment caused LINE-1 hypomethylation. M, methylated sequences and U, unmethylated sequences. B, 5-Aza-dC treatment to LNCaP cells caused severe cytotoxicity as evident by morphological changes whereas with10μg/ml concentration of GTP did not cause cytotoxicity. C, S100P mRNA expression by RT-PCR analysis after treatment with 5-Aza-dC and GTP. A significant decrease in S100P mRNA expression was observed after GTP treatment but not with 5-Aza-dC. Graph represents mRNA levels normalized to GAPDH. Results are mean ± SD of 3 different experiments. **P<0.001, compared to control. The details are described in ‘Materials and Methods’ section.
GTP causes decrease in the expression of pro-metastatic genes in human prostate cancer LNCaP cells
Another consequence of nucleoside inhibitor-induced DNA hypomethylation is activation of tumor promoting genes and oncogenes.34 DNA hypomethylation as a consequence of 5-Aza-dC treatment has been shown to trigger the expression of prometastatic genes in prostate cancer.22–24 Exposure of LNCaP cells to 10μM 5-Aza-dC for 3 days induced higher levels of S100P mRNA. In contrast, exposure of cells to 10μg/mL of GTP resulted in a time-dependent decrease in levels of S100P mRNA (Fig. 6C). These results suggest that GTP has the potential to inhibit gene-specific DNA hypermethylation, thereby reactivating tumor suppressor genes, but without inducing global hypomethylation and thereby inactivating metastasis-specific genes.
DISCUSSION
The present study demonstrates that restoration of GSTP1 expression in LNCaP cells treated with GTP is accompanied by reversal of epigenetic silencing at two levels which include both promoter demethylation and histone modification. This is the first report where the dual role played by GTP in the reactivation of an epigenetically silenced gene has been reported.
Reversal of epigenetic modifications by classical DNMT inhibitors represents an experimental strategy with great promise in reactivation of epigenetically silenced genes.15 One of the limitations of the use of nucleoside analogs in clinical trials has been the occurrence of side effects, such as thrombocytopenia and neutropenia, which are attributed to cytotoxic effects associated with drug incorporation into the DNA, independent of the DNA hypomethylation effect.36 These problems were addressed by the development of zebularine, a chemically stable cytidine analogue, which is less toxic than the 5-Aza-dC nucleosides, but which also utilizes a covalent trapping mechanism for the inhibition of DNMT.37 Due to the inherent cytotoxicity of such nucleoside analogs, it is important to identify agents that can block DNMT without destabilizing the genome. Our studies demonstrate that GTP has the ability to inhibit DNMT1 expression both at message and protein levels causing a reversal in GSTP1 CpG island hypermethylation and restoration of GSTP1 expression in human prostate cancer LNCaP cells. The epigenetic modifications which occurred after 7 days of GTP-treatment were much higher than that observed after 3 days of 5-Aza-dC treatment. Previous studies have demonstrated that methylation of the outer cytosine in the triplet CpGpG in the Sp1 site inhibits GSTP1 transcription in LNCaP cells, suggesting that this may be a mechanism which initiates hypermethylation of CpG islands.10 Methyl-specific sequencing performed on the proximal and distal promoter clones indicate that GTP demethylates the GSTP1 promoter in a temporal- and site- specific manner, in particular, the CpGpG triplet at the Sp1 binding site, a process that correlates with GSTP1 reactivation. Further studies have demonstrated that GTP and EGCG inhibit DNMT activity in extracts from various human cancer cells.28 In particular, EGCG has been shown to inhibit DNMT activity by blocking the entry of cytosine nucleotide into the active site, by forming hydrogen bonds with two critical residues, Glu1265 and Pro1223, in the catalytic pocket of DNMT. We have additionally demonstrated that GTP not only inhibit catalytic activity of DNMT1, but also down-regulate its protein level. A recent study has demonstrated the affinity of green tea catechins for binding to DNA and RNA.38 Our study indicates that these nucleic acid targets may be more specific than previously thought since real-time studies indicate that GTP are able to down-regulate mRNA levels of DNMT1. Further studies are required to reveal whether this binding of GTP to nucleic acid is sequence specific or random.
Enhancement of epigenetic gene silencing after promoter hypermethylation further involves the activity of methyl-CpG binding domain (MBD) proteins, which specifically bind to methylated DNA.30 The physical association of MBDs with methylated DNA cause stearic hindrance for transcription factor binding. Our present studies demonstrate that at least 3 members of the MBD family viz. MBD1, MeCP2 and MBD4 are expressed in human prostate cancer cells and may be involved in gene repression. Prolonged GTP exposure significantly reduces the expression of these proteins. Additionally, our studies demonstrate that GTP inhibits binding of MBD2 to the GSTP1 gene promoter, a phenomenon which is associated to the repression of the GSTP1 in various tumor cells.31, 32 The presence of HDACs in the MBD repressor complexes remodels the chromatin structure by modulating the acetylome.39 In prostate cancer, chromatin changes on the GSTP1 promoter has been shown to be associated with its inactivation.40 Our studies demonstrate that GTP treatment caused inhibition of HDACs 1–3 activity and expression. Furthermore, GTP exposure resulted in a time-dependent increase in levels of acetylated histone H3 at lysine 9 and 18, and in overall H4 acetylation, and thus may influence GSTP1 promoter access by facilitating chromatin unfolding for transcriptional activation. However, additional mechanistic studies are required to substantiate these findings.
Genome-wide DNA hypomethylation has been shown to be pronounced in several prostatic carcinomas particularly those of higher grade and stage.41 Repetitive sequences such as non-long-terminal repeat (non-LTR) retrotransposons, and LINE-1 elements tend to be hypomethylated, and these abnormalities have been linked to chromosomal aberrations. A correlation between demethylation of LINE-1 sequences and prostate cancer progression has been demonstrated.42, 43 Our studies demonstrate an increase in unmethylated LINE-1 sequences after treatment with 5-Aza-dC, whereas the levels of unmethylated LINE-1 sequences are decreased after treatment with GTP. Furthermore, the use of catalytic inhibitors of DNMT may potentially result in of global hypomethylation, an event that might trigger the expression of pro-metastatic genes.34 It has been demonstrated that prolonged exposure of human prostate cancer cells to 5-Aza-dC induces urokinase plasminogen activator (uPA), S100P and S1004, all of which play critical roles in the development of metastases.22–24 Our studies demonstrated that in contrast to the effects observed with 5-Aza-dC exposure, GTP inhibits the expression of the pro-metastatic gene S100P at the transcription level. Additional studies are required to understand GTP mechanism of action.
Many tumor suppressor and receptor genes have been reported to be hypermethylated and transcriptionally silenced during the development of various human cancers, including prostate cancer.44 It is likely that inhibition of DNMT and histone modification can facilitate reactivation of the silenced genes, and this inhibition could contribute to the prevention or perhaps even to the treatment of cancer.45, 46 The delay in the development of prostate cancer by administration of 5-Aza-dC to TRAMP mice has been recently demonstrated as a ‘proof of principle’ that cancer prevention may be achieved through epigenetic modifications.20 Our previous studies have shown that regular consumption of green tea polyphenols, in amounts comparable to those readily achievable in humans, inhibits prostate tumorigenesis in TRAMP mice.47 The findings in the current study suggest that green tea polyphenols may provide cancer preventive activity through modification of the epigenetic process of gene silencing. The doses of GTP and EGCG used in these studies are physiologically achievable in humans. It should be noted that the epigenetic effects observed in our studies are a consequence of prolonged exposure to green tea polyphenols, emphasizing the epidemiologic observation that regular consumption of green tea reduces the risk of several human cancers.48–50 Although it is at least hypothetically possible that prolonged use of green tea might cause severe inhibition of DNMT activity, resulting in global hypomethylation and genomic instability, our current studies and epidemiologic studies provide no evidence for such an adverse effect related to prolonged exposure to green tea; however, exposure to very high doses might theoretically produce such an effect. Based on our mechanistic findings it is probable that green tea polyphenols hold considerable promise for effective chemoprevention of prostate cancer in humans.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by grants from United States Public Health Services RO1 CA115491 and R21 CA109424 We are thankful to Dr Yukihiko Hara at Mitsui Norin Ltd., Japan for kindly providing Polyphenon E® and EGCG for the study and Dr David Schultz for his assistance with ChIP assays. The authors also acknowledge technical assistance provided by Diane Kocha in the Real-time PCR core facility, Shymali from ABI in data analysis of real-time PCR, Simone Edelheit in the sequencing core, Meenakshi Shukla with the MS-sequencing analysis and Ata Abbas for technical assistance at the Case Western Reserve University.
Footnotes
Novelty: Green tea polyphenols, at non-toxic doses, mediates dual action to remodel chromatin and alters DNA methylation patterns in the GSTP1 gene promoter.
Impact: Green tea polyphenols has dual potential to alter DNA methylation and chromatin modeling, the two global epigenetic mechanisms of gene regulation, and their lack of toxicity makes them excellent candidates for the chemoprevention of prostate cancer.
References
- 1.Lin X, Tascilar M, Lee WH, Vles WJ, Lee BH, Veeraswamy R, Asgari K, Freije D, van Rees B, Gage WR, Bova GS, Isaacs WB, et al. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am J Pathol. 2001;159:1815–26. doi: 10.1016/S0002-9440(10)63028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singal R, van Wert J, Bashambu M. Cytosine methylation represses glutathione S-transferase P1 (GSTP1) gene expression in human prostate cancer cells. Cancer Res. 2001;61:4820–6. [PubMed] [Google Scholar]
- 3.Henderson CJ, McLaren AW, Moffat GJ, Bacon EJ, Wolf CR. Pi-class glutathione S-transferase: regulation and function. Chem Biol Interact. 1998;111–112:69–82. doi: 10.1016/s0009-2797(97)00176-2. [DOI] [PubMed] [Google Scholar]
- 4.Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21–6. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 5.Gsur A, Haidinger G, Hinteregger S, Bernhofer G, Schatzl G, Madersbacher S, Marberger M, Vutuc C, Micksche M. Polymorphisms of glutathione-S-transferase genes (GSTP1, GSTM1 and GSTT1) and prostate-cancer risk. Int J Cancer. 2001;95:152–5. doi: 10.1002/1097-0215(20010520)95:3<152::aid-ijc1026>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 6.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci U S A. 1998;95:5275–80. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen KJ, Newkirk KA, Schumaker LM, Aldosari N, Rone JD, Haddad BR. Glutathione S-transferase pi amplification is associated with cisplatin resistance in head and neck squamous cell carcinoma cell lines and primary tumors. Cancer Res. 2003;63:8097–102. [PubMed] [Google Scholar]
- 8.Chen CL, Sheen TS, Lou IU, Huang AC. Expression of multidrug resistance 1 and glutathione-S-transferase-Pi protein in nasopharyngeal carcinoma. Hum Pathol. 2001;32:1240–4. doi: 10.1053/hupa.2001.28950. [DOI] [PubMed] [Google Scholar]
- 9.Moffat GJ, McLaren AW, Wolf CR. Sp1-mediated transcriptional activation of the human Pi class glutathione S-transferase promoter. J Biol Chem. 1996;271:1054–60. doi: 10.1074/jbc.271.2.1054. [DOI] [PubMed] [Google Scholar]
- 10.Millar DS, Ow KK, Paul CL, Russell PJ, Molloy PL, Clark SJ. Detailed methylation analysis of the glutathione S-transferase pi (GSTP1) gene in prostate cancer. Oncogene. 1999;18:1313–24. doi: 10.1038/sj.onc.1202415. [DOI] [PubMed] [Google Scholar]
- 11.Vidanes GM, Paton V, Wallen E, Peehl DM, Navone N, Brooks JD. Silencing of pi-class glutathione S-transferase in MDA PCa 2a and MDA PCa 2b cells. Prostate. 2002;51:225–30. doi: 10.1002/pros.10093. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem. 2004;91:540–52. doi: 10.1002/jcb.10740. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama M, Bennett CJ, Hicks JL, Epstein JI, Platz EA, Nelson WG, De Marzo AM. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am J Pathol. 2003;163:923–33. doi: 10.1016/s0002-9440(10)63452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins TG, Burns PA, Routledge MN. DNA methylation of GSTP1 as biomarker in diagnosis of prostate cancer. Urology. 2007;69:11–6. doi: 10.1016/j.urology.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008;48:257–76. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- 16.Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim Pol. 2006;53:245–56. [PubMed] [Google Scholar]
- 17.Nelson WG, Yegnasubramanian S, Agoston AT, Bastian PJ, Lee BH, Nakayama M, De Marzo AM. Abnormal DNA methylation, epigenetics, and prostate cancer. Front Biosci. 2007;12:4254–66. doi: 10.2741/2385. [DOI] [PubMed] [Google Scholar]
- 18.Morey SR, Smiraglia DJ, James SR, Yu J, Moser MT, Foster BA, Karpf AR. DNA methylation pathway alterations in an autochthonous murine model of prostate cancer. Cancer Res. 2006;66:11659–67. doi: 10.1158/0008-5472.CAN-06-1937. [DOI] [PubMed] [Google Scholar]
- 19.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 20.McCabe MT, Low JA, Daignault S, Imperiale MJ, Wojno KJ, Day ML. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006;66:385–92. doi: 10.1158/0008-5472.CAN-05-2020. [DOI] [PubMed] [Google Scholar]
- 21.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91:11797–801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Williamson M, Bott S, Brookman-Amissah N, Freeman A, Nariculam J, Hubank MJ, Ahmed A, Masters JR. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–5. doi: 10.1038/sj.onc.1210472. [DOI] [PubMed] [Google Scholar]
- 23.Rehman I, Goodarzi A, Cross SS, Leiblich A, Catto JW, Phillips JT, Hamdy FC. DNA methylation and immunohistochemical analysis of the S100A4 calcium binding protein in human prostate cancer. Prostate. 2007;67:341–7. doi: 10.1002/pros.20401. [DOI] [PubMed] [Google Scholar]
- 24.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17:1081–8. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 25.Chen D, Milacic V, Chen MS, Wan SB, Lam WH, Huo C, Landis-Piwowar KR, Cui QC, Wali A, Chan TH, Dou QP. Tea polyphenols, their biological effects and potential molecular targets. Histol Histopathol. 2008;23:487–96. doi: 10.14670/hh-23.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol(−)-epigallocatechin-3-gallate. Cancer Res. 2006;66:2500–5. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, Ahmad N, Nieminen AL, Mukhtar H. Growth inhibition, cell-cycle dysregulation, and induction of apoptosis by green tea constituent (−)-epigallocatechin-3-gallate in androgen-sensitive and androgen-insensitive human prostate carcinoma cells. Toxicol Appl Pharmacol. 2000;164:82–90. doi: 10.1006/taap.1999.8885. [DOI] [PubMed] [Google Scholar]
- 28.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–70. [PubMed] [Google Scholar]
- 29.Millar DS, Paul CL, Molloy PL, Clark SJ. A distinct sequence (ATAAA)n separates methylated and unmethylated domains at the 5′-end of the GSTP1 CpG island. J Biol Chem. 2000;275:24893–9. doi: 10.1074/jbc.M906538199. [DOI] [PubMed] [Google Scholar]
- 30.Prokhortchouk E, Hendrich B. Methyl-CpG binding proteins and cancer: are MeCpGs more important than MBDs? Oncogene. 2002;21:5394–9. doi: 10.1038/sj.onc.1205631. [DOI] [PubMed] [Google Scholar]
- 31.Lin X, Nelson WG. Methyl-CpG-binding domain protein-2 mediates transcriptional repression associated with hypermethylated GSTP1 CpG islands in MCF-7 breast cancer cells. Cancer Res. 2003;63:498–504. [PubMed] [Google Scholar]
- 32.Bakker J, Lin X, Nelson WG. Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J Biol Chem. 2002;277:22573–80. doi: 10.1074/jbc.M203009200. [DOI] [PubMed] [Google Scholar]
- 33.Patra SK, Patra A, Dahiya R. Histone deacetylase and DNA methyltransferase in human prostate cancer. Biochem Biophys Res Commun. 2001;287:705–13. doi: 10.1006/bbrc.2001.5639. [DOI] [PubMed] [Google Scholar]
- 34.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, Torres A. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–23. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 36.Schwartsmann G, Schunemann H, Gorini CN, Filho AF, Garbino C, Sabini G, Muse I, DiLeone L, Mans DR. A phase I trial of cisplatin plus decitabine, a new DNA-hypomethylating agent, in patients with advanced solid tumors and a follow-up early phase II evaluation in patients with inoperable non-small cell lung cancer. Invest New Drugs. 2000;18:83–91. doi: 10.1023/a:1006388031954. [DOI] [PubMed] [Google Scholar]
- 37.Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321:591–9. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuzuhara T, Sei Y, Yamaguchi K, Suganuma M, Fujiki H. DNA and RNA as new binding targets of green tea catechins. J Biol Chem. 2006;281:17446–56. doi: 10.1074/jbc.M601196200. [DOI] [PubMed] [Google Scholar]
- 39.Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496–503. doi: 10.1038/sj.onc.1205602. [DOI] [PubMed] [Google Scholar]
- 40.Okino ST, Pookot D, Majid S, Zhao H, Li LC, Place RF, Dahiya R. Chromatin changes on the GSTP1 promoter associated with its inactivation in prostate cancer. Mol Carcinog. 2007;46:839–46. doi: 10.1002/mc.20313. [DOI] [PubMed] [Google Scholar]
- 41.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–77. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 42.Florl AR, Steinhoff C, Müller M, Seifer HH, Hader C, Engers R, Ackermann R, Schulz WA. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91:985–94. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brothman AR, Swanson G, Maxwell TM, Cui J, Murphy KJ, Herrick J, Speights VO, Isaac J, Rohr LR. Global hypomethylation is common in prostate cancer cells: a quantitative predictor for clinical outcome? Cancer Genet Cytogenet. 2005;156:31–6. doi: 10.1016/j.cancergencyto.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Karpf AR. Epigenomic reactivation screening to identify genes silenced by DNA hypermethylation in human cancer. Curr Opin Mol Ther. 2007;9:231–41. [PubMed] [Google Scholar]
- 45.Li LC, Carroll PR, Dahiya R. Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J Natl Cancer Inst. 2005;97:103–15. doi: 10.1093/jnci/dji010. [DOI] [PubMed] [Google Scholar]
- 46.Kopelovich L, Crowell JA, Fay JR. The epigenome as a target for cancer chemoprevention. J Natl Cancer Inst. 2003;95:1747–57. doi: 10.1093/jnci/dig109. [DOI] [PubMed] [Google Scholar]
- 47.Gupta S, Hastak K, Ahmad N, Lewin JS, Mukhtar H. Inhibition of prostate carcinogenesis in TRAMP mice by oral infusion of green tea polyphenols. Proc Natl Acad Sci U S A. 2001;98:10350–5. doi: 10.1073/pnas.171326098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurahashi N, Sasazuki S, Iwasaki M, Inoue M, Tsugane S JPHC Study Group. Green tea consumption and prostate cancer risk in Japanese men: a prospective study. Am J Epidemiol. 2008;167:71–7. doi: 10.1093/aje/kwm249. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, Holman CD, Huang JP, Xie X. Green tea and the prevention of breast cancer: a case-control study in Southeast China. Carcinogenesis. 2007;28:1074–8. doi: 10.1093/carcin/bgl252. [DOI] [PubMed] [Google Scholar]
- 50.Larsson SC, Wolk A. Tea consumption and ovarian cancer risk in a population-based cohort. Arch Intern Med. 2005;165:2683–6. doi: 10.1001/archinte.165.22.2683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.