Abstract
Arf family proteins are ≈21 kDa GTP-binding proteins that are critical regulators of membrane traffic and the actin cytoskeleton. Studies examining the complex signaling pathways underlying Arf action have relied on recombinant proteins comprised of Arf fused to epitope tags or proteins such as glutathione-S-transferase or green fluorescent protein for both cell-based mammalian cell studies and bacterially expressed recombinant proteins for biochemical assay. However, the effects of such protein fusions on the biochemical properties relevant to the cellular function have been only incompletely studied at best. Here, we have characterized the effect of C-terminal tagging of Arf1 on (i) function in S. cerevisiae, (ii) in vitro nucleotide exchange, and (iii) interaction with guanine nucleotide exchange factors and GTPase-activating proteins. We found that the tagged Arfs were substantially impaired or altered in each assay, compared to the wild type protein, and these changes are certain to alter actions in cells. We discuss the results related to the interpretation of experiments using these reagents and we propose that authors and editors consistently adopt a few simple rules for describing and discussing results obtained with Arf family members that can be readily applied to other proteins.
Keywords: Arf, ArfGAP, ArfGEF, GST, GFP
Introduction
The Arf family is one of at least four distinct families within the Ras superfamily of regulatory GTP binding proteins. It is among the earliest arising families in the evolution of the GTP-binding proteins (1) and has retained very high levels of primary sequence conservation. The Arf family in mammals can be divided into three groups of proteins; the six Arfs share extensive biochemical and cellular functions and >60% sequence identity, the 22 Arf-like (Arl) proteins are much more divergent in functions and are 40–60% identical to each other or to Arfs, and the two Sar proteins are the best defined functionally and are 20–30% identical to other family members. We focus here on the Arf proteins, though preliminary data indicate that the issues raised here are also relevant to the Arl proteins.
Research aimed at defining the functions, mechanisms of action, and degree of functional conservation or redundancies in the evolution of these essential regulators has typically employed combinations of genetic, biochemical, structural and cell biological approaches. For example, Arfs are essential in the yeast, S. cerevisiae, but lethality can be rescued by any of the human Arfs (2–4). Human Arf1 and S. cerevisiae Arf1 are each 181 residues in length and 74% identical. Genetic studies in yeast have led to some of the earliest evidence for the role of Arfs in membrane traffic (4–6) and the relationship of ArfGEFs with the Sec7 domain (7), and have confirmed the importance of N-myristoylation in cells (5). A multidisciplinary approach is particularly important in studies of the Arfs as they regulate a large number of cellular processes at several locations and interact with large numbers of regulators and effectors (8–12). For example, the Arfs can use at least 16 different guanine nucleotide exchange factors (ArfGEFs; that each share the Sec7 domain (13–15), at least 23 GTPase activating proteins (ArfGAPs; that contain an ArfGAP domain (16,17), and a growing list of >15 effectors with no conserved Arf binding domains (8,10,12). Models for the actions of the Arfs in cells are still incomplete in key details, even for the best established and most studied function as regulators of adaptor recruitment to sites of carrier biogenesis in membrane traffic (8, 12,18,19).
It is widely accepted that activation of Arfs (GTP-binding) is tightly coupled to translocation from cytosol to membranes and is catalyzed by ArfGEFs (8,9). GTP binding by Arfs results in activation of a “myristoyl switch” that exposes the covalently attached myristate and N-terminal, amphipathic helix to the aqueous environment and consequent interaction of each of these components with biological membranes. But the sources of specificity among the Arf isoforms or between Arfs and their GEFs, GAPs, and effectors are poorly understood. This is largely due to the complexity of Arf signaling in cells but also to the high level of similarities in primary sequences (e.g., mammalian Arf1, Arf2, and Arf3 each share ~97% identity). This has led to extensive use of epitope tags and fusion proteins in efforts to distinguish among these closely related proteins in cells. Arf proteins (and a subset of Arls) are covalently modified at the N-terminus by functionally essential N-myristoylation. As a result, epitope tags, fluorescent proteins or other fusions are almost universally added to the C-terminus of Arf family members. Although useful tools, the biochemistry of the modified Arfs has not been well characterized.
We are fortunate to have several structures of Arfs, Arf complexes, and Arf related proteins to aid us in the design of optimal deletion or fusion protein mutants for biochemical and cell biological or even genetic studies. These structures consistently document close proximity of the N- and C-termini (20–26)(Figure 1). We previously reported that changing charged residues in the C-terminus of Arf1 affects function (27). The recent NMR structure of myrArf1 also indicated that modifying the C-terminus may influence function of Arf (20). C-terminal residues L170, L173 and L177 associate directly with the myristate in the GDP-bound conformation and, therefore, must have different contacts when the protein is in the GTP-bound conformation, with myristate exposed to solvent or membranes. A further indication of the functional importance of the C-terminus was the finding that deletion of just 3 amino acids from the C-terminus of Arf1 abrogated binding to PICK1 (28). Interference with membrane association of Arf6 resulting from the C-terminal fusion with GFP has been documented by Schwartz and colleagues (29) and led them to generate an internal fusion protein (site of insertion indicated by red arrow in Figure 1B). The structure of Sar1 and the predicted apposition of the C-terminus to membranes (30–33) also raises concerns that fusion of a protein or a highly negatively charged tag, such as the FLAG or HA epitopes, could interfere with function of Arf proteins by either interfering with membrane association, altering orientation on the membrane or access of a second protein to a surface of Arf, or by interacting with the positively charged residues in the C- and N-termini. We have rendered some hypothetical structural relationships between fused proteins and epitope tags with Arf1 in Figure 1.
Figure 1.
A. Schematic of recombinant human Arf proteins used in this paper. B. Modeled structures of recombinant proteins. 1) Ribbon diagram of Arf1 with bound GDP (orange). The N-terminal 17 residues are blue and the C-terminal residues 165–170 are yellow, 171–175 orange, 176–179 red, and 180–181 pink corresponding to the yeast Arf1-CΔ17, CΔ11, CΔ6 and CΔ2 constructs. The Switch 1 region (S1) is shown in dark gray, and the Switch 2 (S2) region in purple. 2) Arf1•GDP with C-terminal HA epitope tag. The backbone ribbon for the HA tag (YPYDVPDYA) is shown in magenta and its two aspartate residues shown in red. The HA tag is positioned near the five lysine and arginine residues, shown in blue, of the Arf1 N- and Cterminal regions. 3) Model of the Arf1-GFP construct with Arf1 shown in green with bound GDP (orange), GFP shown in magenta, and engineered linker shown in light gray. 4) Model of the GST-Arf1 construct with GST shown in magenta, Arf1 shown in green with bound GDP (orange), and engineered linker shown in light gray. C. SDS-PAGE analysis of purified recombinant Arf1 preparations. Arf1, Arf1-HA, Arf1-GFP and GST-Arf1 (3.2 μg protein) were loaded onto lane 1, 2, 3, and 4, respectively, of a 15% polyacrylamide gel. Their molecular weights are 20.7, 21.6, 47.8, and 48.9 kDa.
We have discovered a number of instances in which changes to the C-termini of Arf proteins result in changes to protein activities, interactions, and functions. We highlight them here because we feel understanding the effect of modifications on protein function is essential to interpreting data generated using the modified proteins.
Mutations at or near the C-terminus alter functions of S. cerevisiae Arf1 in vivo
Three studies in the yeast S. cerevisiae led us to question the assumption that the C-terminus of Arf proteins can be modified without consequences to protein interactions or biological activities. The first of these was the identification of a novel allele of ARF1 (arf1-26) that was found in a screen for suppressors of the temperature sensitivity of cdc1-1 by members of the Garrett lab at Duke (unpublished observations). This allele is likely a partial loss of function mutation as it confers a cold sensitive phenotype in a CDC1 background that is dominant to ARF2 but is suppressed by CEN-ARF1.
When arf1-26 was cloned and sequenced we found it contains a 12 nucleotide deletion, resulting in loss of residues Y167EGL170, which are only 11 residues from the C-terminus. The resulting truncation mutant is similar to wild type Arf1 in that (i) its increased expression is toxic to cells, (ii) it supports vegetative growth in arf2− strains, and (iii) it suppresses lethality of arf1− cells resulting from growth on rich (YPD) media containing 40 mM fluoride when present on a CEN plasmid (4). Why Arf deficient yeast are supersensitive to fluoride is not known but it has proven to be a very specific and useful means of selection and phenotype for characterization of Arf functionality in this organism (4, 34). However, the fact that arf1-26 has at least one phenotype distinct from the wild type protein suggests that some residues in the deleted region, or perhaps the overall length of the C-terminal helix, are important to function.
We next designed a series of truncation mutants at the very C-terminus to determine how close some key residues may be from the end. We generated plasmids directing expression of Arf1 under either the ARF1 or stronger GAL1 promoters and shortened by 2, 6, 11, or 17 residues. The analogous amino acids in human Arf1 are indicated by pink, red, orange and yellow shading in Figure 1B, structure #1. The resulting proteins were then assayed for function using a number of activities previously defined for ARF1 in live cells. RT367 is a haploid strain deleted for chromosomal copies of ARF1 and ARF2 and carrying a low copy (CEN) plasmid with ARF1 under regulation by the GAL1 promoter. Because ARF1 is essential for growth, RT367 will grow only on galactose media and the ARF1 plasmid is stably maintained. Introduction of an ARF1 gene under its own promoter supports vegetative growth of cells on glucose only if the introduced ARF1 allele is functional. Wild type ARF1, arf1-CΔ2, or arf1-CΔ6 were each capable of supporting growth in this assay, while arf1-CΔ11 or arf1-CΔ17 could not.
Similarly, over-expression of Arf1 from the strong GAL1 promoter in cells carrying chromosomal ARF1 is toxic to cells. CEN plasmids directing expression of galactose inducible over-expression of Arf1, Arf1-CΔ2, or Arf1-CΔ6 were each lethal while expression of Arf1-CΔ11 or Arf1-CΔ17 were not. Note that the internal deletion found in arf1-26 is loss of residues 167–170, which is just upstream of the C-terminus of Arf1-C Δ11. We conclude from these results that the C-terminal six residues (176–181) are not essential to any of these activities of Arf1 in yeast cells and that the length of the C-terminus may be shortened by as many as six residues without loss of these functions. However, one or more residues immediately N-terminal to this, residues 171–175, and likely also 167–170 is required for at least some Arf1 functions.
In another study aimed at monitoring localization of Arf1 under different conditions we designed a mutant of ARF1 that carries a short epitope tag at the C-terminus (arf1-myc). This tag adds 19 residues (LVPRGSSMEQKLISEEDLN) to the C-terminus and contains a thrombin site and myc epitope. We generated the arf1-myc plasmid and, separately, strains carrying arf1-myc at the ARF1 locus, using the gene replacement strategy (35,36). The constructs were confirmed by PCR amplification of genomic DNA. CEN plasmids bearing arf1-myc behaved like ARF1 in each of the above cell based assays of toxicity or vegetative growth (over-expression is toxic, rescues arf1−/arf2−, suppresses killing by 40 mM NaF in arf2− background). However, diploid strains bearing arf1-myc as the only ARF genes (arf1-myc/arf1-myc arf2−/arf2−) had completely lost the ability to sporulate (Table I). This was true in both our standard lab strain background and also after crossing the arf1-myc allele into the highly sporulation-competent strain amp179 (the generous gift of Aaron Mitchell; Table I). A role for ARF1 in sporulation was further supported by the observation that a different allele, arf1-3 ([T32I]Arf1), was also found to be sporulation incompetent and rescued by ARF1 but only very weakly by arf1-26 (Table I). Note that T32 is adjacent to T31, the residue most commonly mutated to generate dominant negative alleles of Arf1 and homologous Ras superfamily members though no sporulation defects have been reported for T31-Arf1 mutants and no other phenotypes, comparable to those of T31 mutants, were observed in arf1-26.. Thus, we describe a novel requirement for ARF1 in the sporulation process. We speculate that this requirement for ARF1 in sporulation is likely linked to its more general role in membrane traffic because of the high demand for membrane reorganization to generate asci. We conclude that addition of a 19 residue extension at the C-terminus of Arf1 in yeast interferes with some, but not all, protein-protein interactions and functions in cells. Thus, short C-terminal truncations or extensions can compromise Arf1 function in cells. The physical location of the C-terminus near the N-terminus and (predicted to be) near the membrane interface when the protein is bound are each potentially consistent with indirect or non-specific effects and so we sought more direct tests of effects of C-terminal mutations on protein-protein interactions.
Table 1. Sporulation frequencies of strains carrying different ARF1 mutations.
All strains were deleted for ARF2 and all plasmids were CEN based plasmids carrying ARF1 alleles behind the ARF1 promoter. C202 and C198 were derived from the highly sporulation competent AMP179 strain, while the other strains are derived from TT104 (59).
| Strain |
ARF1 |
Plasmid |
Sporulation (%) |
|---|---|---|---|
| C139 | +/+ | -- | 28.4 |
| C135 | myc/myc | -- | 0.0 |
| RT461 | myc/myc | ARF1 | 10.8 |
| RT462 | myc/myc | arf1-myc | 0.7 |
| RT463 | myc/myc | arf1-CΔ6 | 12.4 |
| C202 | +/+ | 50.6 | |
| C198 | myc/myc | 0.0 | |
| RT465 | myc/myc | ARF1 | 10.2 |
| RT467 | myc/myc | arf1-myc | 0.2 |
| C163 | 1-3/1-3 | 0.0 | |
| RT473 | 1-3/1-3 | ARF1 | 13.3 |
| RT475 | 1-3/1-3 | arf1-myc | 1.4 |
Biochemical assays of Arf interactions with protein modulators
We generated bacterial expression vectors for human Arf1, Arf1-HA, Arf1-GFP and GST-Arf1 (constructs depicted in Figure 1A and B). The recombinant Arf1-HA and Arf1-GFP were generated by PCR with templates of mammalian expression vectors that are commonly used in the research community and, therefore, the exact sequences commonly used to study Arf in mammalian cells. Each of these recombinant proteins was purified from bacteria as described in “Materials and Methods” and the purity of each preparation analyzed by SDS-PAGE (Figure 1C). We also prepared myristoylated proteins by co-expressing the recombinant Arf proteins with N-myristoyltransferase, as previously described (37,38). However, we were unable to obtain homogeneous preparations of either Arf1-HA or Arf1-GFP. In addition, yields of acylated Arf1-HA were less than 5% of the yield of myristoylated Arf1 in similar preparations. To avoid the confounding factor of variable fatty acid modification, we used only non-myristoylated but full-length proteins for the experiments reported in this paper.
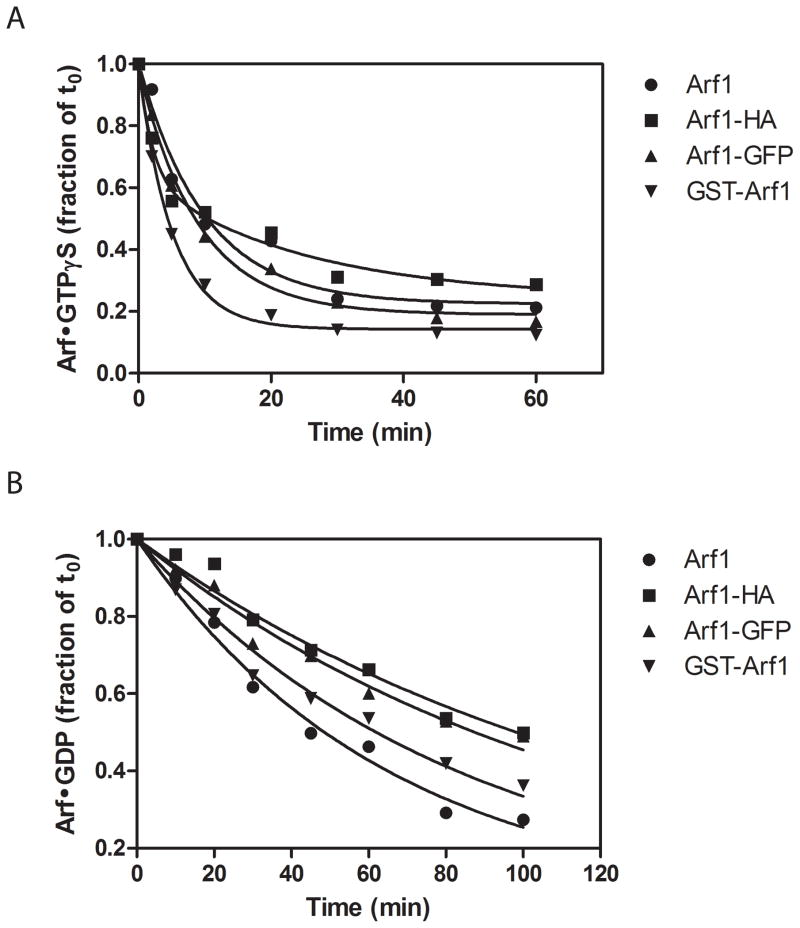
The nucleotide binding and hydrolysis properties of the modified Arfs were characterized for the purified recombinant Arf proteins, in the presence of the Arf GEF ARNO, or different ArfGAPs. We started by examining the rates of GDP and GTPγS dissociation of the purified proteins (Figure 2 and Table 2). In these assays, [Mg2+] was buffered to ≈ 1 μM to increase the rate of nucleotide exchange. As previously described (39), GTPγS dissociated from wild type Arf1 more rapidly than did GDP. Modifications to Arf1 increased the difference between GTP and GDP dissociation rates. Specifically, GTPγS dissociated more rapidly from Arf1-HA and GST-Arf1 than from Arf1, while GDP dissociated more slowly from Arf1-HA, Arf1-GFP and GST-Arf1. The observed differences were typically on the order of two-fold, which are predicted to be well within the range for a physiologically relevant alteration.
Figure 2. Effect of modifying Arf1 on nucleotide dissociation.
The time dependence of GTPγS (A) or GDP (B) dissociation from Arf1, Arf1-HA, Arf1-GFP or GST-Arf1 was determined as described in “Materials and Methods”. The data are expressed as a fraction of radiolabeled nucleotide bound to Arf at the time the reaction was initiated and were fit to single exponential decay equation, except GTPγS dissociation from Arf1-HA which was fit to a double exponential curve. The derived values for the dissociation rate are presented in Table 2. Data shown are one representative of three or four experiments.
Table 2. Effect of modifications of Arf1 on nucleotide dissociation rates.
Nucleotide dissociation rates from Arf1 or tagged Arf1 were determined as described in “Materials and Methods”. The data are the mean ± S.E. of three or four experiments. GTPγS dissociation from Arf1-HA could not be fit to a single exponential, but did fit a double exponential decay equation. The fast component comprised 58 ± 0.04% of the signal. The decay rates are given as k1 and k2, and a weighted average of the two is given for the sake of comparison with wild type and other modified Arf1, whose dissociation time course fit single exponential decay equations.
| koff (min−1) | ||||
|---|---|---|---|---|
| Arf1 | Arf1-HA | Arf1-GFP | GST-Arf1 | |
| GTPγS | 0.098 ± 0.008 | 0.19 ± 0.01** | 0.11 ± 0.01 | 0.18 ± 0.01** |
| k1 | 0.31 ± 0.08 | |||
| k2 | 0.029± 0.007 | |||
| GDP | 0.015 ± 0.001 | 0.0083 ± 0.0001** | 0.0084 ± 0.0002** | 0.010 ± 0.0009** |
p<0.01 compared to wild type.
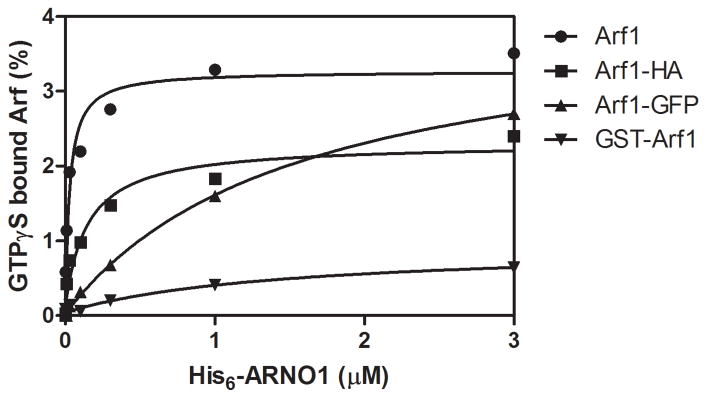
Modification of either the N- or C-terminus led to larger changes in the rate of ARNO-catalyzed nucleotide exchange (Figure 3 and Table 3). For these experiments, ARNO was titrated into a binding reaction that contained recombinant Arf1 preparations and an equimolar concentration of GTPγS. The amount of ARNO required for 50% maximal exchange at 3 min is reported as the C50 in Table 3. Very little activity was observed using GST-Arf1 as a substrate. Exchange on Arf1-GFP required ≈ 50-fold more ARNO than nucleotide exchange on Arf1. Arf1-HA required about 3-fold more ARNO for exchange than Arf1. We also found that Arf1-HA bound less nucleotide than did either Arf1 or Arf1-GFP.
Figure 3. Effect of modifying Arf1 on ARNO1 catalyzed nucleotide exchange.
His6-ARNO1 was titrated into a GTPγS binding reaction as described in “Materials and Methods”. The data were fit to a hyperbola to estimate the amount of ARNO required to reach 50% of the maximal observed exchange in 5 min (C50), which is reported in Table 3. Data shown are representative of four experiments.
Table 3. Effect of modifications of Arf1 on ARNO1 catalyzed nucleotide exchange.
His6-ARNO1 was titrated into a GTPγS binding reaction as described in “Materials and Methods”. The amount of ARNO1 required for 50% maximal exchange at 3 min is reported as the C50. The data are the mean ± S.E. of three experiments.
p<0.05 compared to wild type.
p<0.01 compared to wild type.
No effect
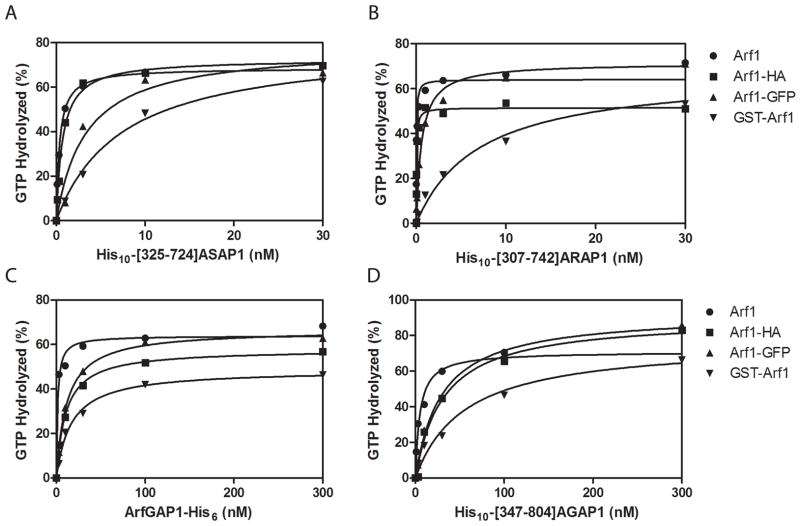
Arf mutations also had differential effects on the ability of Arf GAPs to promote GTP hydrolysis (Figure 4 and Table 4). We titrated purified recombinant Arf GAPs or Arf GAP domains from representative members of 4 subtypes of Arf GAPs (His10-[325-724]ASAP1, His10-[307-742]ARAP1, Arf GAP1-His6 and His10-[347-804]AGAP1) into reactions containing Arf1 loaded with [α32P]GTP. The amount of GAP required for 50% hydrolysis is presented as the C50 in Table 4. These four Arf GAP preparations varied by >150 in their C50 for Arf1, reflecting the differences in enzymatic power of the Arf GAPs that have been previously described (38,40–42). The effect of fusing GFP to Arf1 ranged from a 7.5-fold increase in C50 for AGAP1 to a 23-fold increase in C50 for ARAP1. The effects of the N-terminal GST fusion were more variable, increasing the C50 of AGAP1 by 7.1-fold and the C50 for ARAP1 by 145-fold. C-terminal HA addition had less of an effect on each GAP. The C50 for ARAP1 was affected the least of the 4 GAPs tested (2.4 fold) and Arf GAP1 the most (7.7-fold).
Figure 4. Effect of modifying Arf1 on the ArfGAP catalyzed GTP hydrolysis.
Purified recombinant His10-[325-724]ASAP1 (A), His10-[307-742]ARAP1 (B), ArfGAP1-His6 (C) and His10-[347-804]AGAP1 (D) were titrated into reactions containing Arf1 or modified Arf1 loaded with [α32P]GTP. The data were fit to a hyperbola to estimate the C50 values, which are reported in Table 4. Data shown are representative of at least three experiments.
Table 4. Effect of modifications of Arf1 on ArfGAP-catalyzed GTP hydrolysis.
C50 values were determined as described (51,52). ASAP1, ARAP1, ArfGAP1 and AGAP1 refer to purified recombinant His10-[325–724]ASAP1, His10-[307–742]ARAP1, ArfGAP1-His6 and His10-[347–804]AGAP1, respectively. The data are the mean ± S.E. of three to four experiments. Data were analyzed by repeated measures of ANOVA followed by the Dunnett multiple comparison test on log transformed data.
| C50 (nM) | ||||
|---|---|---|---|---|
| Arf1 | Arf1-HA | Arf1-GFP | GST-Arf1 | |
| ASAP1 | 0.26 ± 0.08 | 0.76 ± 0.13* | 3.70 ± 0.61** | 8.77 ± 1.37** |
| ARAP1 | 0.029 ± 0.008 | 0.071 ± 0.039 | 0.66 ± 0.07** | 4.21 ± 1.57** |
| ArfGAP1 | 1.33 ± 0.42 | 10.3 ± 1.0** | 19.75 ± 8.44** | 30.60 ± 16.62** |
| AGAP1 | 4.79 ± 0.84 | 26.2 ± 6.4* | 36.01 ± 3.87** | 34.43 ± 11.08** |
p<0.05
p<0.01.
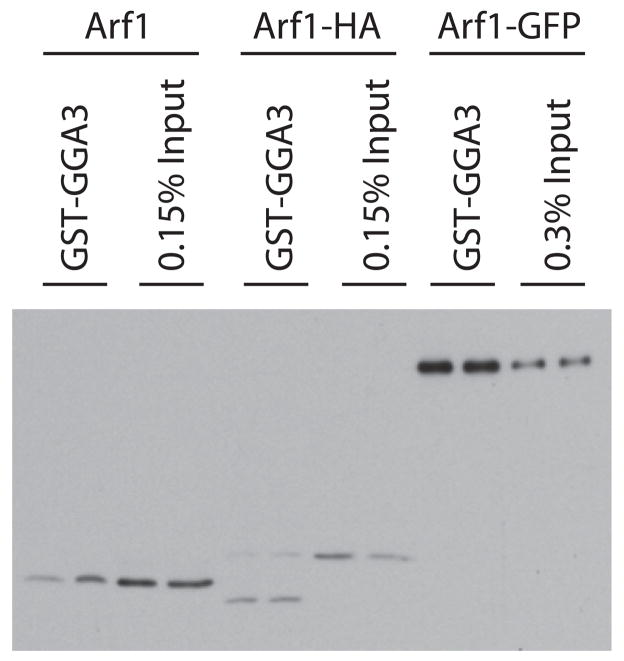
We next determined if the amount of activated Arf1-GFP (Arf1-GFP•GTP) relative to total Arf1-GFP, or Arf1-HA•GTP relative to total Arf1-HA in cells was different than that of Arf1•GTP to total cellular Arf1. HEK293T cells expressing similar levels of recombinant Arf1 or Arf1-GFP were grown and homogenates prepared as described under Methods. GST-[1-313]GGA3 was added to a concentration of at least 100 μM, to selectively precipitate the GTP-bound forms of the proteins (43). The amount of GST-[1-313]GGA3-bound Arf was compared to the amount of Arf in total homogenates using immunoblotting (Figure 5). The level of Arf1•GTP and Arf1-HA•GTP was less than 0.15% of the total Arf1 and Arf1-HA in the sample. In contrast, Arf1-GFP•GTP was greater than 0.3% of the total, yielding an estimated 4- to 8-fold difference in the fractional activation. These results are consistent with the tags affecting interaction with exchange factors and/or GAPs in vivo, as predicted from our in vitro results.
Figure 5. Relative amounts of Arf1•GTP and Arf1-GFP•GTP in HEK293T cells.
Arf1•GTP, Arf1-HA•GTP and Arf1-GFP•GTP levels were determined by selective precipitation of activated Arf from cell lysates using GST-[1-313]GGA3-coated beads. Arf was detected in the precipitates and the indicated amount of cell lysate by immunoblotting. Total cell lysates from Arf1 expressing cells (0.15% of Arf1 and Arf1-HA and 0.3% of Arf1-GFP expressing cells) were loaded to estimate the fraction of activated Arf1, Arf1-HA or Arf1-GFP. The experiment shown here is representative of four independent experiments.
Conclusions
Arfs were discovered ~25 years ago (44–46) and the molecular basis of Arf action and its regulation are still very active and growing areas of research. Complexity in Arf signaling, redundancies in functions between different Arf isoforms, and the high level of structural conservation have slowed the development of detailed molecular models of Arf actions. The increasing use of live cell imaging, proteomics, and localization of proteins in fixed cells and tissues are essential techniques that will continue to be needed in dissecting cell signaling by Arf family members. Epitope tagged Arf proteins and Arf-GFP fusion proteins are valuable tools used for functional cell-based and in vitro studies that have led to important advances. However, as is true of many proteins studied by these techniques, the effects of modifying Arfs on their biochemistry and cellular functions have not been characterized in detail. Here, we summarize a number of studies that indicate tags affect Arf activities and functions in unexpected, multiple and in some cases very dramatic ways, and should be considered when interpreting data generated with tagged Arfs. Consideration of the effects of modifying the termini of Arf1 on its function is relevant at least to other Arf family members; e.g., we have found that addition of small epitope tags to the C-terminus of Arl2 compromises its ability to be imported into mitochondria (47), relative to the untagged protein (C. Zhou and R.A.K., unpublished observations) and Arf6-GFP has been found to have diminished membrane binding compared to wild type Arf6 (29).
Use of yeast as a model system for the study of Arf functions that are relevant to mammalian Arfs is supported by the high degree of structural (74% identity) and functional conservation (each of the five human or one Giardia lamblia Arfs can rescue lethality of arf1− arf2− in yeast)(2–4,48). A number of phenotypes have been described previously and used here to test for functionality of new Arf constructs. These results document a novel role for ARF1 in sporulation and reveal that residues near the C-terminus are critical to some, but not all, Arf1 functions in yeast.
We found that modifications to either terminus of human Arf1 affected spontaneous nucleotide exchange, slowing GDP dissociation and accelerating GTPγS dissociation. The effects on nucleotide exchange can be explained by structural features (20,23–26, 49). The N-terminus of Arf dissociates from the protein core when GDP is exchanged for GTP and the C-terminus is closely apposed to the N-terminus in Arf1•GDP (20). Also, the C-terminus of Arf may make contact with the membrane when Arf is bound to GTP, as suggested by previous biochemical studies (27) and similar to Sar1 (33,30), an Arf family member. Modification of either terminus by fusion of a peptide or protein or changes in charged residues could affect nucleotide exchange by hindering membrane association due to steric hindrance from a bulky group or by charge repulsion.
The mutations of human Arf1 studied here each reduced interactions with Arf regulators and altered nucleotide exchange kinetics. We were not even able to detect ARNO-catalyzed nucleotide exchange when GST was present at the N-terminus. Arf1-GFP was ≈50-fold less efficient a substrate for this GEF than Arf1, and acylated Arf1-GFP was >30-fold less efficient than acylated Arf1 (not shown). Although it is possible that a homogenously myristoylated Arf1-GFP will be a better substrate than the preparation with heterogeneity in the acyl group, the results with the non-acylated proteins together with the acylated proteins lead us to conclude that Arf1-GFP expressed in cells would not be as efficiently activated as endogenous protein by ARNO and likely other exchange factors. Thus, we believe that the presence of the myristate may alter the magnitude of the changes described here but will not change our conclusions on the impact of C-terminal mutations on the biology of the protein. Similarly, we note that a variety of different in vitro and in vivo assays have been used for Arf GEF and Arf GAP by different labs and although not explored here we expect that the magnitude of the observed differences suggest that our conclusions will hold under a variety of different assay conditions. The HA tag had a more modest effect, but did alter activity, with a reduced binding stoichiometry and decreased sensitivity to ARNO. These mutations of Arf1 also affected interaction with Arf GAPs. The effects of a particular modification varied according to the Arf GAP. For instance, the largest effect of the HA tag was on Arf GAP1 whereas the largest effect of GFP was on ARAP1. GFP also affected the amount of the GTP-bound form of the proteins that was recovered from cells, consistent with interfering with exchange factors and GAPs.
We found that the magnitude of the effect was dependent on which Arf GAP was examined and which tag was used. This may also be true for the exchange factors. The total effect would be dependent on the relative contributions of each Arf GEF and Arf GAP and possible effects of the tags on binding to effectors. The relatively large effects of the protein fusions on protein protein interactions could be a consequence of altered orientation of Arf1 on the membrane or of steric hindrance. Because of the very high degree of conservation among different Arfs (both orthologs and paralogs) we also believe that our concerns regarding the unintended effects of C-terminal mutations of Arf1 are relevant to those other proteins.
Predicting the consequences of the changes described here to functionality in cells is difficult because we sampled only a small subset of Arf1 binding partners and because we still do not understand all the sources of specificity in localization and actions of Arfs at different membranes. But given the selective pressure during eukaryotic evolution to develop sensitive and dynamic signaling systems we believe that even a two-fold change in affinity may both directly lead to loss of some signaling and paradoxically could indirectly increase other signaling. This would happen any time an Arf interacts with more than one partner at any location and the affinities for the different partners are differentially affected by the tag. These alterations can be both subtle and unexpected (e.g., the sporulation defect in yeast). But tagging of Arfs is too important and pervasive to completely discontinue in practice. Rather, we argue here for both full disclosure and regular discussions of the issues raised by our results.
We initiated some of the work described above to identify the most suitable recombinant Arf to study the effects of Arf GAPs in cells and were surprised to find the extent to which the affinities for regulatory proteins were affected. While a review of our own published data did not reveal any examples that require either retraction or major changes to models or hypotheses it is likely that the issues raised here impact a number of observations. For example, our difficulty in assessing the isoform specificity of the Arf GAPs in vivo may have been influenced by the tags as well as difficulty in determining whether specific isoforms affect particular biological processes. The effect of GFP fusion at the C-terminus of Arf6 on its membrane association led Schwartz and colleagues to insert a fluorescent protein into an loop of Arf6 (indicated by red arrow in Figure 1B1) to study temporal and spatial activation of Arf6 (29). The issues raised here may be particularly important for live cell kinetic studies. With differential effects of GFP on Arf1 interaction with GEFs, GAPs and effectors, the cycle of GTP binding and hydrolysis by Arf1-GFP is necessarily uncoupled from coat protein dynamics in cells containing endogenous wild type Arf1. Although not directly addressed in this study it is reasonable to also conclude that both the location and kinetics of Arf fusion proteins may be altered from the endogenous Arf in cells as a consequence of tagging, in part because the translocation of Arfs onto membranes is so tightly linked to activation status but also because affinities for binding partners are affected.
In summary, modified Arf proteins are valuable tools but also have some important shortcomings as models for the wild type protein. While researchers in general are well aware of the potential dangers in the use of epitope or GFP tags, the Arf family members are particularly susceptible to being altered in function by the tags as they are (i) small, (ii) undergo large conformational changes, (iii) interact in a regulated and reversible fashion with membranes, (iv) interact with a large number of different proteins that are affected to different degrees by changes, (v) require intact N- and C-terminal residues and structures for functions, and (vi) both termini lie close to the membrane interface. To minimize confusion and misinterpretations, we encourage authors to refer to the modified Arf proteins by their true names at all times and editors to insist upon this rule being consistently applied. If a study is performed with Arf1-GFP the text and figures should always refer to Arf1-GFP and not shorten it to Arf1. If GFP is added to the N-terminus, it should only be referred to as GFP-Arf1 and not Arf1-GFP. If a truncation mutant, e.g., Δ17Arf1, is used in your studies it should only be referred to as Δ17Arf1, and both the arguments supporting extrapolations to the full length protein as well as the dangers should be included in the discussion. A closely related issue is the use of isolated domains, e.g., Sec7 Arf GEF or Arf GAP domains. It should be clear that any time an isolated domain is used as a stand in for the full length protein, functionally important residues and regulatory elements that may be key to biological regulation are likely lost and as a result the observations are strictly only true for the truncation mutation itself and extrapolations to the full length protein should be identified as such. When discussing the results from the use of any such (truncation or tagged) mutants, it would always be helpful to include a brief discussion of the limitations of the use of modified Arf or domain, in comparison to the untagged full length protein and the possibility of unanticipated consequences. At the same time, reviewers of manuscripts should interpret such statements as constructive and intellectually honest and not use them as an excuse to dismiss the study outright as perhaps not being representative of the native protein. These simple guidelines are intended to both simplify and clarify the literature and to help maintain a focus on discovery of the molecular mechanisms of this incredibly complex and important family of cell regulators. We predict that the same guidelines could be constructively employed by researchers and editors for a wide variety of proteins.
Materials and Methods
Yeast constructs and assays
The strains used in this study are shown in Table I. All yeast media used (YPD, YPGal, SD, pre-sporulation, and sporulation) were described previously (50). The S. cerevisiae Arf1-myc protein consists of the full-length Arf1 followed by a six amino acid thrombin cleavage site and thirteen residue myc epitope. The gene encoding this construct was introduced into the genome of TT104 (4) via gene replacement and the resulting strain, RT321, was then crossed to make the diploid C134. Segregants of C134 were later crossed to each other to generate C135, a diploid strain that is homozygous for arf1-myc and arf2−. The same strategies were used to generate other strains shown in Table I.
C-terminal truncations of S. cerevisiae Arf1 were generated by PCR amplification of the open reading frame of wild type Arf1 and cloned into vectors for DNA sequencing using NdeI and XbaI sites. Each was then sub-cloned into pBM272 (CEN4 URA3 pGAL1-10). One set of plasmids inserted the open reading frames directly downstream of the GAL1 promoter. A parallel set of truncation mutants was generated in which the GAL1 promoter was replaced by the ARF1 promoter.
Vegetative growth, lethality from protein over-expression and sensitivity to NaF (40 mM) assays were performed as previously described (4). Sporulation frequencies were determined by counting asci. Strains were grown in pre-sporulation medium at 30°C overnight. They were switched to sporulation medium at equal cell densities (0.8 A600 units/ml), and incubated with shaking for 3–5 days. At least 1,000 cells were counted for each frequency determination and only those asci with three or four spores were considered to have sporulated. Similar frequencies were obtained when sporulation was performed on plates or liquid medium. Each condition was assayed in at least two independent experiments
Recombinant Arf1
A bacterial expression vector for full length Arf1 has been described (50). The mammalian expression vectors for full length Arf1 fused through its C terminus directly to the HA epitope “YPYDVPDYA” (Arf1-HA) (52), and Arf1fused to GFP at Arf residue 178 through the linker “KDPPVAT” (Arf1-GFP)(53), both generous gifts from Dr. J.G. Donaldson, were used as PCR templates and the amplified products were cloned into pET17b (5′ NdeI, 3′ XhoI). For GST-Arf1, the PCR fragment of Arf1 was cloned into pGEX-4T2 (5′ BamHI, 3′ XhoI). The cDNA encoded GST fused through the linker “SDLVPRGS” to the initiating methionine of Arf1. Both strands of PCR products were sequenced to confirm no mutations were introduced. Recombinant Arf1-HA was purified using the same procedure as for Arf1 as described previously (51). The purification of Arf1-GFP was also similar, with a minor modification. After the bacterial cells expressing Arf1-GFP were lysed in 20mM Tris, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol, the lysate was applied to a 5 ml HiTrapQ column (GE Healthcare Life Sciences). Instead of collecting the flow-through as for Arf1 and Arf1-HA, a 25 ml gradient (100 to 600 mM NaCl in lysis buffer) was used to elute Arf1-GFP. The fractions containing Arf1-GFP (green in color) were pooled and further separated by size exclusion chromatography with HiLoad 26/60 Superdex 75 (GE Healthcare Life Sciences). GST-Arf1 was purified with glutathione Sepharose 4B according to the manufacturer’s protocol (GE Healthcare Life Sciences).
Nucleotide binding
Binding reactions were performed as previously described (39), except the reactions contained 0.5 mM large unilamellar vesicles (LUVs) instead of L-α-dimyristoylphosphatidylcholine (DMPC) and cholate. LUVs were prepared by extrusion with lipids purchased from Avanti Polar Lipids. They contained 40% phosphatidylcholine (PC), 25% phosphatidylethanolamine (PE), 15% phosphatidylserine (PS), 9% phosphatidylinositol (PI), 1% phosphatidylinositol 4,5-biphosphate (PIP2), and 10% cholesterol.
Nucleotide dissociation
The method used to determine the dissociation rates of GTPγS and GDP have been described (39), except that the binding of nucleotide was carried out in the presence of LUVs instead of DMPC/cholate. There was an additional modification for GDP dissociation. Instead of incubating with [α32P]GDP directly, Arf1 or tagged Arf1 was incubated with [α32P]GTP for 2 hr in the nucleotide exchange buffer. Then 500 nM ASAP1 PZA was added to the reactions for 10 min to convert the Arf-bound [α32P]GTP to [α32P]GDP. A sample was taken to confirm complete conversion. Data were fit to a single or double exponential decay equations, as indicated in the text, using GraphPad Prism.
ARNO1 GEF activity
His6-ARNO1 was expressed in and purified from bacteria. His6-ARNO1 was titrated into a GTPγS binding reaction in the nucleotide exchange buffer, but with 2 mM MgCl2, 1mM EDTA (instead of 0.5 mM MgCl2, 1 mM EDTA). Less than 0.07% of the Arf exchanged nucleotide in the absence of His6-ARNO with MgCl2 at the higher concentration. The reactions also contained LUVs with a composition of 40% PC, 25% PE, 15% PS, 7.5% PI, 2.5% PIP2, and 10% cholesterol at a total lipid concentration of 0.5 mM. The reactions were initiated by addition of His6-ARNO and stopped after 5 min at 30°C.
GAP Assays
GAP assays were performed as described(37,54,55). Briefly, Arf1 or modified Arf1 was loaded with [α32P]GTP, and His10-[325-724]ASAP1, His10-[307-742]ARAP1, Arf GAP1-His6 and His10-[347-804]AGAP1, prepared as described (38,41,55), were titrated into reactions containing LUVs of a composition appropriate for the particular GAP (For ASAP1, ArfGAP1 and AGAP1, LUVs were composed of 40% PC, 25% PE, 15% PS, 9% PI, 1% PIP2, 10% cholesterol. For ARAP1, the LUVs were composed of 40% PC, 25% PE, 15% PS, 7% PI, 2.5% PIP2, 0.5% PIP3, 10% cholesterol) in 25 mM HEPES, pH7.5, 100 mM NaCl, 1mM DTT, 2mM MgCl2, 1mM GTP. Reactions were initiated by the addition of the substrate Arf1•[α32P]GTP and terminated after 3 min.
Determination of in vivo Arf1•GTP
HEK293T cells were transfected with human Arf1/pcDNA3.1 (for expressing Arf1) and human Arf1/pEGFPN1 (for expressing Arf1-GFP). Cells were harvested and lysed 24 hour after transfection. GST-[1-313]GGA3 was used to capture Arf•GTP as previously described (43). Arf•GTP and total Arf were detected by immunoblotting with monoclonal anti-Arf antibody 1D9 (Abcam)(56).
Modeling
The Arf1 model structure was generated from GDP-bound Arf1 taken from its complex with brefeldin and a sec7 domain (pdb 1R8Q)(22), with the structure of Switch 2 obtained by superposition with yeast GDP-bound, non-myristoylated Arf1 in the free form (provided by J. Prestegard, personal communication), since Switch 2 is altered in the complex. The Arf-HA, Arf-GFP and GST-Arf constructs were generated with Maestro (Schroedinger, Inc. NY, NY) using the structures for green fluorescent protein (pdb 1GFL (57) and glutathione S-transferase (the GST portion of pdb 1B5G, (58), and using engineered linkers and HA-tag generated in random conformations.
Miscellaneous
Statistical analyses were performed with GraphPad Prism®. Data were log transformed prior to analysis to normalize variances when appropriate.
Acknowledgments
We thank Stephen Garrett (UMDNJ-New Jersey Medical School) for generously sharing his unpublished data and providing strains and plasmids containing arf1-26. James Prestegard (Univ of Georgia), Stephen Garret, Julie Donaldson (National Heart, Lung and Blood Institute) and Jonathan Goldberg (Memorial Sloan-Kettering) provided insightful and helpful discussions. We are also indebted to Julie Donaldson, James E. Casanova (University of Virginia) and Elizabeth Sztul (University of Alabama at Birmingham) for reading and critiquing drafts of the manuscript. The work was supported by the Intramural Program of the National Cancer Institute, National Institutes of Health (JMG, XJ, MC, and PR) and by extramural support from NIGMS (GM-61268 and GM-67226; RAK).
Reference List
- 1.Dong JH, Wen JF, Tian HF. Homologs of eukaryotic Ras superfamily proteins in prokaryotes and their novel phylogenetic correlation with their eukaryotic analogs. Gene. 2007;396:116–124. doi: 10.1016/j.gene.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Kahn RA, Kern FG, Clark J, Gelmann EP, Rulka C. Human Adp-Ribosylation Factors -A Functionally Conserved Family of Gtp-Binding Proteins. J Biol Chem. 1991;266:2606–2614. [PubMed] [Google Scholar]
- 3.Lee FJS, Moss J, Vaughan M. Human and Giardia ADP-Ribosylation Factors (Arfs) Complement Arf Function in Saccharomyces-Cerevisiae. J Biol Chem. 1992;267:24441–24445. [PubMed] [Google Scholar]
- 4.Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP Ribosylation Factor Is An Essential Protein in Saccharomyces-Cerevisiae and Is Encoded by 2 Genes. Mol Cell Biol. 1990;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahn RA, Clark J, Rulka C, Stearns T, Zhang CJ, Randazzo PA, Terui T, Cavenagh M. Mutational Analysis of Saccharomyces-Cerevisiae Arf1. J Biol Chem. 1995;270:143–150. doi: 10.1074/jbc.270.1.143. [DOI] [PubMed] [Google Scholar]
- 6.Stearns T, WILLINGHAM MC, Botstein D, Kahn RA. ADP-Ribosylation Factor Is Functionally and Physically Associated with Golgi-Complex. Proc Nat’l Acad Sci USA. 1990;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 8.Gillingham Alison K, Munro Sean. The Small G Proteins of the Arf Family and Their Regulators. Ann Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 9.Nie ZZ, Randazzo PA. Arf GAPs and membrane traffic. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- 10.Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 11.Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 12.Kahn Richard A. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–3879. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 14.Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 15.Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn RA, Bruford E, Inoue H, Logsdon JM, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, Cassel D. Consensus nomenclature for the human ArfGAP domain containing proteins. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cukierman E, Huber I, Rotman M, Cassel D. THE Arf1 GTPase-activating protein - Zinc-finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 18.Logsdon JM, Kahn RA. The Arf Family Tree. In: Kahn RA, editor. Arf family GTPases. Vol. 1. Kluwer; The Netherlands: 2003. pp. 1–21. [Google Scholar]
- 19.Burd CG, Strochlic TI, Setty SRG. Arf-like GTPases: not so Arf-like after all. Trends in Cell Biology. 2004;14:687–694. doi: 10.1016/j.tcb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Liu YZ, Kahn RA, Prestegard JH. Structure and Membrane Interaction of Myristoylated ARF1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menetrey J, Macia E, Pasqualato S, Franco M, Cherfils J. Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nature Structural Biology. 2000;7:466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- 22.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 23.Pasqualato S, Menetrey J, Franco M, Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Reports. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg J. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 25.Amor JC, Horton JR, Zhu XJ, Wang Y, Sullards C, Ringe D, Cheng XD, KAHN RA. Structures of yeast ARF2 and ARL1 - Distinct roles for the N terminus in the structure and function of ARF family GTPases. J Biol Chem. 2001;276:42477–42484. doi: 10.1074/jbc.M106660200. [DOI] [PubMed] [Google Scholar]
- 26.Amor JC, Harrison DH, Kahn RA, Ringe D. Structure Of The Human ADP-Ribosylation Factor-1 Complexed With GDP. Nature. 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- 27.Randazzo PA. Functional interaction of ADP-ribosylation factor 1 with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1997;272:7688–7692. [PubMed] [Google Scholar]
- 28.Takeya R, Takeshige K, Sumimoto H. Interaction of the PDZ domain of human PICK1 with class I ADP-ribosylation factors. Biochem Biophys Res Comm. 2000;267:149–155. doi: 10.1006/bbrc.1999.1932. [DOI] [PubMed] [Google Scholar]
- 29.Hall B, Mclean MA, Davis K, Casanova JE, Sligar SG, Schwartz MA. A fluorescence resonance energy transfer activation sensor for Arf6. Anal Biochem. 2008;374:243–249. doi: 10.1016/j.ab.2007.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi XP, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 31.Fath S, Mancias JD, Bi XP, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 32.Bickford LC, Mossessova E, Goldberg J. A structural view of the COPII vesicle coat. Current Opinion in Structural Biology. 2004;14:147–153. doi: 10.1016/j.sbi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Bi XP, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23 center dot Sar1 complexed with the active fragment of sec31. Developmental Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang CJ, Cavenagh MM, Kahn RA. A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:19792–19796. doi: 10.1074/jbc.273.31.19792. [DOI] [PubMed] [Google Scholar]
- 35.Rothstein RJ. One-Step Gene Disruption in Yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 36.Orrweaver TL, Szostak JW, Rothstein RJ. Genetic Applications of Yeast Transformation with Linear and Gapped Plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 37.Luo RB, Ahvazi B, Amariei D, Shroder D, Burrola B, Losert W, Randazzo PA. Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem J. 2007;402:439–447. doi: 10.1042/BJ20061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo RB, Randazzo PA. Kinetic analysis of Arf GAP1 indicates a regulatory role for coatomer. J Biol Chem. 2008;283:21965–21977. doi: 10.1074/jbc.M802268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Randazzo PA, Terui T, Sturch S, Fales HM, Ferridge AG, Kahn RA. The Myristoylated Amino-Terminus of ADP-Ribosylation Factor-1 Is A Phospholipid-Sensitive and GTP-Sensitive Switch. J Biol Chem. 1995;270:14809–14815. doi: 10.1074/jbc.270.24.14809. [DOI] [PubMed] [Google Scholar]
- 40.Luo RB, Jacques K, Ahvazi B, Stauffer S, Premont RT, Randazzo PA. Mutational analysis of the Arf1 center dot GTP/Arf GAP interface reveals an Arf1 mutant that selectively affects the Arf GAP ASAP1. Curr Biol. 2005;15:2164–2169. doi: 10.1016/j.cub.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 41.Nie ZZ, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Takei J, Randazzo PA. AGAP1, an endosome-associated, phosphoinositide-dependent ADP- ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biol Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 42.Jian XY, Brown P, Schuck P, Gruschus JM, Balbo A, Hinshaw JE, Randazzo PA. Autoinhibition of Arf GTPase-activating Protein Activity by the BAR Domain in ASAP1. J Biol Chem. 2009;284:1652–1663. doi: 10.1074/jbc.M804218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon HY, Bonifacino JS, Randazzo PA. In vitro assays of Arf1 interaction with GGA proteins. Gtpases Regulating Membrane Dynamics. 2005;404:316–332. doi: 10.1016/S0076-6879(05)04028-0. [DOI] [PubMed] [Google Scholar]
- 44.Kahn RA, Gilman AG. The Protein Cofactor Necessary for ADP-Ribosylation of Gs by Cholera-Toxin Is Itself a GTP Binding-Protein. J Biol Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- 45.Kahn RA, Gilman AG. Purification of A Protein Cofactor Required for Adp-Ribosylation of the Stimulatory Regulatory Component of Adenylate-Cyclase by Cholera-Toxin. J Biol Chem. 1984;259:6228–6234. [PubMed] [Google Scholar]
- 46.Sewell JL, Kahn RA. Sequences of the Bovine and Yeast ADP-Ribosylation Factor and Comparison to Other GTP-Binding Proteins. Proc Nat’l Acad Sci USA. 1988;85:4620–4624. doi: 10.1073/pnas.85.13.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharer JD, Shern JF, Van Valkenburgh H, Wallace DC, Kahn RA. ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol Biol Cell. 2002;13:71–83. doi: 10.1091/mbc.01-05-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Reports. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherman F. Getting Started with Yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 51.Randazzo PA, Weiss O, Kahn RA. Preparation of Recombinant ADP-Ribosylation Factor. Methods in Enzymology. 1992;219:362–369. doi: 10.1016/0076-6879(92)19036-6. [DOI] [PubMed] [Google Scholar]
- 52.Teal SB, Hsu VW, Peters PJ, Klausner RD, Donaldson JG. An activating mutation in Arf1 stabiliizes coatomer binding to Golgi membranes. J Biol Chem. 1994;269:3135–3138. [PubMed] [Google Scholar]
- 53.Presley JF, Ward TH, Pfeifer AC, Siggia FD, Phair JF, Lippincott-schwartz J. Dissecion of COPI an dArf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 54.Che MM, Nie ZZ, Randazzo PA. Assays and properties of the Arf GAPs AGAP1, ASAP1, and Arf GAP1. Methods Enzymol. 2005;404:147–163. doi: 10.1016/S0076-6879(05)04015-2. [DOI] [PubMed] [Google Scholar]
- 55.Randazzo PA, Miura K, Jackson TR. Assay and purification of phosphoinositide-dependent ADP- ribosylation factor (ARF) GTPase activating proteins. Methods Enzymol. 2001;329:343–354. doi: 10.1016/s0076-6879(01)29096-x. [DOI] [PubMed] [Google Scholar]
- 56.Cavenagh MM, Whitney JA, Carroll K, Zhang CJ, Boman AL, ROSENWALD AG, Mellman I, KAHN RA. Intracellular distribution of Arf proteins in mammalian cells -Arf6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- 57.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nature Biotechnology. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 58.St Charles R, Matthews JH, Zhang EL, Tulinsky A. Bound structures of novel P3-P1 prime; beta-strand mimetic inhibitors of thrombin. J Med Chem. 1999;42:1376–1383. doi: 10.1021/jm980052n. [DOI] [PubMed] [Google Scholar]
- 59.Zhang CJ, Bowzard JB, Anido A, Kahn RA. Four ARF GAPs in Saccharomyces cerevisiae have both overlapping and distinct functions. Yeast. 2003;20:315–330. doi: 10.1002/yea.966. [DOI] [PubMed] [Google Scholar]