Abstract
Personality traits are summarized by five broad dimensions with pervasive influences on major life outcomes, strong links to psychiatric disorders, and clear heritable components. To identify genetic variants associated with each of the five dimensions of personality we performed a genome wide association (GWA) scan of 3,972 individuals from a genetically isolated population within Sardinia, Italy. Based on analyses of 362,129 single nucleotide polymorphisms (SNPs) we found several strong signals within or near genes previously implicated in psychiatric disorders. They include the association of Neuroticism with SNAP25 (rs362584, P = 5 × 10−5), Extraversion with BDNF and two cadherin genes (CDH13 and CDH23; Ps < 5 × 10−5), Openness with CNTNAP2 (rs10251794, P = 3 × 10−5), Agreeableness with CLOCK (rs6832769, P = 9 × 10−6), and Conscientiousness with DYRK1A (rs2835731, P = 3 × 10−5). Effect sizes were small (less than 1% of variance), and most failed to replicate in the follow-up independent samples (N up to 3,903), though the association between Agreeableness and CLOCK was supported in two of three replication samples (overall P = 2 × 10−5). We infer that a large number of loci may influence personality traits and disorders, requiring larger sample sizes for the GWA approach to identify significant genetic variants.
Keywords: personality, genome wide association, founder population, psychiatry, five-factor model
Introduction
Behavior genetic studies reveal that personality traits, like psychiatric disorders, have a genetic basis. Twin, adoption, and family studies indicate that personality factors are heritable, with about 50% of the variance of the underlying components accounted for by additive and non-additive genetic factors (1-3). However, identifying the genetic variants associated with personality traits is challenging. A large number of studies have tested several candidate genes, especially for the Neuroticism factor, but these studies have produced largely inconclusive results. Similarly, genetic linkage studies, usually based on 400-500 microsatellite markers, have suggested loci for Neuroticism (4-7), but only a few genomic regions (e.g., 12q) have been reported in multiple studies. Difficulties in identifying specific loci suggest that, as has been observed for a number of quantitative traits, genetic influences on these complex traits are likely attributable to many genes, each with a small effect size. To detect such small genetic effects, there is a growing interest in high throughput genotyping technologies that examine large numbers (e.g., 500,000) of single nucleotide polymorphisms (SNPs) densely mapped across the entire genome. Using these more incisive genome-wide association (GWA) scans, recent studies have reliably identified common variants associated with complex traits and diseases that include BMI (8, 9), height (10, 11), inflammatory bowel disease (12, 13), and type 1 and type 2 diabetes (13, 14). So far, only one GWA study of a personality trait has been published (15). That study used DNA pools from about 2000 individuals with extreme scores, but the results were limited to Neuroticism, only one of the five broad dimensions of personality.
Personality profiles assessed with the NEO-PI-R questionnaire can be conveniently summarized by five major dimensions (16). Neuroticism (N), the tendency to experience negative emotions such as anxiety, anger, and depression; Extraversion (E), the tendency to be sociable, warm, active, assertive, cheerful, and in search of stimulation; Openness to Experience (O), the tendency to be imaginative, creative, unconventional, emotionally and artistically sensitive; Agreeableness (A), the dimension of interpersonal relations, characterized by altruism, trust, modesty, and cooperativeness; and Conscientiousness (C), a tendency to be organized, strong-willed, persistent, reliable, and a follower of rules and ethical principles. Consistent with their biological basis, these five dimensions can be assessed in all cultures tested so far and the five-factor structure can be clearly replicated in most samples (17). Sex differences and maturational trends are other universal features of personality, with women everywhere generally scoring higher on Neuroticism and Agreeableness (18), and with younger people generally scoring higher on Neuroticism, Extraversion, and Openness, but lower on Agreeableness and Conscientiousness in most cultures (19). Furthermore, the five factors are predictors of important life outcomes (20, 21), including well-being (22), academic performance (23), vocational interests (24), marital stability and satisfaction (25), health risk behaviors (26, 27), and longevity (28). All five factors are related to personality disorders (29), and several researchers advocate a dimensional model in the upcoming DSM-V to reorganize the conceptualization and diagnosis of personality disorders (30). More generally, personality traits are thought of as risk factors, diagnostic indicators, and predictors of onset, severity, and outcome for most psychiatric disorders (31-33). These phenotypic links are supported at the genetic level; twin studies indicate that personality traits share a large proportion of genetic variance with depression and other disorders (31, 34-36). Personality traits are increasingly recognized as endophenotypes in genetic studies of mental disorders (7, 37).
The present study provides the first GWA results for all five dimensions of personality, as measured by the Revised NEO Personality Inventory (NEO-PI-R)(38). This study is part of the SardiNIA project (3), which has targeted a highly interrelated population from the isolated Ogliastra region of Sardinia, Italy (3). Common variants associated with several complex traits (8, 11, 39-41) have been successfully identified in this sample, and the results replicated across diverse populations. The studies of population isolates are advantageous because these groups are highly homogeneous, reducing the risk of spurious associations due to population stratification. Furthermore, population isolates have more extensive stretches of linkage disequilibrium (LD) compared to outbred populations (42). The wider level of LD increases the genome-wide coverage of standard SNP arrays in cohorts such as the Sardinian. The advantages of having reduced variability in a founder population come at the cost of lower power to replicate the effects in more heterogeneous populations. However, several findings from this Sardinian cohort have been replicated in other populations (8, 11, 39-41).
Here we report associations of the five personality factors with 362,129 SNPs in 3,972 Sardinians. SNPs most strongly associated with each factor were genotyped in independent samples to look at the replicability of the findings in other populations.
Method
Sample description
We recruited 6,148 individuals, about 62% of the population aged 14 to 102 years, from a cluster of four towns in the Lanusei Valley (3). Subjects are native-born, and at least 95% are known to have all grandparents born in the same province (3). Valid personality data was obtained from 5,669 subjects (43), of which 3,972 were genotyped. The sample was composed of 2,250 women and 1,722 men (43.3%). Age ranged from 14 to 94 (M = 42.8, SD = 17).
Personality assessment
Personality traits were assessed using the Italian version of the Revised NEO Personality Inventory (NEO-PI-R), a 240-item measure of the five dimensions of personality (38). The domain scores are computed by summing up the six facets that compose each factor. Items are answered on a five-point Likert scale, from strongly disagree to strongly agree, and scales are roughly balanced to control for the effect of acquiescence. The NEO-PI-R has a robust factor structure that has been replicated in Italy (44) and in more than 50 cultures (17). Scales have shown longitudinal stability (45), cross-observer agreement, and convergent and discriminant validity in a large body of studies (16). Trained Sardinian psychologists administered the tests. In the Sardinian sample, the NEO-PI-R showed good psychometric properties, with internal consistency reliabilities for the five factors ranging from 0.80 to 0.87, and a factor structure that replicated the American normative structure at the phenotypic and the genetic level (3, 43, 46).
Genotyping and imputation
DNA was extracted from blood samples. In the Sardinian cohort, 3,329 and 1,412 individuals were genotyped with the Affymetrix 10K and Affymetrix 500K Mapping array set, respectively, with 436 individuals generating an overlapping dataset. We took advantage of the relatedness among individuals in our sample to reduce study costs. Using a modified Lander-Green algorithm, full genotypes on the 2,893 individuals typed with only the 10K panel were imputed based on stretches of shared haplotype, permitting analyses on 4,305 individuals, of which 3,972 had personality data (47). For individuals who had genotype data available at the SNP being tested, we coded genotypes as 0, 1, or 2, depending on the number of copies of an arbitrary reference allele for each SNP. For individuals with missing genotype data, we used the Lander-Green algorithm to estimate the number of copies of the allele carried by each individual (based on the genotypes of family members) and assigned each individual a score ranging between 0 and 2 (47). This estimate incorporates allele frequency information, the genotypes of relatives for the SNP of interest, and flanking marker data. For computational efficiency, the Lander-Green algorithm was applied to sub-pedigrees, each including no more than 20-25 individuals, resulting in a dataset where the average analysis unit consisted of a family with 12.3 members and 3.2 generations.
GWA analysis
Of the combined 500K and 10K Mapping array sets, the association analyses focused on 362,129 SNPs that passed quality control checks (48, 49). The remaining SNPs failed quality checks (∼2.9% of SNPs failed checks for data completeness, Hardy-Weinberg equilibrium, and Mendelian incompatibilities) or had a minor allele frequency of <5% (∼25.7% of SNPs had low minor allele frequencies). Although the five factor scores are approximately normally distributed, to avoid inflated type I error rates an inverse normal transformation was applied to all phenotype variables prior to analysis. Association analyses were carried out as described elsewhere (8, 47). The additive genetic effect was estimated for each SNP in the context of a variance component model that accounts for resemblance among related individuals (47). We analyzed each of the five factor scores in turn, including sex, age, and age2 as covariates.
Our analytical approach considers all observed or estimated genotypes (rather than focusing on alleles transmitted from heterozygous parents) and thus is not immune to effects of population stratification. In homogenous populations, this type of analysis is expected to be more powerful (50, 51). To adjust for the effects of population structure and cryptic relatedness among sampled individuals, we used the genomic control method to adjust our test statistics for each trait separately (52). We checked the genomic control value for our genome-wide association analyses (52), and carried out principal component analysis of genome-wide SNP data in a subset of unrelated individuals (53). Neither analysis suggested evidence for population substructure or genetic outliers in the sample.
To evaluate association on the X chromosome, we modeled a polygenic variance component shared according to an X-linked kinship coefficient in addition to the usual autosomal polygenic variance component (3, 47). Further, we assumed that average phenotypic values for hemizygous males would be the same as for homozygous females (3, 47).
Meta-Analysis
We use meta-analysis to summarize the results from the Sardinia and replication samples. The overall z-statistic and the corresponding P-value were calculated as a weighted average, where weights were proportional to the square root of the number of individuals examined in each sample and selected such that the squared weights summed to 1.
Replication samples
We attempted to replicate six top signals for each of the five factors in two independent samples. The first replication sample was from Tecumseh, Michigan, USA. Complete data were obtained for 923 individuals (age: M = 44.3, SD = 15.7; 58% Female), which consists of 110 unrelated individuals and 813 individuals clustering in 424 families. Personality traits were assessed using the NEO-PI, an earlier version of the NEO-PI-R, which assesses the Agreeableness and Conscientiousness factors using 18-item scales instead of the NEO-PI-R's 48-item scales. Twenty five SNPs were successfully genotyped using Taqman® SNP genotyping assays (Applied Biosystems, Foster City, CA). The second replication sample came from the Netherlands and consisted of participants from the Netherlands Twin Register. The analyses for this study were conducted on 1,158 individuals from 418 families (age: M = 45.3, SD = 14.6; 61% Female). Personality traits were assessed using a validated Dutch version of the NEO-FFI, a short version of the NEO-PI-R that assesses each factor using 12 items scale. SNPs were genotyped using Sequenom technology.
The results from the GWA analyses and the two replication samples prompted us to examine a single SNP in one additional sample. The third follow-up sample tested for a single SNP consisted of the 1,822 participants of the Erasmus Rucphen Family study in whom the SNP had been genotyped and for whom personality data were available (age: M = 48.0, SD = 14.5; 58% female). Personality was assessed using the NEO-FFI and the SNP was genotyped using Taqman®.
Results
We present the results of a GWA scan for five broad personality factors in a founder population from Sardinia, Italy. Although none of the initial results reaches genome-wide significance using the conservative Bonferroni threshold, several interesting candidate genes map near SNPs exhibiting strong evidence of association - prompting us to examine SNPs with the strongest signals in additional samples. Even when none of the SNPs examined reach genome wide significance in the original scan, we expect that sets of SNPs showing nominally strong associations will be enriched for truly associated SNPs (54, 55).
SNPs with P-values lower or equal to 10−5 are presented in supplementary Table A (45 SNPs associated with Neuroticism, 54 with Extraversion, 59 with Openness, 112 with Agreeableness, and 33 with Conscientiousness). Table 1 presents the top six SNPs that we sought to replicate in two independent samples. For each trait, we selected six top non-redundant SNPs and excluded those that were both outside of any gene and not in LD with any other surrounding SNPs. Table 1 provides P-values and a standardized measure of effect size (Z), with the sign (+ or −) indicating the direction of the effect.
Table 1.
Top association identified in the SardiNIA GWA analyses and tested in two follow-up samples.
| SNP | Gene | Chr | Position | All ele | Sardinia | Replication: US | Replication: Netherlands | Combined replication | Combined all | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | Z | P | Z | P | Z | P | P | |||||
| Note: Sample size: Sardinia N = 3,972; US N = 923; Netherlands N = 1158. Follow-up samples and combined replication p-values are one-tail test. All positions refer to May 2004 genome assembly. The effect size “Z” is measured in standard deviation units (so that an effect of 0.1 indicates that each additional copy of the allele increases trait values by 0.1 standard deviations, on average), with the sign (+ or −) indicating the direction of the effect. | ||||||||||||
| Neuroticism | ||||||||||||
| rs6047641 | - | 20 | 21739854 | G | 6.54E-06 | +0.197 | 0.153 | −0.096 | 0.338 | −0.032 | 0.160 | 2.16E-03 |
| rs1159275 | - | 1 | 192380899 | T | 8.67E-06 | −0.175 | 0.453 | −0.007 | 0.372 | +0.018 | 0.435 | 4.53E-04 |
| rs7329003 | - | 13 | 106571549 | A | 8.99E-06 | −0.156 | NA | 0.226 | +0.037 | 3.86E-04 | ||
| rs2039528 | PTPRF | 1 | 43629623 | A | 1.60E-05 | +0.140 | NA | 0.227 | −0.033 | 5.81E-04 | ||
| rs1849710 | TMEM16D | 12 | 100024546 | C | 2.29E-05 | −0.167 | 0.018 | +0.146 | 0.179 | −0.052 | 0.239 | 2.58E-03 |
| rs362584 | SNAP25 | 20 | 10202475 | G | 5.03E-05 | +0.134 | 0.064 | +0.081 | 0.353 | +0.017 | 0.097 | 5.22E-05 |
| Extraversion | ||||||||||||
| rs644148 | ZNF180 | 19 | 49662775 | G | 8.03E-06 | +0.136 | * | NA | 8.03E-06 | |||
| rs17635977 | CDH23 | 10 | 73016270 | A | 1.14E-05 | +0.137 | 0.237 | +0.033 | 0.473 | −0.003 | 0.335 | 1.42E-04 |
| rs4783307 | CDH13 | 16 | 81634135 | G | 1.70E-05 | +0.130 | 0.473 | +0.004 | 0.244 | −0.029 | 0.318 | 1.34E-03 |
| rs904208 | - | 5 | 143451120 | C | 1.77E-05 | +0.132 | 0.120 | +0.057 | 0.164 | +0.043 | 0.065 | 1.27E-05 |
| rs2813838 | - | 7 | 23922532 | G | 2.10E-05 | −0.127 | 0.048 | +0.08 | 0.183 | +0.038 | 0.037 | 1.63E-02 |
| rs8056579 | CDH13 | 16 | 81380925 | G | 2.53E-05 | −0.212 | 0.462 | +0.007 | 0.055 | +0.087 | 0.105 | 7.43E-03 |
| rs928114 | DAPK1 | 9 | 87408360 | C | 3.83E-05 | +0.162 | 0.262 | −0.043 | 0.362 | +0.021 | 0.436 | 1.19E-03 |
| Openness | ||||||||||||
| rs644148 | ZNF180 | 19 | 49662775 | G | 9.44E-07 | +0.145 | * | NA | 9.44E-07 | |||
| rs6610953 | FUNDC1 | X | 44156440 | G | 1.73E-06 | +0.154 | 0.300 | +0.029 | NA | 5.74E-06 | ||
| rs17819128 | CREBL2 | 12 | 12652926 | G | 3.02E-06 | +0.173 | 0.202 | +0.052 | 0.263 | −0.036 | 0.466 | 1.27E-04 |
| rs9291420 | MIST | 4 | 10169156 | G | 3.39E-06 | +0.152 | 0.429 | −0.011 | 0.125 | −0.068 | 0.164 | 1.42E-03 |
| rs1037791 | TSPAN13 | 7 | 16597902 | A | 3.94E-06 | +0.149 | NA | 0.440 | +0.008 | 3.59E-05 | ||
| rs586281 | - | 1 | 182931141 | G | 6.18E-06 | +0.178 | 0.118 | +0.072 | 0.387 | −0.015 | 0.283 | 6.38E-05 |
| Agreeableness | ||||||||||||
| rs1380251 | - | 1 | 217957570 | G | 1.64E-06 | −0.235 | NA | 0.375 | −0.023 | 1.25E-05 | ||
| rs2540226 | THUMPD2 | 2 | 39870711 | T | 3.85E-06 | +0.130 | 0.120 | +0.050 | * | 2.98E-06 | ||
| rs6832769 | CLOCK | 4 | 56139122 | A | 8.71E-06 | −0.141 | 0.050 | −0.077 | 0.110# | −0.055 | 0.022 | 1.74E-06## |
| rs602041 | - | 11 | 59535439 | T | 1.28E-05 | +0.155 | 0.473 | −0.005 | 0.256 | +0.035 | 0.329 | 1.48E-04 |
| rs9940706 | CDH13 | 16 | 82256207 | C | 1.45E-05 | −0.290 | 0.101 | +0.088 | 0.424 | −0.009 | 0.240 | 1.94E-03 |
| rs7637878 | BFSP2 | 3 | 134677154 | G | 1.85E-05 | +0.148 | 0.323 | +0.025 | 0.178 | −0.045 | 0.351 | 1.17E-03 |
| Conscientiousness | ||||||||||||
| rs11626232 | SMOC1 | 14 | 69557149 | C | 4.82E-06 | −0.175 | 0.028 | −0.134 | 0.473 | +0.004 | 0.112 | 9.95E-06 |
| rs10953555 | LAMB1 | 7 | 107175297 | C | 1.20E-05 | +0.140 | 0.155 | +0.050 | 0.346 | −0.017 | 0.166 | 3.86E-05 |
| rs17006841^ | MRPS18C | 4 | 84734382 | C | 1.56E-05 | −0.361 | NA | NA | 1.56E-05 | |||
| rs2835731 | DYRK1A | 21 | 37718598 | C | 2.81E-05 | −0.208 | 0.262 | +0.051 | 0.495 | +0.002 | 0.332 | 1.70E-03 |
| rs13070781 | EIF4E3 | 3 | 71836676 | A | 2.82E-05 | +0.141 | 0.201 | −0.043 | 0.070 | −0.068 | 0.048 | 1.56E-02 |
| rs10945200 | COL19A1 | 6 | 70948461 | G | 3.05E-05 | +0.141 | 0.174 | −0.044 | * | 8.12E-04 | ||
In the US sample, for technical reasons we did not genotype the ZNF180 SNP rs644148 but the nearby rs565819 (with E: P = 0.12; with O: P = 0.44). In the Dutch sample instead of THUMPD2 rs2540226 we genotyped the nearby rs1861243 (with A: P = 0.39) and instead of COL19A1 rs10945200 we genotyped the nearby rs3806052 (with C: P = 0.27).
In the Dutch sample, the association of rs6832769 and Agreeableness was evaluated using imputed genotypes estimated including 44 surrounding microsatellites.
The combined all value in the Table includes the SardiNIA GWA and the two follow-up samples. When the third follow-up sample is combined (N = 7,875), the association is reduced to P = 8.6 × 10−5.
rs17006841 was merged into rs3182340.
Neuroticism
Of the SNPs that showed the strongest association with Neuroticism, rs362584 (P = 5.03 × 10−5) is attractive, because it is within intron 1 of the gene synaptosomal-associated protein of 25 kD (SNAP25). SNAP25 plays a critical role in neurotransmitter release, axonal growth, and synaptic plasticity (56). Deletion of the region containing the SNAP-25 gene in the Coloboma mouse causes neurological abnormalities, including hyperactivity. Such a phenotype is consistent with a role for SNAP25 in attention deficit hyperactivity disorder, which has been tested in a number of studies with mostly positive results (57, 58). Furthermore, abnormalities in the level of SNAP25 have been linked to other psychiatric disorders (59-61), and genetic variants in SNAP25 have been associated with cognitive ability (62). In addition to its intrinsic interest for a personality trait, the association between SNAP-25 and Neuroticism is relevant to several psychiatric disorders for which Neuroticism is an intermediate phenotype/endophenotype. In the follow-up samples we found a trend, in the same direction, for the association of SNAP25 with Neuroticism (P = .097). A SNP in the gene TMEM16D (rs1849710; P = 2.29 × 10−5), maps in the 12q region relatively close to the marker D12S346, which showed the strongest linkage peak in a extremely discordant and concordant sibling pairs study, particularly among female pairs (4). To examine sex-specific effects in the Sardinian sample, we conducted additional association analyses in women and men separately (sex-specific P-values for the SNPs in Table 1 are presented in supplementary Table B). Results indicate that rs1849710 on 12q was associated with Neuroticism in females (P = 4.34 × 10−4) and also males (P = 4.39 × 10−3). No sex-specific effects reached genome-wide significance using the stringent Bonferroni correction.
Extraversion
In an interesting pattern for this factor, multiple independent SNPs within two Cadherin genes (CDH13 and CDH23) have the strongest association with Extraversion, though none of these effects were replicated in the two independent samples. The Cadherin genes encode for cell-cell adhesion proteins that form complexes crucial in regulating synapse formation, function, and plasticity (63-65). CDH23 is expressed in neurosensory epithelium and linked to some instances of deafness (66). CDH13 is expressed in the heart and several brain tissues, where it is thought to act as a regulator of neural cell growth (67). For rs904208 we found a trend (P = .065) in the replication samples consistent with the effect found in Sardinia. For rs2813838 there was a consistent effect in the two follow-up samples, but unfortunately the effect in the Sardinia GWA was in the opposite direction (see signs of Z scores in Table 1). In addition to the SNPs shown in Table 1, a further interesting association is with rs11030064 (P = 8.05 × 10−5), a SNP lying close to the brain-derived neurotrophic factor (BDNF) gene, and an association was also found with the extensively studied variation at Val66Met (rs6265; P = 0.0016). Several signals were also seen in the region of RAB3GAP1 (rs16831315; P = 8.05 × 10−5), which encodes a protein implicated in the exocytosis of neurotransmitters and hormones; in GFRA1, the gene for a glial cell line-derived neurotrophic factor, (rs4562724; P = 9.45 × 10−5); and in DCAMKL1 (rs17786591; P = 2.97 × 10−5), encoding a doublecortin and CaM kinase-like protein.
Openness
The most significant effect across the five factors was found between rs644148 and Openness (P = 9.44 × 10−7). This same SNP was strongly associated with Extraversion as well. Although Openness and Extraversion are correlated, we observed only a modest overlap between association results for the two traits. For example, among markers that were associated with Extraversion at p < .001, less than 5% were also associated with Openness at a similar significance level (and vice-versa). The overlap was even more reduced at more stringent significance levels, indicating that the association between rs644148 and the two traits is quite exceptional. We were unable to type this SNP in the replication samples, but a nearby SNP (rs565819; LD =1) was genotyped in the US sample and no association was found. Among the other high ranked associations for Openness there is an intriguing association with rs10251794 (P = 3.43 × 10−5), an intronic SNP in CNTNAP2, which encodes for the member of the neurexin family that has been linked with autism (68-70) and a complex phenotype of schizophrenia, epilepsy, and cognitive impairment (71). Other genes with strong signals and plausible biological relevance are the brain-specific angiogenesis inhibitor 3 (BAI3; rs9342730; P = 1.22 × 10−5) and the myelin oligodendrocyte glycoprotein (MOG; rs16895223; P = 4.60 × 10−5).
Agreeableness
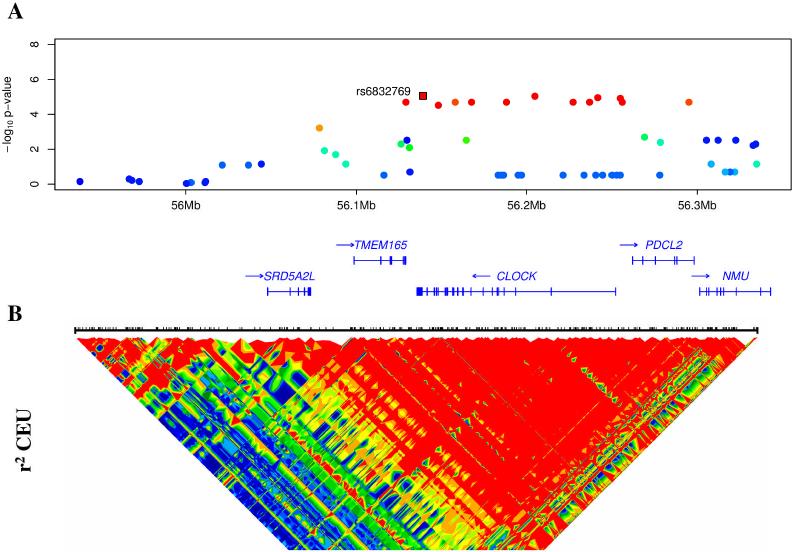
The most notable finding is the association of Agreeableness with several SNPs within or close to the CLOCK gene (see Figure 1). The strongest signal was with rs6832769 (P = 8.71 × 10−6). The two follow-up samples showed similar effects that reached statistical significance when meta-analytically combined (P = .022). Based on the initial replication, we genotyped rs6832769 in an additional sample of 1,822 individuals. In this third follow-up sample we failed to replicate the association of the rs6832769 CLOCK variant with Agreeableness (P = 0.47). Still, in the combined SardiNIA GWA and the three follow-up samples (N = 7,875) the P value was 2 × 10−5. CLOCK encodes for a transcription factor that is essential for circadian rhythm (72, 73), which in turn has large influences on human behavior, cognition, and emotion. Agreeableness has been linked to morningness preference (74), and have similar maturational trends. Younger adults tend to be more evening type and antagonistic, and older people are more agreeable (75) and more likely to be “morning types” (76). Mixed findings have been reported for a CLOCK variant known as 3111 T/C (rs1801260) with numerous traits and disorders, including morningness-eveningness preferences (77), sleep and mood disorders (78, 79), schizophrenia (80), and lower body weight among individuals with eating disorders (81). The 3111 T/C variant is weakly linked to rs6832769 (r2 = 0.212; physical distance = 3,175bp). To relate our finding to previous studies we used a hidden Markov program (11) to impute results for the 3111 T/C SNP. The imputed genotype was significantly associated with Agreeableness (rs1801260, P = .0067, effect = .08), with the T allele associated with higher Agreeableness scores. This is consistent with the findings of the T allele associated with morning preference and lower risk of mental disorders found in previous studies (77-81).
Figure 1. Association with Agreeableness and LD pattern in the CLOCK region.
SNPs showing evidence of association with Agreeableness in the SardiNIA GWA scan. P-values are plotted against genomic position in NCBI Build 35. The SNP (rs6832769) showing the strongest association, and tested in the replication samples, is highlighted. Other SNPs are colored according to their degree of disequilibrium with rs6832769, ranging from high (red), to intermediate (green), to low (blue). Transcripts in the region are indicated at the bottom of the graph, with an arrow indicating the direction of transcription. Bottom panel presents patterns of linkage disequilibrium (r2) for the CLOCK gene region in the Hap Map CEPH population.
Beyond the SNPs reported in Table 1, other strong signals in genes with plausible biological relevance are OPCML (rs11223249; P = 3.52 × 10−5) an opioid binding protein; CTNNA2 (rs2861913; P = 6.95 × 10−5), the alpha-N-catenin that interacts with cadherin proteins in essential brain functions (63-65); and IKBKAP (rs10118853; P = 5.11 × 10−5), a gene that causes familial dysautonomia, a sensory and autonomic neuropathy.
Conscientiousness
The strongest signal for Conscientiousness was with a SNP within the gene SMOC1 (rs11626232; P = 4.82 × 10−6), which also showed a meta-analytic trend in the replication samples (P = .11). Conscientiousness was strongly associated with rs2835731 (P = 2.81 × 10−5) and other SNPs within the gene DYRK1A, which is thought to have an effect on brain development. However, the association was not supported in the follow-up samples. DYRK1A maps to the Down Syndrome critical region on chromosome 21, and several other lines of evidence, including observations in a transgenic mouse model (82), suggest that DYRK1A is involved in mental retardation associated with Down Syndrome (83). Furthermore, DYRK1A has been associated with Alzheimer disease (84). Being persistent, organized, and self-controlled are central traits of Conscientiousness, and deficiencies along these dimensions are clinical features associated with the neurodegenerative diseases for which DYRK1A has been implicated. There are several studies that support the links of Conscientiousness with Alzheimer disease (85, 86).
Discussion
We have presented the results of this first GWA study of all five major dimensions of personality assessed with the NEO-PI-R, a comprehensive, reliable, and widely used measure of the Five-Factor Model. Compared with the existing literature (and especially with the candidate gene studies), a major strength is the sample size of about 4,000 individuals included in the GWA analyses. A further contribution to increased signal/noise ratio is provided by the relative homogeneity of this sample from a founder population in Sardinia. The GWA approach offers a new opportunity for a systematic search of the genetic underpinnings of personality traits. These data might become even more valuable with the accumulation of GWA results from multiple samples, as for other traits and diseases (11, 87, 88). To provide some initial evidence of association beyond our Sardinian cohort, a selected number of top signals from the GWA analyses were typed in two independent samples. Although replication attempts clearly failed to find consistent effects for most SNPs we examined, a few provided more convincing evidence. In particular, the association of Agreeableness with the CLOCK gene variant was consistent across the Sardinia and the two follow-up samples, but it was not in a third follow-up sample. For Neuroticism, the top SNPs in Sardinia were not among the top signals identified in the previous GWA scan (15), but we found some evidence for a role of SNAP25, and another SNP, rs1849710, maps in the 12q region where one of the most convincing linkage peaks has been reported (4, 7).
Given the large number of statistical tests performed, the likelihood of false positives is high. To address the problem of multiple testing we evaluated the significance level against the overly-stringent Bonferroni threshold (it is questionable whether the Bonferroni correction is appropriate (89), and whether the number of tests are truly independent, given that many SNPs are in high LD). Using the Bonferroni threshold, none of the associations between the 5 personality factors and the individual SNPs tested reached the genome-wide significance. Nevertheless, we believe our results provide useful insights into the genetic architecture of personality traits. For example, when we used simulations that took into account the specific structure of the SardiNIA pedigrees, the availability of phenotype data and the pattern individuals genotyped with the 10K and 500K arrays in each family to evaluate the power of experimental design, we estimated ∼88% power to detect alleles that account for 1.5% or more of the variance in one of the five main NEO-PI-R personality dimensions (at p < 1×10−5). We expect that we would have nearly 100% power to replicate these associations in samples of >2000 unrelated individuals. Unfortunately, we had only low power (∼50% or less) to replicate smaller effects even in our larger follow-up panel of about 2,000 individuals. Since we did not observe any association signals in the SardiNIA sample that replicate consistently in the follow-up samples, it seems unlikely that alleles with large effects on personality exist. Researchers interested in the genetics of personality should not be discouraged because for many other quantitative traits with a definite genetic basis, such as height, it is now clear that most associated common alleles have only modest effects (11, 90).
Caution is also required in rejecting the role of a SNP based on a failed replication attempt (91). In our case, the use of the relatively homogeneous founder population might have facilitated the detection of associations, but some of the identified SNPs may be particularly difficult to replicate in more heterogeneous populations. For example, the SNPs we identified might be in LD with the functional variant in the Sardinia but not in other populations. Some of the associations we identified might also depend on population-specific genetic or environmental background (gene-gene and gene-environment interactions). The replication samples also differed in recruitment strategies and used shorter and slightly less reliable phenotypic measures, which might have reduced power and contributed to differences across samples. In fact, heterogeneity among candidate gene studies has been linked to differences in the instrument used (92), but when we scored the SardiNIA data for the shorter questionnaires versions we found little differences in the results. Furthermore, given the universality of the five factors (17) and the commonality of genetic factors for each of a variety of other complex traits that have been studied in many populations, the major limitations are almost certainly the numbers of individuals studied in relation to the small effect sizes observed. Increasing power would likely require much larger initial and follow-up scans. As with other quantitative traits, meta-analysis of genome-wide association scans may provide an effective means to dissect these small effects (11, 41).
In addition to the five broad dimensions, future research should examine the specific facets (or lower-order traits) that compose each factor. The facets describe more specific and narrow phenotypes, which might be more easily linked to genetic variants. In fact, although facets tend to covary, a high score on the broad factor can result from the effect of different facets across individuals. For example, among those who score high on Neuroticism, some might score high on anxiety but not depression, whereas other might score high on depression but not anxiety. This phenotypic variability increases noise and reduces the likelihood of identifying genetic variants. There is less phenotypic variability associated with the narrower phenotype assessed by facets, which should provide more power for GWA scans. However, analyses of multiple facets increase the number of tests and the risk of false positives.
Our results are consistent with most GWA studies of other quantitative traits in identifying SNPs that explain very small amounts of variance, generally less than 1%, and even these estimates for any particular trait/SNP are likely to be inflated (the “winner's curse”). However, even a SNP that explains a very small amount of variance can guide our understanding of the biological underpinning of complex phenotypes and diseases. Indeed, genetic association tests across the five personality factors point to several SNPs within genes known for their functions in the brain and their effects on behavior and mental disorders (e.g., SNAP-25, CDH13, CDH23, BDNF, CNTNAP2, CLOCK, CTNNA2, IKBKAP, DYRK1A). These findings seem to reflect the phenotypic links between personality and psychiatric disorders. If confirmed in future studies, these findings might also advance our understanding of the continuum between normal and abnormal personality phenotypes. Given the high degree of comorbidity (93, 94) and other limitations of categorical systems (95), a dimensional approach to molecular psychiatry might well provide greater power to detect genetic variants associated with psychiatric disorders, and also provide possible points for eventual pharmaceutical intervention.
Acknowledgments
We thank the individuals who participated in this study; The SardiNIA team thanks Monsignore Piseddu (Bishop of Ogliastra), the mayors of the four Sardinian towns (Lanusei, Ilbono, Arzana and Elini), and the head of the Public Health Unit ASL4 for cooperation. We thank Prof. Antonio Cao for his leadership of the SardiNIA project.
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Dr. Margit Burmeister's laboratory was supported by the National Institute of Health NIH - NIMH R21 MH070793 and the National Alliance for Research on Schizophrenia and Depression (NARSAD), Independent Investigator Award (MB) and Young Investigator Award (SV).
Footnotes
Financial disclosure Paul T. Costa, Jr. receives royalties from the Revised NEO Personality Inventory. The authors declare that they have no other competing interests.
Supplementary Material
References
- 1.Bouchard TJ, Loehlin JC. Genes, evolution, and personality. Behavior Genetics. 2001;31:243–273. doi: 10.1023/a:1012294324713. [DOI] [PubMed] [Google Scholar]
- 2.Jang KL, McCrae RR, Angleitner A, Riemann R, Livesley WJ. Heritability of facet-level traits in a cross-cultural twin sample: Support for a hierarchical model of personality. Journal of Personality and Social Psychology. 1998 Jun;74(6):1556–1565. doi: 10.1037//0022-3514.74.6.1556. [DOI] [PubMed] [Google Scholar]
- 3.Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, et al. Heritability of Cardiovascular and Personality Traits in 6,148 Sardinians. PloS Genetics. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fullerton J, Cubin M, Tiwari H, Wang C, Bomhra A, Davidson S, et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. American Journal of Human Genetics. 2003 Apr;72(4):879–890. doi: 10.1086/374178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash MW, Huezo-Diaz P, Williamson RJ, Sterne A, Purcell S, Hoda F, et al. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet. 2004 Oct 1;13(19):2173–2182. doi: 10.1093/hmg/ddh239. [DOI] [PubMed] [Google Scholar]
- 6.Neale BM, Sullivan PF, Kendler KS. A genome scan of neuroticism in nicotine dependent smokers. Am J Med Genet B Neuropsychiatr Genet. 2005 Jan 5;132(1):65–69. doi: 10.1002/ajmg.b.30095. [DOI] [PubMed] [Google Scholar]
- 7.Kuo PH, Neale MC, Riley BP, Patterson DG, Walsh D, Prescott CA, et al. A genome-wide linkage analysis for the personality trait neuroticism in the Irish affected sib-pair study of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2007 Jun 5;144(4):463–468. doi: 10.1002/ajmg.b.30478. [DOI] [PubMed] [Google Scholar]
- 8.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007 Jul 20;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007 May 11;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007 Oct;39(10):1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008 Jan 13; doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006 Dec 1;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun 7;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006 Mar;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 15.Shifman S, Bhomra A, Smiley S, Wray NR, James MR, Martin NG, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2007 Jul 31; doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCrae RR, Costa PT., Jr. Personality in adulthood: A Five-Factor Theory perspective. 2nd edn. Guilford Press; New York: 2003. [Google Scholar]
- 17.McCrae RR, Terracciano A. 78 Members of the Personality Profiles of Cultures Project. Universal features of personality traits from the observer's perspective: Data from 50 cultures. Journal of Personality and Social Psychology. 2005;88:547–561. doi: 10.1037/0022-3514.88.3.547. [DOI] [PubMed] [Google Scholar]
- 18.Costa PT, Jr., Terracciano A, McCrae RR. Gender differences in personality traits across cultures: Robust and surprising findings. Journal of Personality and Social Psychology. 2001;81:322–331. doi: 10.1037/0022-3514.81.2.322. [DOI] [PubMed] [Google Scholar]
- 19.McCrae RR, Costa PT, Jr., de Lima MP, Simões A, Ostendorf F, Angleitner A, et al. Age differences in personality across the adult life span: Parallels in five cultures. Developmental Psychology. 1999;35:466–477. doi: 10.1037//0012-1649.35.2.466. [DOI] [PubMed] [Google Scholar]
- 20.Paunonen SV. Big five factors of personality and replicated predictions of behavior. Journal of Personality and Social Psychology. 2003 Feb;84(2):411–424. [PubMed] [Google Scholar]
- 21.Ozer DJ, Benet-Martinez V. Personality and the prediction of consequential outcomes. Annual Review of Psychology. 2006;57:8.1–8.21. doi: 10.1146/annurev.psych.57.102904.190127. [DOI] [PubMed] [Google Scholar]
- 22.Costa PT, Jr., McCrae RR. Influence of Extraversion and Neuroticism on subjective well-being: Happy and unhappy people. Journal of Personality and Social Psychology. 1980;38:668–678. doi: 10.1037//0022-3514.38.4.668. [DOI] [PubMed] [Google Scholar]
- 23.Chamorro-Premuzic T, Furnham A. Personality Traits and Academic Examination Performance. European Journal of Personality. 2003;17:237–250. [Google Scholar]
- 24.Gottfredson GD, Jones EM, Holland JL. Personality and vocational interests: The relation of Holland's six interest dimensions to the five robust dimensions of personality. Journal of Counseling Psychology. 1993;40:518–524. [Google Scholar]
- 25.Kelly EL, Conley JJ. Personality and compatibility: A prospective analysis of marital stability and marital satisfaction. Journal of Personality and Social Psychology. 1987;52:27–40. doi: 10.1037//0022-3514.52.1.27. [DOI] [PubMed] [Google Scholar]
- 26.Terracciano A, Lockenhoff CE, Crum RM, Bienvenu OJ, Costa PT., Jr. Five-Factor Model personality profiles of drug users. BMC Psychiatry. 2008;8:22. doi: 10.1186/1471-244X-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trobst KK, Herbst JH, Masters HL, III, Costa PT., Jr. Personality pathways to unsafe sex: Personality, condom use, and HIV risk behaviors. Journal of Research in Personality. 2002;36:117–133. [Google Scholar]
- 28.Terracciano A, Lockenhoff CE, Zonderman AB, Ferrucci L, Costa PT., Jr. Personality predictors of longevity: activity, emotional stability, and conscientiousness. Psychosom Med. 2008;70(6):621–627. doi: 10.1097/PSY.0b013e31817b9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widiger TA, Costa PT, Jr., McCrae RR. A proposal for Axis II: Diagnosing personality disorders using the Five-Factor Model. In: Costa PT Jr., Widiger TA, editors. Personality disorders and the Five-Factor Model of personality. 2nd edn. American Psychological Association; Washington, DC: 2002. pp. 431–456. [Google Scholar]
- 30.Widiger TA, Trull TJ. Plate tectonics in the classification of personality disorder: shifting to a dimensional model. Am Psychol. 2007 Feb-Mar;62(2):71–83. doi: 10.1037/0003-066X.62.2.71. [DOI] [PubMed] [Google Scholar]
- 31.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A longitudinal twin study of personality and major depression in women. Archives of General Psychiatry. 1993;50(11):853–862. doi: 10.1001/archpsyc.1993.01820230023002. [DOI] [PubMed] [Google Scholar]
- 32.Dinzeo TJ, Docherty NM. Normal personality characteristics in schizophrenia: a review of the literature involving the FFM. J Nerv Ment Dis. 2007 May;195(5):421–429. doi: 10.1097/01.nmd.0000253795.69089.ec. [DOI] [PubMed] [Google Scholar]
- 33.Bagby RM, Rector NA, Bindseil K, Dickens SE, Levitan RD, Kennedy SH. Self-report ratings and informant ratings of personalities of depressed outpatients. American Journal of Psychiatry. 1998;155(437-438) doi: 10.1176/ajp.155.3.437. [DOI] [PubMed] [Google Scholar]
- 34.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006 Oct;63(10):1113–1120. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 35.Fanous A, Gardner CO, Prescott CA, Cancro R, Kendler KS. Neuroticism, major depression and gender: a population-based twin study. Psychol Med. 2002 May;32(4):719–728. doi: 10.1017/s003329170200541x. [DOI] [PubMed] [Google Scholar]
- 36.Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006 May;163(5):857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- 37.Savitz JB, Cupido CL, Ramesar RS. Trends in suicidology: personality as an endophenotype for molecular genetic investigations. PLoS Med. 2006 May;3(5):e107. doi: 10.1371/journal.pmed.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa PT, Jr., McCrae RR. Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) professional manual. Psychological Assessment Resources; Odessa, FL: 1992. [Google Scholar]
- 39.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of {beta}-thalassemia. Proc Natl Acad Sci USA. 2008 Feb 1; doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, et al. The GLUT9 Gene Is Associated with Serum Uric Acid Levels in Sardinia and Chianti Cohorts. PLoS Genet. 2007 Nov 9;3(11):e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008 Feb;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006 May;38(5):556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 43.Costa PT, Jr., Terracciano A, Uda M, Vacca L, Mameli C, Pilia G, et al. Personality traits in Sardinia: testing founder population effects on trait means and variances. Behav Genet. 2007 Mar;37(2):376–387. doi: 10.1007/s10519-006-9103-6. [DOI] [PubMed] [Google Scholar]
- 44.Terracciano A. The Italian version of the NEO PI-R: conceptual and empirical support for the use of targeted rotation. Personality and Individual Differences. 2003 Dec;35(8):1859–1872. doi: 10.1016/S0191-8869(03)00035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terracciano A, Costa PTJ, McCrae RR. Personality Plasticity After Age 30. Personality and Social Psychology Bulletin. 2006;32:999–1009. doi: 10.1177/0146167206288599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamagata S, Suzuki A, Ando J, Ono Y, Kijima N, Yoshimura K, et al. Is the genetic structure of human personality universal? A cross-cultural twin study from North America, Europe, and Asia. J Pers Soc Psychol. 2006 Jun;90(6):987–998. doi: 10.1037/0022-3514.90.6.987. [DOI] [PubMed] [Google Scholar]
- 47.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007 Nov;81(5):913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005 May;76(5):887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005 Aug 15;21(16):3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 50.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000 Jan;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999 Jan;64(1):259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999 Dec;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 53.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 54.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006 Feb;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 55.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007 Aug 2;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osen-Sand A, Catsicas M, Staple JK, Jones KA, Ayala G, Knowles J, et al. Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature. 1993 Jul 29;364(6436):445–448. doi: 10.1038/364445a0. [DOI] [PubMed] [Google Scholar]
- 57.Kustanovich V, Merriman B, McGough J, McCracken JT, Smalley SL, Nelson SF. Biased paternal transmission of SNAP-25 risk alleles in attention-deficit hyperactivity disorder. Mol Psychiatry. 2003 Mar;8(3):309–315. doi: 10.1038/sj.mp.4001247. [DOI] [PubMed] [Google Scholar]
- 58.Feng Y, Crosbie J, Wigg K, Pathare T, Ickowicz A, Schachar R, et al. The SNAP25 gene as a susceptibility gene contributing to attention-deficit hyperactivity disorder. Mol Psychiatry. 2005 Nov;10(11):998–1005. doi: 10.1038/sj.mp.4001722. 1973. [DOI] [PubMed] [Google Scholar]
- 59.Honer WG, Falkai P, Bayer TA, Xie J, Hu L, Li HY, et al. Abnormalities of SNARE mechanism proteins in anterior frontal cortex in severe mental illness. Cereb Cortex. 2002 Apr;12(4):349–356. doi: 10.1093/cercor/12.4.349. [DOI] [PubMed] [Google Scholar]
- 60.Fatemi SH, Earle JA, Stary JM, Lee S, Sedgewick J. Altered levels of the synaptosomal associated protein SNAP-25 in hippocampus of subjects with mood disorders and schizophrenia. Neuroreport. 2001 Oct 29;12(15):3257–3262. doi: 10.1097/00001756-200110290-00023. [DOI] [PubMed] [Google Scholar]
- 61.Scarr E, Gray L, Keriakous D, Robinson PJ, Dean B. Increased levels of SNAP-25 and synaptophysin in the dorsolateral prefrontal cortex in bipolar I disorder. Bipolar Disord. 2006 Apr;8(2):133–143. doi: 10.1111/j.1399-5618.2006.00300.x. [DOI] [PubMed] [Google Scholar]
- 62.Gosso MF, de Geus EJ, van Belzen MJ, Polderman TJ, Heutink P, Boomsma DI, et al. The SNAP-25 gene is associated with cognitive ability: evidence from a family-based study in two independent Dutch cohorts. Mol Psychiatry. 2006 Sep;11(9):878–886. doi: 10.1038/sj.mp.4001868. [DOI] [PubMed] [Google Scholar]
- 63.Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002 Jul 3;35(1):77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 64.Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol Cell Neurosci. 2004 Dec;27(4):509–521. doi: 10.1016/j.mcn.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirano S, Suzuki ST, Redies C. The cadherin superfamily in neural development: diversity, function and interaction with other molecules. Front Biosci. 2003 Jan 1;8:d306–355. doi: 10.2741/972. [DOI] [PubMed] [Google Scholar]
- 66.Siemens J, Kazmierczak P, Reynolds A, Sticker M, Littlewood-Evans A, Muller U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc Natl Acad Sci USA. 2002 Nov 12;99(23):14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, et al. Expression of T-cadherin (CDH13, H-Cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000 Apr;74(4):1489–1497. doi: 10.1046/j.1471-4159.2000.0741489.x. [DOI] [PubMed] [Google Scholar]
- 68.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008 Jan;82(1):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008 Jan;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008 Jan;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2007 Jul 24; doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 72.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997 May 16;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steeves TD, King DP, Zhao Y, Sangoram AM, Du F, Bowcock AM, et al. Molecular cloning and characterization of the human CLOCK gene: expression in the suprachiasmatic nuclei. Genomics. 1999 Apr 15;57(2):189–200. doi: 10.1006/geno.1998.5675. [DOI] [PubMed] [Google Scholar]
- 74.DeYoung CG, Hasher L, Djikic M, Criger B, Peterson JB. Morning people are stable people: Circadian rhythm and the higher-order factors of the Big Five. Personality and Individual Differences. 2007;43:267–276. [Google Scholar]
- 75.Terracciano A, McCrae RR, Brant LJ, Costa PT., Jr. Hierarchical linear modeling analyses of NEO-PI-R scales in the Baltimore Longitudinal Study of Aging. Psychology and Aging. 2005;20:493–506. doi: 10.1037/0882-7974.20.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paine SJ, Gander PH, Travier N. The epidemiology of morningness/eveningness: influence of age, gender, ethnicity, and socioeconomic factors in adults (30-49 years) J Biol Rhythms. 2006 Feb;21(1):68–76. doi: 10.1177/0748730405283154. [DOI] [PubMed] [Google Scholar]
- 77.Mishima K, Tozawa T, Satoh K, Saitoh H, Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005 Feb 5;133(1):101–104. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- 78.Benedetti F, Dallaspezia S, Fulgosi MC, Lorenzi C, Serretti A, Barbini B, et al. Actimetric evidence that CLOCK 3111 T/C SNP influences sleep and activity patterns in patients affected by bipolar depression. Am J Med Genet B Neuropsychiatr Genet. 2007 Jul 5;144(5):631–635. doi: 10.1002/ajmg.b.30475. [DOI] [PubMed] [Google Scholar]
- 79.Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, et al. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet B Neuropsychiatr Genet. 2003 Nov 15;123(1):23–26. doi: 10.1002/ajmg.b.20038. [DOI] [PubMed] [Google Scholar]
- 80.Takao T, Tachikawa H, Kawanishi Y, Mizukami K, Asada T. CLOCK gene T3111C polymorphism is associated with Japanese schizophrenics: a preliminary study. Eur Neuropsychopharmacol. 2007 Mar;17(4):273–276. doi: 10.1016/j.euroneuro.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Tortorella A, Monteleone P, Martiadis V, Perris F, Maj M. The 3111T/C polymorphism of the CLOCK gene confers a predisposition to a lifetime lower body weight in patients with anorexia nervosa and bulimia nervosa: a preliminary study. Am J Med Genet B Neuropsychiatr Genet. 2007 Dec 5;144(8):992–995. doi: 10.1002/ajmg.b.30508. [DOI] [PubMed] [Google Scholar]
- 82.Altafaj X, Dierssen M, Baamonde C, Marti E, Visa J, Guimera J, et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum Mol Genet. 2001 Sep 1;10(18):1915–1923. doi: 10.1093/hmg/10.18.1915. [DOI] [PubMed] [Google Scholar]
- 83.Dowjat WK, Adayev T, Kuchna I, Nowicki K, Palminiello S, Hwang YW, et al. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007 Feb 8;413(1):77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kimura R, Kamino K, Yamamoto M, Nuripa A, Kida T, Kazui H, et al. The DYRK1A gene, encoded in chromosome 21 Down syndrome critical region, bridges between beta-amyloid production and tau phosphorylation in Alzheimer disease. Hum Mol Genet. 2007 Jan 1;16(1):15–23. doi: 10.1093/hmg/ddl437. [DOI] [PubMed] [Google Scholar]
- 85.Dawson DV, Welsh-Bohmer KA, Siegler IC. Premorbid personality predicts level of rated personality change in patients with Alzheimer disease. Alzheimer Disease & Associated Disorders. 2000 Jan-Mar;14(1):11–19. doi: 10.1097/00002093-200001000-00002. [DOI] [PubMed] [Google Scholar]
- 86.Wilson RS, Schneider JA, Arnold SE, Bienias JL, Bennett DA. Conscientiousness and the incidence of Alzheimer disease and mild cognitive impairment. Arch Gen Psychiatry. 2007 Oct;64(10):1204–1212. doi: 10.1001/archpsyc.64.10.1204. [DOI] [PubMed] [Google Scholar]
- 87.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008 Mar 18; doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, et al. Wholegenome association study of bipolar disorder. Mol Psychiatry. 2008 Mar 4; doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998 Apr 18;316(7139):1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008 May;40(5):584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP. Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci USA. 2008 Jan 15;105(2):617–622. doi: 10.1073/pnas.0705554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sen S, Burmeister M, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet. 2004 May 15;127(1):85–89. doi: 10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- 93.Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, et al. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Arch Gen Psychiatry. 1994 Jan;51(1):8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- 94.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005 Jun;62(6):617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Costa PT Jr., Widiger TA, editors. Personality disorders and the Five-Factor Model of personality. American Psychological Association; Washington, DC: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.